Multi-Omics Analysis Revealed Increased De Novo Synthesis of Serine and Lower Activity of the Methionine Cycle in Breast Cancer Cell Lines
Abstract
1. Introduction
2. Results
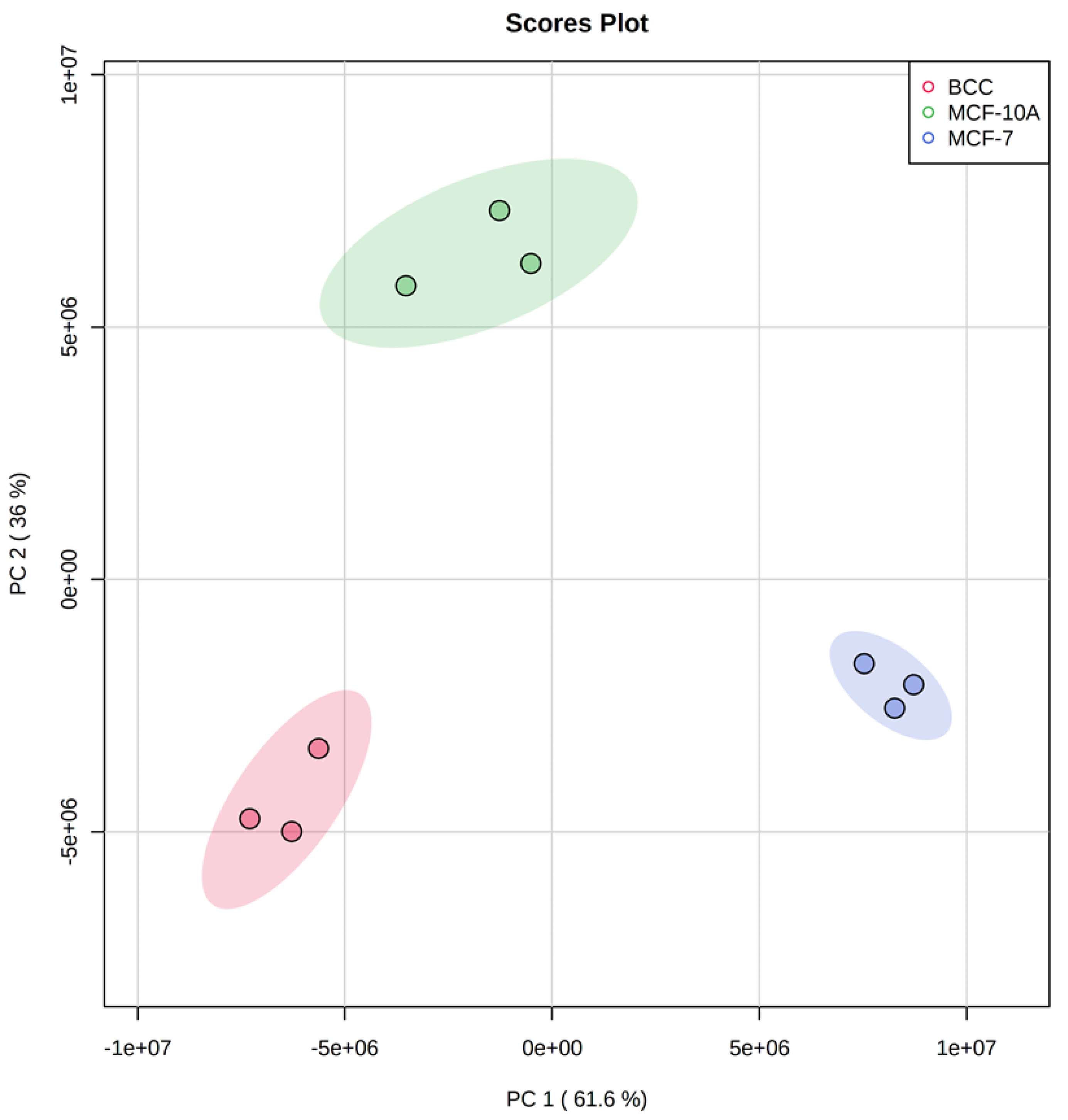
2.1. Clustering of Cell Types Based on Metabolite Concentrations
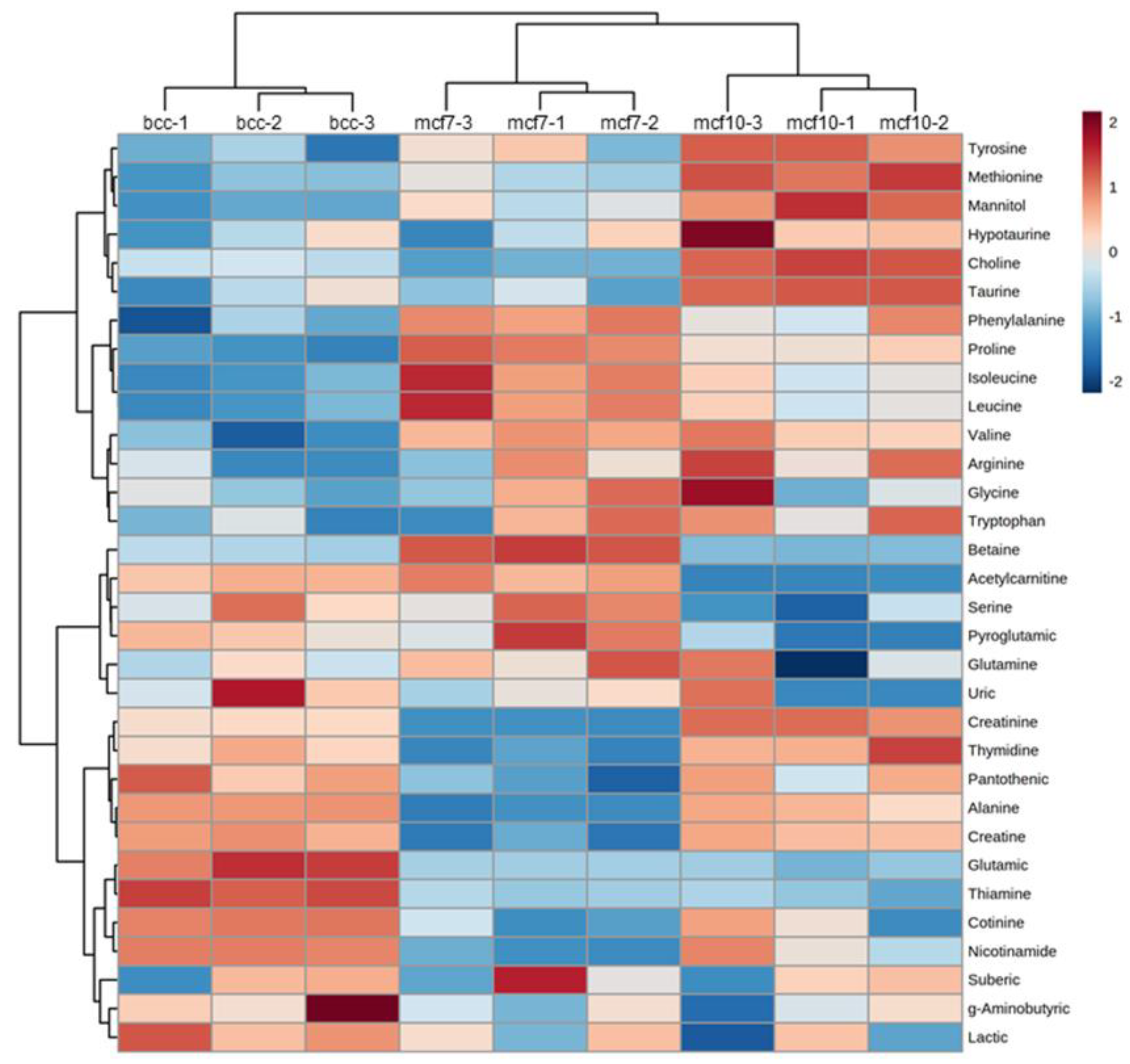
2.2. Clustering of Metabolites Based on Their Concentration Patterns
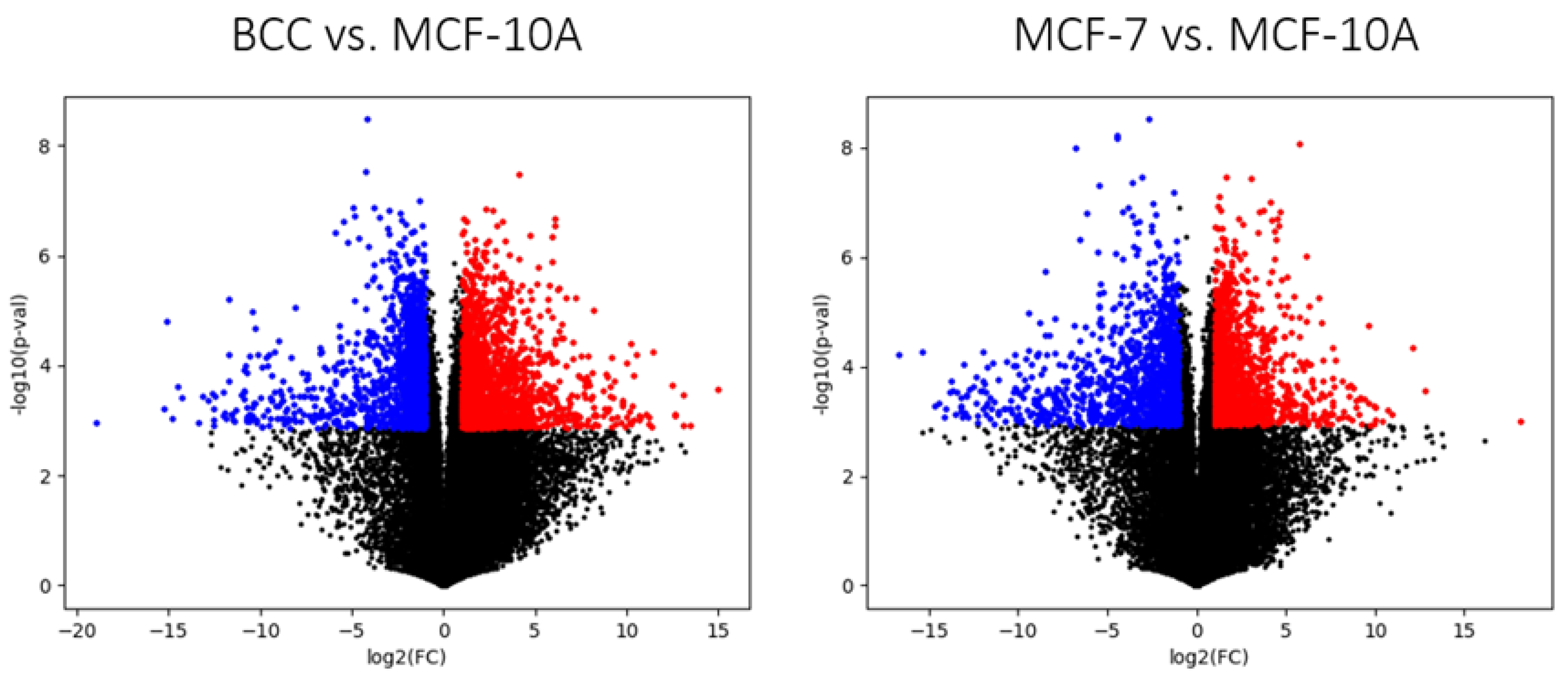
2.3. Differential Gene Expression of the Three Breast-Derived Cell Lines
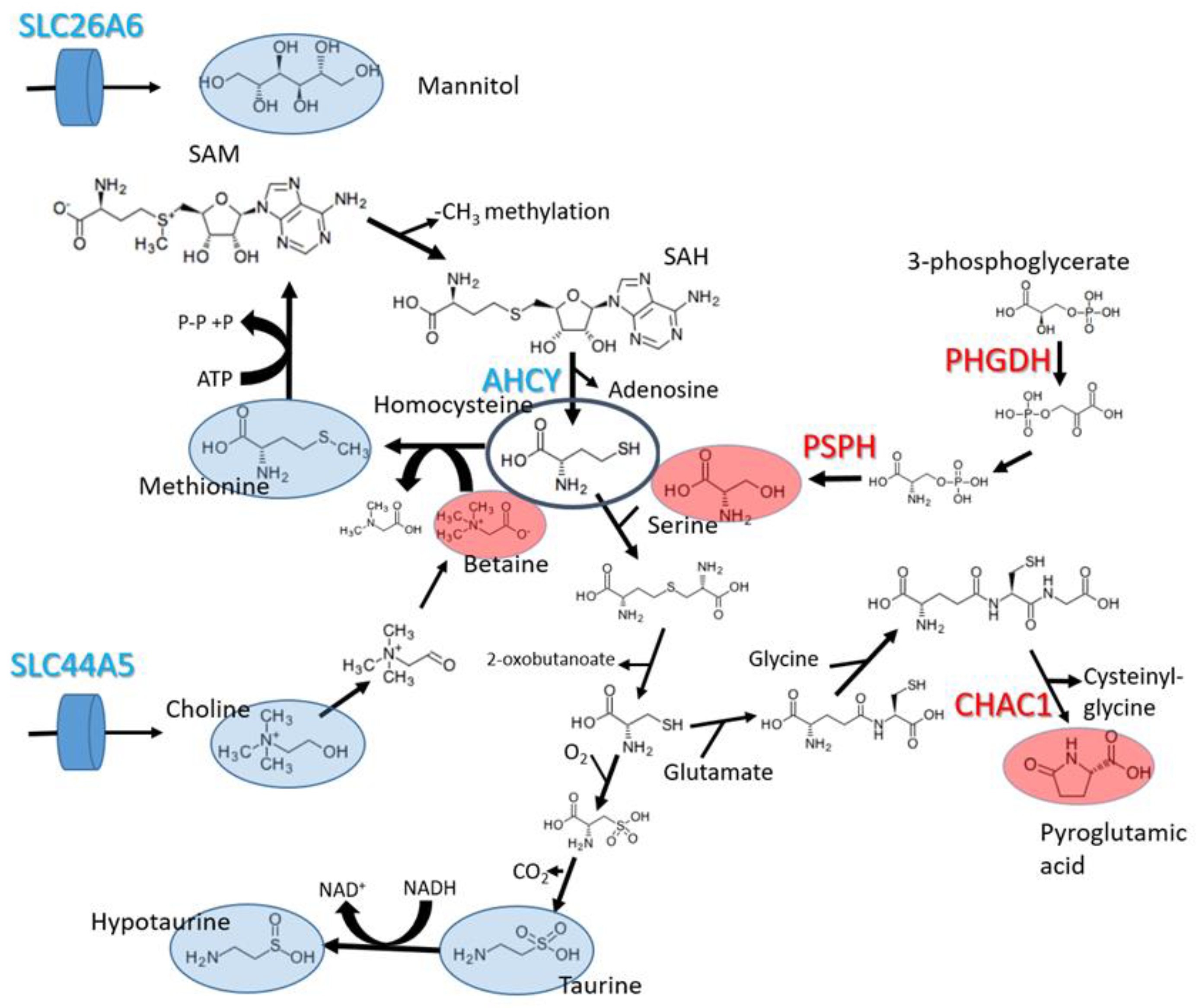
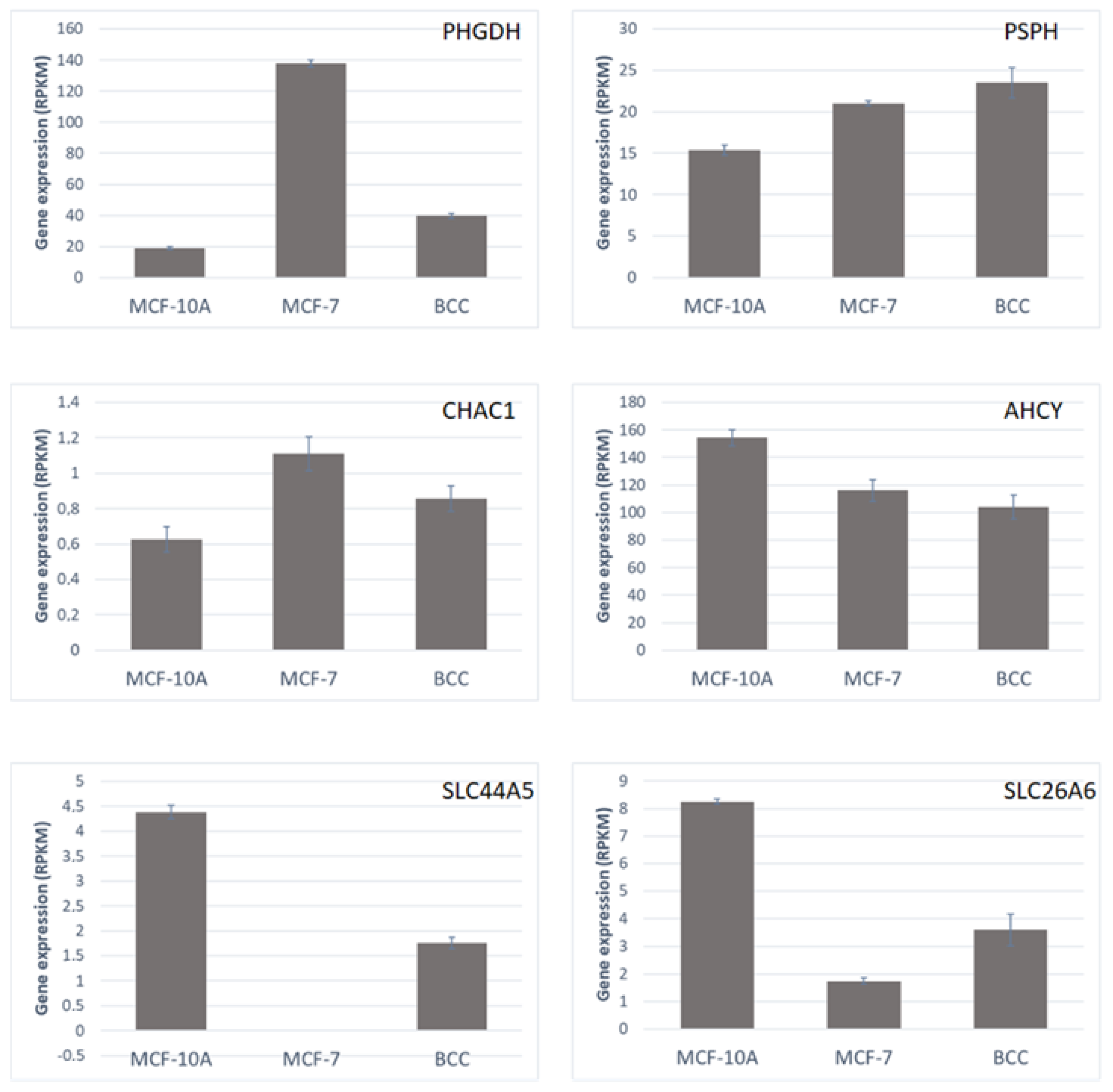
2.4. Relationships between Perturbed Metabolites and Expression of Metabolic Genes
2.5. Results of Flux Balance Analysis (FBA)
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture Medium
4.2. UPLC-ESI-MS Conditions
4.3. Analysis of Metabolomics Data
4.4. RNA Sequencing
4.5. Analysis of RNA-Seq Data
4.6. Integrated Data Analysis Using pyTARG
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Dang, C.V. Links between metabolism and cancer. Genes Dev. 2012, 26, 877–890. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, T.N.; Shelton, L.M. Cancer as a metabolic disease. Nutr. Metab. 2010, 7, 7. [Google Scholar] [CrossRef]
- Vander Heiden, M.G. Targeting cancer metabolism: A therapeutic window opens. Nat. Rev. Drug Discov. 2011, 10, 671–684. [Google Scholar] [CrossRef]
- Ward, P.S.; Thompson, C.B. Metabolic reprogramming: A cancer hallmark even Warburg did not anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef]
- Thiele, I.; Swainston, N.; Fleming, R.M.; Hoppe, A.; Sahoo, S.; Aurich, M.K.; Haraldsdottir, H.; Mo, M.L.; Rolfsson, O.; Stobbe, M.D.; et al. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 2013, 31, 419–425. [Google Scholar] [CrossRef]
- Raškevičius, V.; Mikalayeva, V.; Antanavičiūtė, I.; Ceslevičienė, I.; Kairys, V.; Bordel, S. Genome scale metabolic models as tools for drug design and personalized medicine. PLoS ONE 2018, 13, e0190636. [Google Scholar] [CrossRef]
- Bordel, S. Constraint based modeling of metabolism allows finding metabolic cancer hallmarks and identifying personalized therapeutic windows. Oncotarget 2018, 9, 19716–19729. [Google Scholar] [CrossRef]
- Antanavičiūtė, I.; Mikalayeva, V.; Ceslevičienė, I.; Milašiūtė, G.; Skeberdis, V.A.; Bordel, S. Transcriptional hallmarks of cancer cell lines reveal an emerging role of branched chain amino acid catabolism. Sci. Rep. 2017, 7, 7820. [Google Scholar] [CrossRef]
- Bordel, S.; Agren, R.; Nielsen, J. Sampling the solution space in genome-scale metabolic networks reveals transcriptional regulation in key enzymes. PLoS Comput. Biol. 2010, 6, e1000859. [Google Scholar] [CrossRef]
- Borgos, S.E.; Bordel, S.; Sletta, H.; Ertesvag, H.; Jakobsen, O.; Bruheim, P.; Ellingsen, T.E.; Nielsen, J.; Valla, S. Mapping global effects of the anti-sigma factor MucA in Pseudomonas fluorescens SBW25 through genome-scale metabolic modeling. BMC Syst. Biol. 2013, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.L.; Bordel, S.; Hong, K.; Nielsen, J. Gcn4p and the Crabtree effect of yeast: Drawing the causal model of the Crabtree effect in Saccharomyces cerevisiae and explaining evolutionary trade-offs of adaptation to galactose through systems biology. FEMS Yeast Res. 2014, 14, 654–662. [Google Scholar] [CrossRef]
- Bordel, S.; Nielsen, J. Identification of flux control in metabolic networks using non-equilibrium thermodynamics. Metab. Eng. 2010, 12, 369–377. [Google Scholar] [CrossRef]
- Mikalayeva, V.; Ceslevičienė, I.; Sarapinienė, I.; Žvikas, V.; Skeberdis, V.A.; Jakštas, V.; Bordel, S. Fatty acid synthesis and degradation interplay to regulate the oxidative stress in cancer cells. Int. J. Mol. Sci. 2019, 20, 1348. [Google Scholar] [CrossRef] [PubMed]
- Mikalayeva, V.; Pankevičiūtė, M.; Žvikas, V.; Skeberdis, V.A.; Bordel, S. Contribution of branched chain amino acids to energy production and mevalonate synthesis in cancer cells. Biochem. Biophys. Res. Commun. 2021, 585, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Antoniewicz, M.R.; Kelleher, J.K.; Stephanopoulos, G. Elementary metabolic units (EMU): A novel framework for modelling isotopic distributions. Metab. Eng. 2007, 9, 68–86. [Google Scholar] [CrossRef]
- Wishart, D.S.; Mandal, R.; Stanislaus, A.; Ramirez-Gaona, M. Cancer Metabolomics and the Human Metabolome Database. Metabolites 2016, 6, 10. [Google Scholar] [CrossRef]
- Ward, P.S.; Patel, J.; Wise, D.R.; Abdel-Wahab, O.; Bennett, B.D.; Coller, H.A.; Cross, J.R.; Fantin, V.R.; Hedvat, C.V.; Perl, A.E.; et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010, 17, 225–234. [Google Scholar] [CrossRef]
- Leal, J.F.; Ferrer, Y.; Blanco-Aparicio, C.; Hernandez-Losa, J.; Ramon y Cajal, S.; Carnero, A.; Lleonart, M.E. S-adenosylhomocysteine hydrolase downregulation contributes to tumorigenesis. Carcinogenesis 2008, 29, 2089–2095. [Google Scholar] [CrossRef]
- Zhao, G.; He, F.; Wu, C.; Li, P.; Li, N.; Deng, J.; Zhu, G.; Ren, W.; Peng, Y. Betaine in Inflammation: Mechanistic Aspects and Applications. Front. Immunol. 2018, 9, 1070. [Google Scholar] [CrossRef]
- Amelio, I.; Cutruzzolá, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Locasale, J.W.; Grassian, A.R.; Melman, T.; Lyssiotis, C.A.; Mattaini, K.R.; Bass, A.J.; Heffron, G.; Metallo, C.M.; Muranen, T.; Sharfi, H.; et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 2011, 43, 869–874. [Google Scholar] [CrossRef]
- Kumar, A.; Tikoo, S.; Maity, S.; Sengupta, S.; Sengupta, S.; Kaur, A.; Kumar Bachhawat, A. Mammalian proapoptotic factor ChaC1 and its homologues function as gamma-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep. 2012, 13, 1095–1101. [Google Scholar] [CrossRef] [PubMed]
- Goebel, G.; Berger, R.; Strasak, A.M.; Egle, D.; Muller-Holzner, E.; Schmidt, S.; Rainer, J.; Presul, E.; Parson, W.; Lang, S.; et al. Elevated mRNA expression of CHAC1 splicing variants is associated with poor outcome for breast and ovarian cancer patients. Br. J. Cancer 2012, 106, 189–198. [Google Scholar] [CrossRef]
- Console, L.; Scalise, M.; Mazza, T.; Pochini, L.; Galluccio, M.; Giangregorio, N.; Tonazzi, A.; Indiveri, C. Carnitine Traffic in Cells. Link With Cancer. Front. Cell Dev. Biol. 2020, 8, 583850. [Google Scholar] [CrossRef] [PubMed]
- Delle Cave, D.; Ilisso, C.P.; Mosca, M.; Pagano, M.; Martino, E.; Porcelli, M.; Cacciapuoti, G. The Anticancer Effects of S-Adenosylmethionine on Breast Cancer Cells. JSM Chem. 2017, 5, 1049. [Google Scholar]
- Newman, A.C.; Maddocks, O.D.K. One-carbon metabolism in cancer. Br. J. Cancer 2017, 116, 1449–1504. [Google Scholar] [CrossRef]
- Chen, W.L.; Sun, H.P.; Li, D.D.; Wang, Z.H.; You, Q.D.; Guo, X.K. G9a-An appealing antineoplastic target. Curr. Cancer Drug Targets. 2017, 17, 555–568. [Google Scholar] [CrossRef]
- Possemato, R.; Marks, K.M.; Shaul, Y.D.; Pacold, M.E.; Kim, D.; Birsoy, K.; Sethumadhavan, S.; Woo, H.K.; Jang, H.G.; Jha, A.K.; et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 2011, 476, 346–350. [Google Scholar] [CrossRef]
- Maddocks, O.D.; Berkers, C.R.; Mason, S.M.; Zheng, L.; Blyth, K.; Gottlieb, E.; Vousden, K.H. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 2013, 493, 542–546. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Tao, Y.; Duran, A.; Llado, V.; Galvez, A.; Barger, J.F.; Castilla, E.A.; Chen, J.; Yajima, T.; Porollo, A.; et al. Control of nutrient stress-induced metabolic reprogramming by PKCζ in tumorigenesis. Cell 2013, 152, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Ravez, S.; Spillier, Q.; Marteau, R.; Feron, O.; Frederick, R. Challenges and opportunities in the development of serine synthetic pathway inhibitors for cancer therapy. J. Med. Chem. 2017, 60, 1227–1237. [Google Scholar] [CrossRef]
- Pacold, M.E.; Brimacombe, K.R.; Chan, S.H.; Rohde, J.M.; Lewis, C.A.; Swier, L.J.; Possemato, R.; Chen, W.W.; Sullivan, L.B.; Fiske, B.P.; et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat. Chem. Biol. 2016, 12, 452–458. [Google Scholar] [CrossRef]
- Sellick, C.A.; Hansen, R.; Stephens, G.M.; Goodacre, R.; Dickson, A.J. Metabolite extraction from suspension-cultured mammalian cells for global metabolite profiling. Nat. Protoc. 2011, 6, 1241–1249. [Google Scholar] [CrossRef]
- Virgiliou, C.; Gika, H.G.; Theodoridis, G.A. Metabolic Profiling: Methods and Protocols. Methods Mol. Biol. 2018, 1738, 65–81. [Google Scholar] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; Morais, D.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucl. Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pankevičiūtė-Bukauskienė, M.; Mikalayeva, V.; Žvikas, V.; Skeberdis, V.A.; Bordel, S. Multi-Omics Analysis Revealed Increased De Novo Synthesis of Serine and Lower Activity of the Methionine Cycle in Breast Cancer Cell Lines. Molecules 2023, 28, 4535. https://doi.org/10.3390/molecules28114535
Pankevičiūtė-Bukauskienė M, Mikalayeva V, Žvikas V, Skeberdis VA, Bordel S. Multi-Omics Analysis Revealed Increased De Novo Synthesis of Serine and Lower Activity of the Methionine Cycle in Breast Cancer Cell Lines. Molecules. 2023; 28(11):4535. https://doi.org/10.3390/molecules28114535
Chicago/Turabian StylePankevičiūtė-Bukauskienė, Monika, Valeryia Mikalayeva, Vaidotas Žvikas, V. Arvydas Skeberdis, and Sergio Bordel. 2023. "Multi-Omics Analysis Revealed Increased De Novo Synthesis of Serine and Lower Activity of the Methionine Cycle in Breast Cancer Cell Lines" Molecules 28, no. 11: 4535. https://doi.org/10.3390/molecules28114535
APA StylePankevičiūtė-Bukauskienė, M., Mikalayeva, V., Žvikas, V., Skeberdis, V. A., & Bordel, S. (2023). Multi-Omics Analysis Revealed Increased De Novo Synthesis of Serine and Lower Activity of the Methionine Cycle in Breast Cancer Cell Lines. Molecules, 28(11), 4535. https://doi.org/10.3390/molecules28114535






