Multivariate Curve Resolution Methodology Applied to the ATR-FTIR Data for Adulteration Assessment of Virgin Coconut Oil
Abstract
1. Introduction
2. Results and Discussion
2.1. Comparison among FTIR Spectra of Pure Virgin Coconut Oil and Adulterants
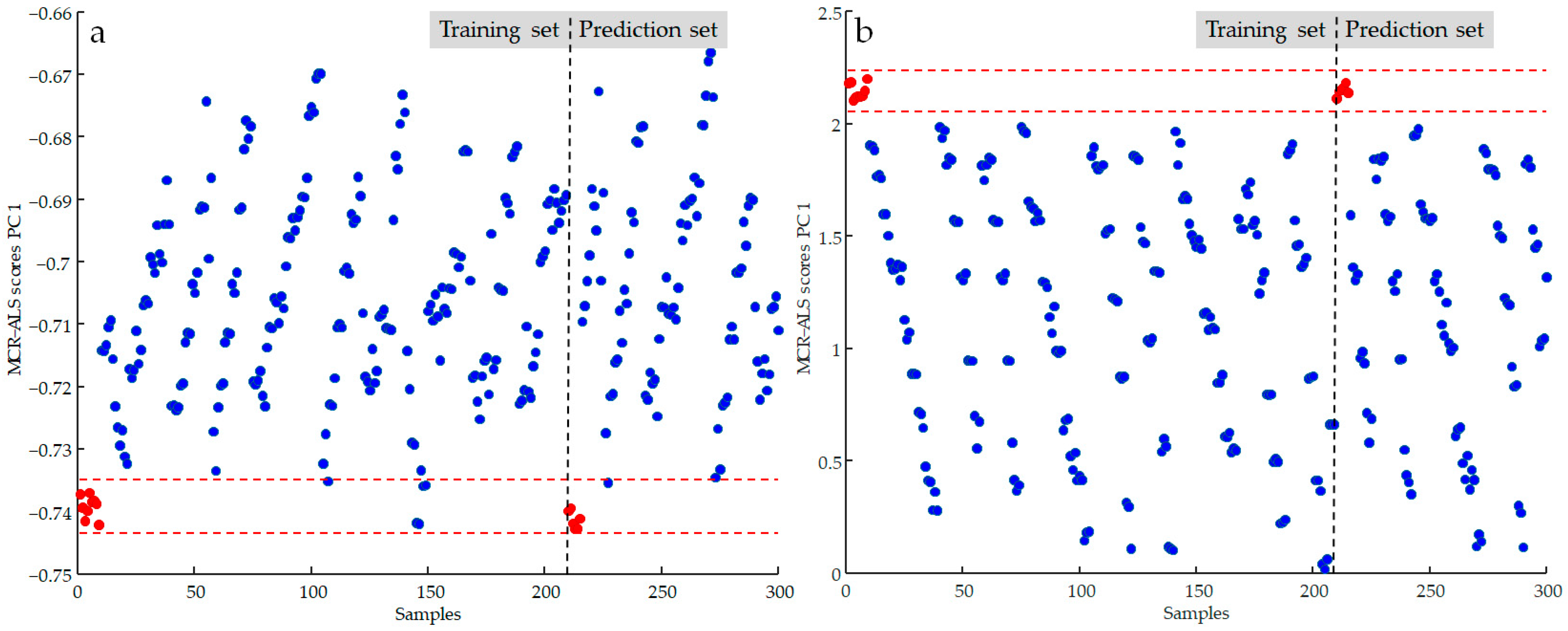
2.2. Adulteration Detection by Multivariate Resolution of Pure and Blended VCO Samples
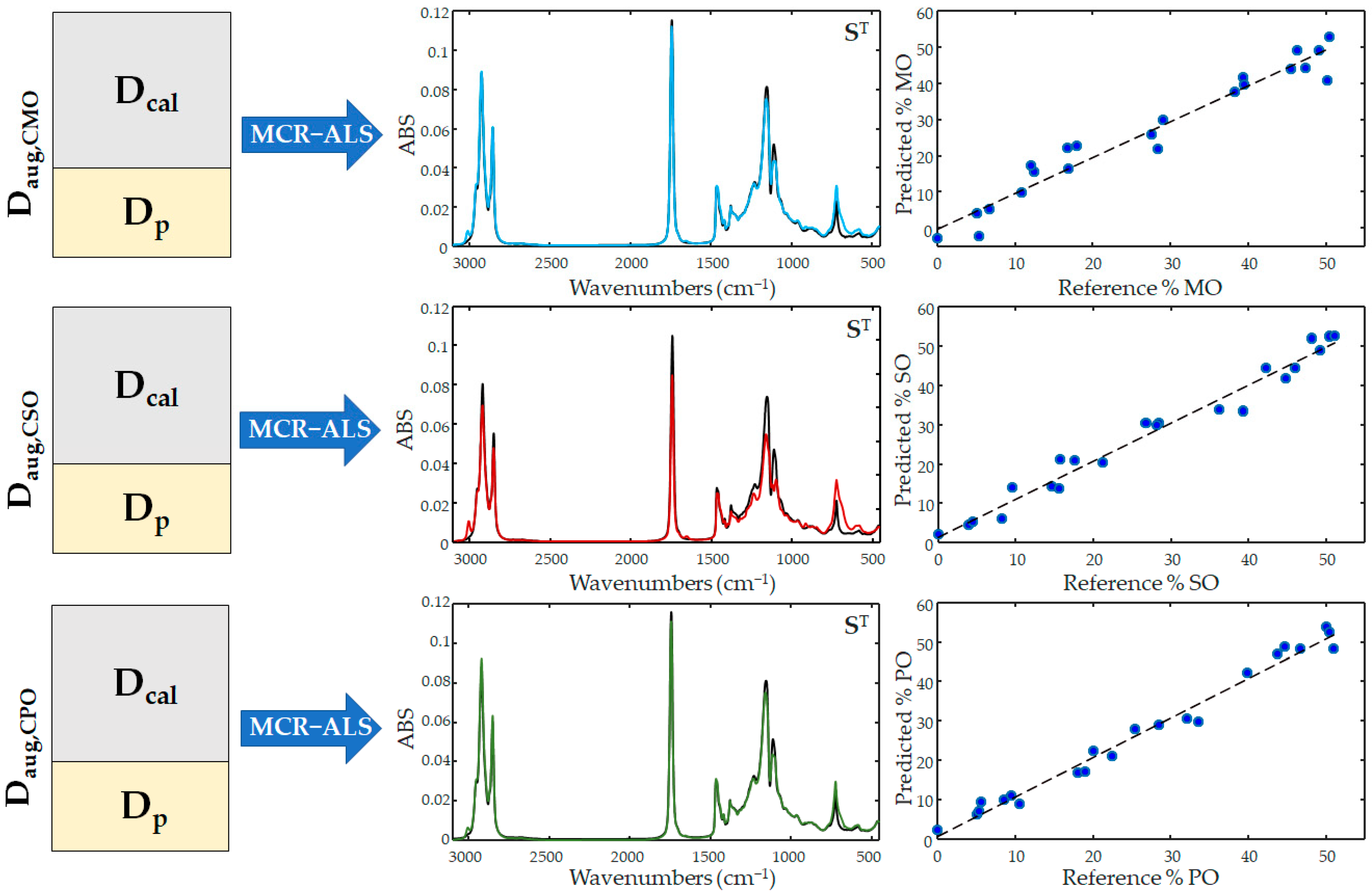
2.3. Quantitative Evaluation of Coconut Oil Adulterations
3. Materials and Methods
3.1. Virgin Coconut Oil Collection and Sample Arrangement
3.2. FTIR-ATR Spectra Acquisition and Treatment
3.3. Chemometric Method
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Swathi, S.K.; Gangwal, J.; Pillai, P.K.; Rathore, K.; Sreejith, E.J.; Yadav, J. A Review on Narikela (Coconut Palm). Int. Int. J. Res. Publ. Rev. 2021, 2, 175–179. [Google Scholar]
- Rohman, A.; Irnawati; Erwanto, Y.; Lukitaningsih, E.; Rafi, M.; Fadzilah, N.A.; Windarsih, A.; Sulaiman, A.; Zakaria, Z. Virgin Coconut Oil: Extraction, Physicochemical Properties, Biological Activities and Its Authentication Analysis. Food Rev. Int. 2021, 37, 46–66. [Google Scholar] [CrossRef]
- Bawalan, D.D.; Chapman, K.R. Virgin Coconut Oil Production Manual for Micro- and Village-Scale Processing; FAC Regional Office for Asia and the Pacific Bangkok: Bangkok, Thailand, 2006. [Google Scholar]
- Marina, A.M.; Che Man, Y.B.; Amin, I. Virgin Coconut Oil: Emerging Functional Food Oil. Trends Food Sci. Technol. 2009, 20, 481–487. [Google Scholar] [CrossRef]
- Ramesh, S.V.; Pandiselvam, R.; Thushara, R.; Manikantan, M.R.; Hebbar, K.B.; Beegum, S.; Mathew, A.C.; Neenu, S.; Shil, S. Engineering Intervention for Production of Virgin Coconut Oil by Hot Process and Multivariate Analysis of Quality Attributes of Virgin Coconut Oil Extracted by Various Methods. J. Food Process Eng. 2020, 43, e13395. [Google Scholar] [CrossRef]
- Asiah, N.; Astuti, R.M.; Cempaka, L.; Setiani, R. Physical and Chemical Characteristic of Virgin Coconut Oil under Mix Culture Fermentation Technique. J. Phys. Conf. Ser. 2019, 1364, 012009. [Google Scholar] [CrossRef]
- Salian, V.; Shetty, P. Coconut Oil and Virgin Coconut Oil: An Insight into Its Oral and Overall Health Benefits. J. Clin. Diagn. Res. 2018, 12, 1–3. [Google Scholar] [CrossRef]
- Mansor, T.S.T.; Man, C.; Afiq, A.; Nurul, K. Physicochemical Properties of Virgin Coconut Oil Extracted from Different Processing Methods. Int. Food Res. J. 2012, 19, 837–845. [Google Scholar]
- Ghani, N.A.A.; Channip, A.-A.; Chok Hwee Hwa, P.; Ja’afar, F.; Yasin, H.M.; Usman, A. Physicochemical Properties, Antioxidant Capacities, and Metal Contents of Virgin Coconut Oil Produced by Wet and Dry Processes. Food Sci. Nutr. 2018, 6, 1298–1306. [Google Scholar] [CrossRef]
- Marina, A.M.; Che Man, Y.B.; Nazimah, S.A.H.; Amin, I. Chemical Properties of Virgin Coconut Oil. J. Am. Oil Chem. Soc. 2009, 86, 301–307. [Google Scholar] [CrossRef]
- Priya, R.B.; Rashmitha, R.; Preetham, G.S.; Chandrasekar, V.; Mohan, R.J.; Sinija, V.R.; Pandiselvam, R. Detection of Adulteration in Coconut Oil and Virgin Coconut Oil Using Advanced Analytical Techniques: A Review. Food Anal. Methods 2022, 15, 2917–2930. [Google Scholar] [CrossRef]
- Rohman, A.; Che Man, Y.B. The Use of Fourier Transform Mid Infrared (FT-MIR) Spectroscopy for Detection and Quantification of Adulteration in Virgin Coconut Oil. Food Chem. 2011, 129, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Rifna, E.J.; Pandiselvam, R.; Kothakota, A.; Subba Rao, K.V.; Dwivedi, M.; Kumar, M.; Thirumdas, R.; Ramesh, S.V. Advanced Process Analytical Tools for Identification of Adulterants in Edible Oils—A Review. Food Chem. 2022, 369, 130898. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Li, P.; Ma, F.; Wang, X.; Matthäus, B.; Chen, R.; Yang, Q.; Zhang, W.; Zhang, Q. Detection of Virgin Coconut Oil Adulteration with Animal Fats Using Quantitative Cholesterol by GC × GC–TOF/MS Analysis. Food Chem. 2015, 178, 128–135. [Google Scholar] [CrossRef]
- Komaram, A.C.; Anjaneyulu, E.; Goswami, K.; Nayak, R.R.; Kanjilal, S. Detection and Quantification of Palmolein and Palm Kernel Oil Added as Adulterant in Coconut Oil Based on Triacylglycerol Profile. J. Food Sci. Technol. 2021, 58, 4420–4428. [Google Scholar] [CrossRef] [PubMed]
- Marina, A.M.; Man, Y.B.C.; Amin, I. Use of the SAW Sensor Electronic Nose for Detecting the Adulteration of Virgin Coconut Oil with RBD Palm Kernel Olein. J. Am. Oil Chem. Soc. 2010, 87, 263–270. [Google Scholar] [CrossRef]
- Mansor, T.S.T.; Man, Y.B.C.; Shuhaimi, M. Employment of Differential Scanning Calorimetry in Detecting Lard Adulteration in Virgin Coconut Oil. J. Am. Oil Chem. Soc. 2012, 89, 485–496. [Google Scholar] [CrossRef]
- Dayrit, F.M.; Buenafe, O.E.M.; Chainani, E.T.; de Vera, I.M.S. Analysis of Monoglycerides, Diglycerides, Sterols, and Free Fatty Acids in Coconut (Cocos nucifera L.) Oil by 31P NMR Spectroscopy. J. Agric. Food Chem. 2008, 56, 5765–5769. [Google Scholar] [CrossRef]
- Terouzi, W.; De Luca, M.; Bolli, A.; Oussama, A.; Patumi, M.; Ioele, G.; Ragno, G. A Discriminant Method for Classification of Moroccan Olive Varieties by Using Direct FT-IR Analysis of the Mesocarp Section. Vib. Spectrosc. 2011, 56, 123–128. [Google Scholar] [CrossRef]
- Castro, R.C.; Ribeiro, D.S.M.; Santos, J.L.M.; Páscoa, R.N.M.J. Comparison of near Infrared Spectroscopy and Raman Spectroscopy for the Identification and Quantification through MCR-ALS and PLS of Peanut Oil Adulterants. Talanta 2021, 230, 122373. [Google Scholar] [CrossRef]
- De Luca, M.; Ioele, G.; Spatari, C.; Caruso, L.; Galasso, M.P.; Ragno, G. Evaluation of human breastmilk adulteration by combining Fourier transform infrared spectroscopy and partial least square modeling. Food Sci. Nutr. 2019, 7, 2194–2201. [Google Scholar] [CrossRef]
- Rohman, A. Infrared Spectroscopy for Quantitative Analysis and Oil Parameters of Olive Oil and Virgin Coconut Oil: A Review. Int. J. Food Prop. 2017, 20, 1447–1456. [Google Scholar] [CrossRef]
- Neves, M.D.G.; Poppi, R.J. Authentication and Identification of Adulterants in Virgin Coconut Oil Using ATR/FTIR in Tandem with DD-SIMCA One Class Modeling. Talanta 2020, 219, 121338. [Google Scholar] [CrossRef] [PubMed]
- Amit; Jamwal, R.; Kumari, S.; Dhaulaniya, A.S.; Balan, B.; Singh, D.K. Application of ATR-FTIR Spectroscopy along with Regression Modelling for the Detection of Adulteration of Virgin Coconut Oil with Paraffin Oil. LWT 2020, 118, 108754. [Google Scholar] [CrossRef]
- Bassbasi, M.; De Luca, M.; Souhassou, S.; Hirri, A.; Berkani, M.; Kzaiber, F.; Ioele, G.; Ragno, G.; Oussama, A. Determination of Milk Adulteration by Sucrose Using FT-MIR Spectroscopy and Chemometrics Methods. Agric. Res. J. 2014, 11, 175–180. [Google Scholar]
- Zicker, M.C.; Craig, A.P.; de Oliveira Ramiro, D.; Franca, A.S.; Labanca, R.A.; Ferreira, A.V.M. Quantitative Analysis of Acidity Level in Virgin Coconut Oils by Fourier Transform Infrared Spectroscopy and Chemometrics. Eur. J. Lipid Sci. Technol. 2016, 118, 1350–1357. [Google Scholar] [CrossRef]
- Amit; Jamwal, R.; Kumari, S.; Dhaulaniya, A.S.; Balan, B.; Kelly, S.; Cannavan, A.; Singh, D.K. Utilizing ATR-FTIR Spectroscopy Combined with Multivariate Chemometric Modelling for the Swift Detection of Mustard Oil Adulteration in Virgin Coconut Oil. Vib. Spectrosc. 2020, 109, 103066. [Google Scholar] [CrossRef]
- Amit; Jamwal, R.; Kumari, S.; Kelly, S.; Cannavan, A.; Singh, D.K. Rapid Detection of Pure Coconut Oil Adulteration with Fried Coconut Oil Using ATR-FTIR Spectroscopy Coupled with Multivariate Regression Modelling. LWT 2020, 125, 109250. [Google Scholar] [CrossRef]
- Pandurangan, M.; Murugesan, S.; Shettu, N.; Gajivaradhan, M. Detection of Adulteration of Coconut Oil Using Fourier Transform Infrared Spectroscopy and Chemometrics. Int. J. Stat. Appl. Math. 2017, 2, 46–51. [Google Scholar]
- De Géa Neves, M.; Poppi, R.J. Monitoring of Adulteration and Purity in Coconut Oil Using Raman Spectroscopy and Multivariate Curve Resolution. Food Anal. Methods 2018, 11, 1897–1905. [Google Scholar] [CrossRef]
- Morozov, A.N.; Kochikov, I.V.; Novgorodskaya, A.V.; Sologub, A.A.; Fufurin, I.L. Statistical Estimation of the Probability of the Correct Substance Detection in Ftir Spectroscopy. Comput. Opt. 2015, 39, 614–621. [Google Scholar] [CrossRef]
- Yuan, L.; Meng, X.; Xin, K.; Ju, Y.; Zhang, Y.; Yin, C.; Hu, L. A Comparative Study on Classification of Edible Vegetable Oils by Infrared, near Infrared and Fluorescence Spectroscopy Combined with Chemometrics. Spectrochim. Acta Mol. Biomol. Spectrosc. 2023, 288, 122120. [Google Scholar] [CrossRef] [PubMed]
- Consonni, V.; Baccolo, G.; Gosetti, F.; Todeschini, R.; Ballabio, D. A MATLAB Toolbox for Multivariate Regression Coupled with Variable Selection. Chemom. Intell. Lab. Syst. 2021, 213, 104313. [Google Scholar] [CrossRef]
- Galvão, R.K.H.; Araujo, M.C.U.; José, G.E.; Pontes, M.J.C.; Silva, E.C.; Saldanha, T.C.B. A Method for Calibration and Validation Subset Partitioning. Talanta 2005, 67, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Jaumot, J.; de Juan, A.; Tauler, R. MCR-ALS GUI 2.0: New Features and Applications. Chemom. Intell. Lab. Syst. 2015, 140, 1–12. [Google Scholar] [CrossRef]



| Adulterant | Maize Oil (MO) | Peanut Oil (PO) | Sunflower Oil (SO) |
| Absorbance data | |||
| N. components | 2 | 2 | 2 |
| RMSEP | 3.8237 | 2.4511 | 2.7890 |
| R2 | 0.9748 | 0.9914 | 0.9870 |
| Rp2 VCO-adulterant | 0.977–0.979 | 0.971–0.929 | 0.915–0.938 |
| RE% | 11.9986 | 7.9505 | 8.6965 |
| Derivative data | |||
| N. components | 2 | 2 | 2 |
| RMSEP | 2.6623 | 2.6754 | 1.7925 |
| R2 | 0.9877 | 0.9906 | 0.9944 |
| Rp2 VCO-adulterant | 0.989–0.988 | 0.956–0.988 | 0.975–0.915 |
| RE% | 8.3540 | 8.6780 | 5.5894 |
| SNV data | |||
| N. components | 2 | 2 | 2 |
| RMSEP | 2.7310 | 1.9991 | 3.1079 |
| R2 | 0.9879 | 0.9947 | 3.1079 |
| Rp2 VCO-adulterant | 0.995–0.987 | 0.991–0.990 | 0.992–0.879 |
| RE% | 8.5699 | 6.4843 | 9.6910 |
| MSC data | |||
| N. components | 2 | 2 | 2 |
| RMSEP | 4.4441 | 2.4318 | 2.8090 |
| R2 | 0.9652 | 0.9928 | 0.9867 |
| Rp2 VCO-adulterant | 0.981–0.880 | 0.987–0.892 | 0.982–0.878 |
| RE% | 13.9452 | 7.8878 | 8.7591 |
| Variable selection optimization procedure GA + PLS | |||
| Adulterant | Maize oil (MO) | Peanut oil (PO) | Sunflower oil (SO) |
| Data set | Derivative | SNV | Derivative |
| PLS factors in GA | 3 | 3 | 2 |
| RMSECV | 1.1747 | 1.1878 | 0.7299 |
| R2 | 0.992 | 0.993 | 0.997 |
| N. of variables | 426 | 426 | 284 |
| Predictive performance of MCR calibration models after variable selection procedure | |||
| N. components | 2 | 2 | 2 |
| RMSEP | 1.1969 | 1.1937 | 1.4702 |
| R2 | 0.9973 | 0.9975 | 0.9962 |
| RE% | 3.7557 | 3.8182 | 4.5843 |
| Pure Sample Set | Mixture Sample Sets |
|---|---|
| VCO a brand 1 (VCO1) = 5 samples | VCO adulterated with MO 5–50%, 10 × 3 = 30 samples for each VCO brand = 90 c CMO b samples |
| VCO brand 2 (VCO2) = 5 samples | VCO adulterated with PO 5–50%, 30 samples for each VCO brand = 90 CPO samples |
| VCO brand 3 (VCO3) = 5 samples | VCO adulterated with SO 5–50%, 30 samples for each VCO brand = 90 CSO samples |
| MO a = 5 samples | |
| PO a = 5 samples | Total samples: 30 pure oil samples + 270 mixture oil samples = 300 samples |
| SO a = 5 samples |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Luca, M.; Ioele, G.; Grande, F.; Occhiuzzi, M.A.; Chieffallo, M.; Garofalo, A.; Ragno, G. Multivariate Curve Resolution Methodology Applied to the ATR-FTIR Data for Adulteration Assessment of Virgin Coconut Oil. Molecules 2023, 28, 4661. https://doi.org/10.3390/molecules28124661
De Luca M, Ioele G, Grande F, Occhiuzzi MA, Chieffallo M, Garofalo A, Ragno G. Multivariate Curve Resolution Methodology Applied to the ATR-FTIR Data for Adulteration Assessment of Virgin Coconut Oil. Molecules. 2023; 28(12):4661. https://doi.org/10.3390/molecules28124661
Chicago/Turabian StyleDe Luca, Michele, Giuseppina Ioele, Fedora Grande, Maria Antonietta Occhiuzzi, Martina Chieffallo, Antonio Garofalo, and Gaetano Ragno. 2023. "Multivariate Curve Resolution Methodology Applied to the ATR-FTIR Data for Adulteration Assessment of Virgin Coconut Oil" Molecules 28, no. 12: 4661. https://doi.org/10.3390/molecules28124661
APA StyleDe Luca, M., Ioele, G., Grande, F., Occhiuzzi, M. A., Chieffallo, M., Garofalo, A., & Ragno, G. (2023). Multivariate Curve Resolution Methodology Applied to the ATR-FTIR Data for Adulteration Assessment of Virgin Coconut Oil. Molecules, 28(12), 4661. https://doi.org/10.3390/molecules28124661











