Riok1, A Novel Potential Target in MSI-High p53 Mutant Colorectal Cancer Cells
Abstract
1. Introduction
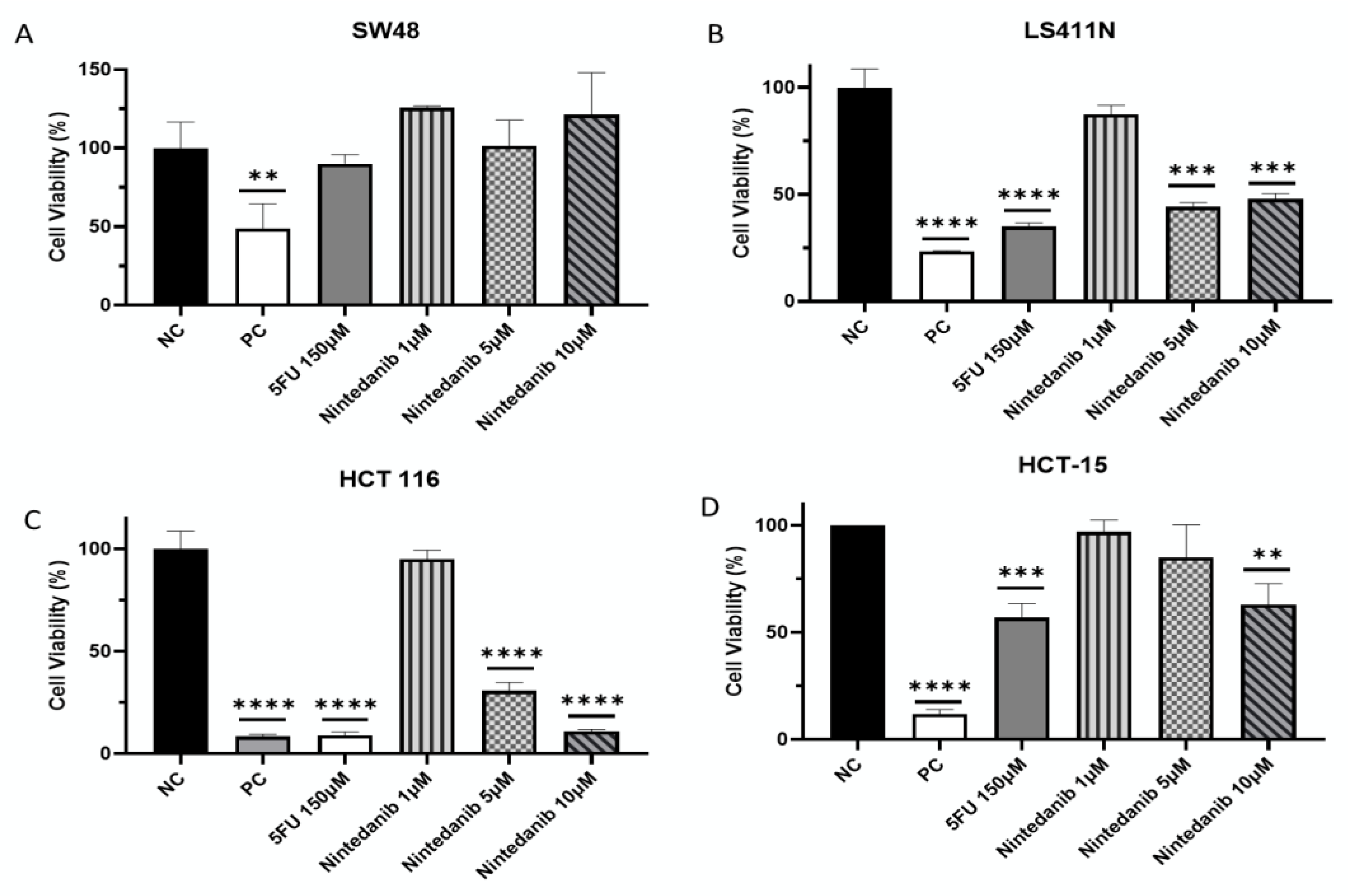
2. Results
3. Materials and Methods
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef] [PubMed]
- Gelsomino, F.; Barbolini, M.; Spallanzani, A.; Pugliese, G.; Cascinu, S. The evolving role of microsatellite instability in colorectal cancer: A review. Cancer Treat. Rev. 2016, 51, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, P.; Zhang, Y. The Devil is in the details for DNA mismatch repair. Proc. Natl. Acad. Sci. USA 2017, 114, 3552–3554. [Google Scholar] [CrossRef]
- Uehara, T.; Ma, D.; Yao, Y.; Lynch, J.P.; Morales, K.; Ziober, A.; Feldman, M.; Ota, H.; Sepulveda, A.R.H. pylori Infection Is Associated with DNA Damage of Lgr5-Positive Epithelial Stem Cells in the Stomach of Patients with Gastric Cancer. Dig. Dis. Sci. 2013, 58, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Ciriano, I.; Lee, S.; Park, W.-Y.; Kim, T.-M.; Park, P.J. A molecular portrait of microsatellite instability across multiple cancers. Nat. Commun. 2017, 8, 15180. [Google Scholar] [CrossRef] [PubMed]
- Aaltonen, L.A.; Peltomäki, P.; Leach, F.S.; Sistonen, P.; Pylkkänen, L.; Mecklin, J.-P.; Järvinen, H.; Powell, S.M.; Jen, J.; Hamilton, S.R.; et al. Clues to the Pathogenesis of Familial Colorectal Cancer. Science 1993, 260, 812–816. [Google Scholar] [CrossRef]
- Peltomäki, P. Role of DNA Mismatch Repair Defects in the Pathogenesis of Human Cancer. J. Clin. Oncol. 2003, 21, 1174–1179. [Google Scholar] [CrossRef] [PubMed]
- Bonneville, R.; Krook, M.A.; Chen, H.-Z.; Smith, A.; Samorodnitsky, E.; Wing, M.R.; Reeser, J.W.; Roychowdhury, S. Detection of Microsatellite Instability Biomarkers via Next-Generation Sequencing. In Biomarkers for Immunotherapy of Cancer; Methods in Molecular Biology; Thurin, M., Cesano, A., Marincola, F., Eds.; Humana: New York, NY, USA, 2020. [Google Scholar]
- Pawlik, T.M.; Raut, C.P.; Rodriguez-Bigas, M.A. Colorectal Carcinogenesis: MSI-H Versus MSI-L. Dis. Markers 2004, 20, 199–206. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas (TCGA) Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Iacopetta, B. TP53 mutation in colorectal cancer. Hum. Mutat. 2003, 21, 271–276. [Google Scholar] [CrossRef]
- Michel, M.; Kaps, L.; Maderer, A.; Galle, P.R.; Moehler, M. The Role of p53 Dysfunction in Colorectal Cancer and Its Implication for Therapy. Cancers 2021, 13, 2296. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jakubowski, M.M.; Hunt, J.L. KRAS Gene Mutation in Colorectal Cancer Is Correlated With Increased Proliferation and Spontaneous Apoptosis. Am. J. Clin. Pathol. 2011, 135, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Hadziavdić, V.; Pavlović-Calić, N.; Eminović, I. Microsatellite Instability and Loss of Heterozygosity of Tumor Suppressor Genes in Bosnian Patients with Sporadic Colorectal Cancer. Bosn. J. Basic Med. Sci. 2008, 8, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Thibodeau, S.N.; Bren, G.; Schaid, D. Microsatellite Instability in Cancer of the Proximal Colon. Science 1993, 260, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Tiong, K.-L.; Chang, K.-C.; Yeh, K.-T.; Liu, T.-Y.; Wu, J.-H.; Hsieh, P.-H.; Lin, S.-H.; Lai, W.-Y.; Hsu, Y.-C.; Chen, J.-Y.; et al. CSNK1E/CTNNB1 Are Synthetic Lethal To TP53 in Colorectal Cancer and Are Markers for Prognosis. Neoplasia 2014, 16, 441–450. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, Y.; Sheng, J.; Wu, F.; Li, K.; Huang, R.; Wang, X.; Jiao, T.; Guan, X.; Lu, Y.; et al. P53-R273H mutation enhances colorectal cancer stemness through regulating specific lncRNAs. J. Exp. Clin. Cancer Res. 2019, 38, 379. [Google Scholar] [CrossRef]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011, 2, 466–474. [Google Scholar] [CrossRef]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef]
- Kwon, Y.; Park, M.; Jang, M.; Yun, S.; Kim, W.K.; Kim, S.; Paik, S.; Lee, H.J.; Hong, S.; Kim, T.I.; et al. Prognosis of stage III colorectal carcinomas with FOLFOX adjuvant chemotherapy can be predicted by molecular subtype. Oncotarget 2017, 8, 39367–39381. [Google Scholar] [CrossRef]
- Sargent, D.J.; Marsoni, S.; Monges, G.; Thibodeau, S.N.; Labianca, R.; Hamilton, S.R.; French, A.J.; Kabat, B.; Foster, N.R.; Torri, V.; et al. Defective Mismatch Repair As a Predictive Marker for Lack of Efficacy of Fluorouracil-Based Adjuvant Therapy in Colon Cancer. J. Clin. Oncol. 2010, 28, 3219–3226. [Google Scholar] [CrossRef]
- Li, K.; Luo, H.; Huang, L.; Luo, H.; Zhu, X. Microsatellite instability: A review of what the oncologist should know. Cancer Cell Int. 2020, 20, 16. [Google Scholar] [CrossRef]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087.e3. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, F.; Reischmann, N.; Fauth, L.; Taromi, S.; Mastroianni, J.; Köhler, M.; Halbach, S.; Becker, A.C.; Deng, N.; Schmitz, T.; et al. The Atypical Kinase RIOK1 Promotes Tumor Growth and Invasive Behavior. Ebiomedicine 2017, 20, 79–97. [Google Scholar] [CrossRef]
- Cho, Y.-H.; Ro, E.J.; Yoon, J.-S.; Mizutani, T.; Kang, D.-W.; Park, J.-C.; Kim, T.I.; Clevers, H.; Choi, K.-Y. 5-FU promotes stemness of colorectal cancer via p53-mediated WNT/β-catenin pathway activation. Nat. Commun. 2020, 11, 5321. [Google Scholar] [CrossRef]
- Liu, Y.; Bodmer, W.F. Analysis of P53 mutations and their expression in 56 colorectal cancer cell lines. Proc. Natl. Acad. Sci. USA 2006, 103, 976–981. [Google Scholar] [CrossRef]
- Mantovani, F.; Collavin, L.; Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019, 26, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.; Pereira, F.; Reis, C.; Oliveira, M.J.; Sousa, M.J.; Preto, A. Crucial Role of Oncogenic KRAS Mutations in Apoptosis and Autophagy Regulation: Therapeutic Implications. Cells 2022, 11, 2183. [Google Scholar] [CrossRef]
- Frejno, M.; Meng, C.; Ruprecht, B.; Oellerich, T.; Scheich, S.; Kleigrewe, K.; Drecoll, E.; Samaras, P.; Hogrebe, A.; Helm, D.; et al. Proteome activity landscapes of tumor cell lines determine drug responses. Nat. Commun. 2020, 11, 3639. [Google Scholar] [CrossRef]
- Fabregat, A.; Korninger, F.; Viteri, G.; Sidiropoulos, K.; Marin-Garcia, P.; Ping, P.; Wu, G.; Stein, L.; D’eustachio, P.; Hermjakob, H. Reactome graph database: Efficient access to complex pathway data. PLoS Comput. Biol. 2018, 14, e1005968. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Marin-Garcia, P.; Ping, P.; Stein, L.; D’eustachio, P.; Hermjakob, H. Reactome diagram viewer: Data structures and strategies to boost performance. Bioinformatics 2018, 34, 1208–1214. [Google Scholar] [CrossRef]
- Gillespie, M.B.; Jassal, R.; Stephan, M.; Milacic, K.; Rothfels, A.; Senff-Ribeiro, J.; Griss, C.; Sevilla, L.; Matthews, C.; Gong, C.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Griss, J.; Viteri, G.; Sidiropoulos, K.; Nguyen, V.; Fabregat, A.; Hermjakob, H. ReactomeGSA—Efficient Multi-Omics Comparative Pathway Analysis. Mol. Cell. Proteom. 2020, 19, 2115–2125. [Google Scholar] [CrossRef]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Sidiropoulos, K.; Viteri, G.; Sevilla, C.; Jupe, S.; Webber, M.; Orlic-Milacic, M.; Jassal, B.; May, B.; Shamovsky, V.; Duenas, C.; et al. Reactome enhanced pathway visualization. Bioinformatics 2017, 33, 3461–3467. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Schaefer, C.F.; Anthony, K.; Krupa, S.; Buchoff, J.; Day, M.; Hannay, T.; Buetow, K.H. PID: The Pathway Interaction Database. Nucleic Acids Res. 2009, 37, D674–D679. [Google Scholar] [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’Ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 2016, baw100. [Google Scholar] [CrossRef] [PubMed]
- Llosa, N.J.; Cruise, M.; Tam, A.; Wicks, E.C.; Hechenbleikner, E.M.; Taube, J.M.; Blosser, R.L.; Fan, H.; Wang, H.; Luber, B.S.; et al. The Vigorous Immune Microenvironment of Microsatellite Instable Colon Cancer Is Balanced by Multiple Counter-Inhibitory Checkpoints. Cancer Discov. 2015, 5, 43–51. [Google Scholar] [CrossRef]
- Xiao, Y.; Freeman, G.J. The Microsatellite Instable Subset of Colorectal Cancer Is a Particularly Good Candidate for Checkpoint Blockade Immunotherapy. Cancer Discov. 2015, 5, 16–18. [Google Scholar] [CrossRef] [PubMed]
- Angelopoulou, K.; Stratis, M.; Diamandis, E.P. Humoral immune response against p53 protein in patients with colorectal car-cinoma. Int. J. Cancer 1997, 70, 46–51. [Google Scholar] [CrossRef]
- Chasov, V.; Zaripov, M.; Mirgayazova, R.; Khadiullina, R.; Zmievskaya, E.; Ganeeva, I.; Valiullina, A.; Rizvanov, A.; Bulatov, E. Promising New Tools for Targeting p53 Mutant Cancers: Humoral and Cell-Based Immunotherapies. Front. Immunol. 2021, 12, 707734. [Google Scholar] [CrossRef] [PubMed]
- Malekzadeh, P.; Pasetto, A.; Robbins, P.F.; Parkhurst, M.R.; Paria, B.C.; Jia, L.; Gartner, J.J.; Hill, V.; Yu, Z.; Restifo, N.P.; et al. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J. Clin. Investig. 2019, 129, 1109–1114. [Google Scholar] [CrossRef]
- Blagih, J.; Buck, M.D.; Vousden, K.H. p53, cancer and the immune response. J. Cell Sci. 2020, 133, jcs237453. [Google Scholar] [CrossRef]
- Marjon, K.; Cameron, M.J.; Quang, P.; Clasquin, M.F.; Mandley, E.; Kunii, K.; McVay, M.; Choe, S.; Kernytsky, A.; Gross, S.; et al. MTAP Deletions in Cancer Create Vulnerability to Targeting of the MAT2A/PRMT5/RIOK1 Axis. Cell Rep. 2016, 15, 574–587. [Google Scholar] [CrossRef]
- Ciardiello, D.; Maiorano, B.; Martinelli, E. Targeting KRASG12C in colorectal cancer: The beginning of a new era. ESMO Open 2022, 8, 100745. [Google Scholar] [CrossRef]
- Ciardiello, F.; Ciardiello, D.; Martini, G.; Napolitano, S.; Tabernero, J.; Cervantes, A. Clinical management of metastatic colorectal cancer in the era of precision medicine. CA Cancer J. Clin. 2022, 72, 372–401. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, S.; Wang, J.; Myeroff, L.; Parsons, R.; Sun, L.; Lutterbaugh, J.; Fan, R.S.; Zborowska, E.; Kinzler, K.W.; Vogelstein, B.; et al. Inactivation of the Type II TGF-β Receptor in Colon Cancer Cells with Microsatellite Instability. Science 1995, 268, 1336–1338. [Google Scholar] [CrossRef]
- Vilar, E.; Gruber, S.B. Microsatellite instability in colorectal cancer—The stable evidence. Nat. Rev. Clin. Oncol. 2010, 7, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Vilar, E.; Mukherjee, B.; Kuick, R.; Raskin, L.; Misek, D.E.; Taylor, J.M.; Giordano, T.J.; Hanash, S.M.; Fearon, E.R.; Rennert, G.; et al. Gene Expression Patterns in Mismatch Repair-Deficient Colorectal Cancers Highlight the Potential Therapeutic Role of Inhibitors of the Phosphatidylinositol 3-Kinase-AKT-Mammalian Target of Rapamycin Pathway. Clin. Cancer Res. 2009, 15, 2829–2839. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.M.; Zilz, N.; Beazer-Barclay, Y.; Bryan, T.M.; Hamilton, S.R.; Thibodeau, S.N.; Vogelstein, B.; Kinzler, K.W. APC mutations occur early during colorectal tumorigenesis. Nature 1992, 359, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Emanuele, M.J.; Li, D.; Creighton, C.J.; Schlabach, M.R.; Westbrook, T.F.; Wong, K.-K.; Elledge, S.J. A Genome-wide RNAi Screen Identifies Multiple Synthetic Lethal Interactions with the Ras Oncogene. Cell 2009, 137, 835–848. [Google Scholar] [CrossRef] [PubMed]
- Read, R.D.; Fenton, T.R.; Gomez, G.G.; Wykosky, J.; Vandenberg, S.R.; Babic, I.; Iwanami, A.; Yang, H.; Cavenee, W.K.; Mischel, P.S.; et al. A Kinome-Wide RNAi Screen in Drosophila Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma. PLoS Genet. 2013, 9, e1003253. [Google Scholar] [CrossRef]
- Roesch, A.; Vogt, T.; Stolz, W.; Dugas, M.; Landthaler, M.; Becker, B. Discrimination between gene expression patterns in the invasive margin and the tumour core of malignant melanomas. Melanoma Res. 2003, 13, 503–509. [Google Scholar] [CrossRef]
- Haug, S.; Schnerch, D.; Halbach, S.; Mastroianni, J.; Dumit, V.I.; Follo, M.; Hasenburg, A.; Köhler, M.; Dierbach, H.; Herzog, S.; et al. Metadherin exon 11 skipping variant enhances metastatic spread of ovarian cancer. Int. J. Cancer 2015, 136, 2328–2340. [Google Scholar] [CrossRef]
- Sarkar, D. AEG-1/MTDH/LYRIC in Liver Cancer. Adv. Cancer Res. 2013, 120, 193–221. [Google Scholar] [CrossRef]
- Burrows, C.; Latip, N.A.; Lam, S.-J.; Carpenter, L.; Sawicka, K.; Tzolovsky, G.; Gabra, H.; Bushell, M.; Glover, D.M.; Willis, A.E.; et al. The RNA binding protein Larp1 regulates cell division, apoptosis and cell migration. Nucleic Acids Res. 2010, 38, 5542–5553. [Google Scholar] [CrossRef]
- Essegian, D.; Khurana, R.; Stathias, V.; Schürer, S.C. The Clinical Kinase Index: A Method to Prioritize Understudied Kinases as Drug Targets for the Treatment of Cancer. Cell Rep. Med. 2020, 1, 100128. [Google Scholar] [CrossRef]
- Lim, H.; He, D.; Qiu, Y.; Krawczuk, P.; Sun, X.; Xie, L. Rational discovery of dual-indication multi-target PDE/Kinase inhibitor for precision anti-cancer therapy using structural systems pharmacology. PLoS Comput. Biol. 2019, 15, e1006619. [Google Scholar] [CrossRef]
- Chung, D.C. The genetic basis of colorectal cancer: Insights into critical pathways of tumorigenesis. Gastroenterology 2000, 119, 854–865. [Google Scholar] [CrossRef]
- Clark, K.J.; Cary, N.R.; Grace, A.A.; Metcalfe, J.C. Microsatellite Mutation of Type II Transforming Growth Factor-β Receptor Is Rare in Atherosclerotic Plaques. Arter. Thromb. Vasc. Biol. 2001, 21, 555–559. [Google Scholar] [CrossRef]
- Li, X.-L.; Zhou, J.; Chen, Z.-R.; Chng, W.-J. p53 mutations in colorectal cancer- molecular pathogenesis and pharmacological reactivation. World J. Gastroenterol. 2015, 21, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, S.; Wan, K.; Yu, L.; Zhao, C.; Deng, H.; Ou, Q.; Qin, J.; Hu, J.; Hou, Z. RIOK1 mediates p53 degradation and radioresistance in colorectal cancer through phosphorylation of G3BP2. Oncogene 2022, 41, 3433–3444. [Google Scholar] [CrossRef]
- Weinberg, F.; Schulze, E.; Fatouros, C.; Schmidt, E.; Baumeister, R.; Brummer, T. Expression pattern and first functional characterization of riok-1 in Caenorhabditis elegans. Gene Expr. Patterns 2014, 15, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Kiburu, I.N.; LaRonde-LeBlanc, N. Interaction of Rio1 Kinase with Toyocamycin Reveals a Conformational Switch That Controls Oligomeric State and Catalytic Activity. PLoS ONE 2012, 7, e37371. [Google Scholar] [CrossRef]
- Kubiński, K.; Masłyk, M. The Link between Protein Kinase CK2 and Atypical Kinase Rio1. Pharmaceuticals 2017, 10, 21. [Google Scholar] [CrossRef]
- Mielecki, M.; Krawiec, K.; Kiburu, I.; Grzelak, K.; Zagórski, W.; Kierdaszuk, B.; Kowa, K.; Fokt, I.; Szymanski, S.; Świerk, P.; et al. Development of novel molecular probes of the Rio1 atypical protein kinase. Biochim. Biophys. Acta 2013, 1834, 1292–1301. [Google Scholar] [CrossRef] [PubMed]
- Hörmann, A.; Hopfgartner, B.; Köcher, T.; Corcokovic, M.; Krammer, T.; Reiser, C.; Bader, G.; Shi, J.; Ehrenhöfer, K.; Wöhrle, S.; et al. RIOK1 kinase activity is required for cell survival irrespective of MTAP status. Oncotarget 2018, 9, 28625–28637. [Google Scholar] [CrossRef]
- McFall, T.; Stites, E.C. A mechanism for the response of KRASG13D expressing colorectal cancers to EGFR inhibitors. Mol. Cell. Oncol. 2020, 7, 1701914. [Google Scholar] [CrossRef]
- Briffa, R.; Um, I.; Faratian, D.; Zhou, Y.; Turnbull, A.K.; Langdon, S.P.; Harrison, D.J. Multi-Scale Genomic, Transcriptomic and Proteomic Analysis of Colorectal Cancer Cell Lines to Identify Novel Biomarkers. PLoS ONE 2015, 10, e0144708. [Google Scholar] [CrossRef]
- Brosh, R.; Rotter, V. When mutants gain new powers: News from the mutant p53 field. Nat. Rev. Cancer 2009, 9, 701–713. [Google Scholar] [CrossRef]
- Muller, P.A.; Vousden, K.H. Mutant p53 in Cancer: New Functions and Therapeutic Opportunities. Cancer Cell 2014, 25, 304–317. [Google Scholar] [CrossRef]
- Kim, D.; Xue, J.Y.; Lito, P. Targeting KRAS(G12C): From Inhibitory Mechanism to Modulation of Antitumor Effects in Patients. Cell 2020, 183, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Debnath, J.; Brugge, J.S. Modelling glandular epithelial cancers in three-dimensional cultures. Nat. Rev. Cancer 2005, 5, 675–688. [Google Scholar] [CrossRef]
- Clarke, J.R.S.; Douglas, L.R.; Duriez, P.J.; Balourdas, D.-I.; Joerger, A.C.; Khadiullina, R.; Bulatov, E.; Baud, M.G.J. Discovery of Nanomolar-Affinity Pharmacological Chaperones Stabilizing the Oncogenic p53 Mutant Y220C. ACS Pharmacol. Transl. Sci. 2022, 5, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.; Thomas, G.; Volarević, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 2018, 18, 51–63. [Google Scholar] [CrossRef] [PubMed]
- D’Cruz, O.J.; Uckun, F.M. Protein kinase inhibitors against malignant lymphoma. Expert Opin. Pharmacother. 2013, 14, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Berginski, M.E.; Moret, N.; Liu, C.; Goldfarb, D.; Sorger, P.K.; Gomez, S.M. The Dark Kinase Knowledgebase: An online compendium of knowledge and experimental results of understudied kinases. Nucleic Acids Res. 2021, 49, D529–D535. [Google Scholar] [CrossRef]
- Gyori, B.M.; Bachman, J.A.; Subramanian, K.; Muhlich, J.L.; Galescu, L.; Sorger, P.K. From word models to executable models of signaling networks using automated assembly. Mol. Syst. Biol. 2017, 13, 954. [Google Scholar] [CrossRef] [PubMed]
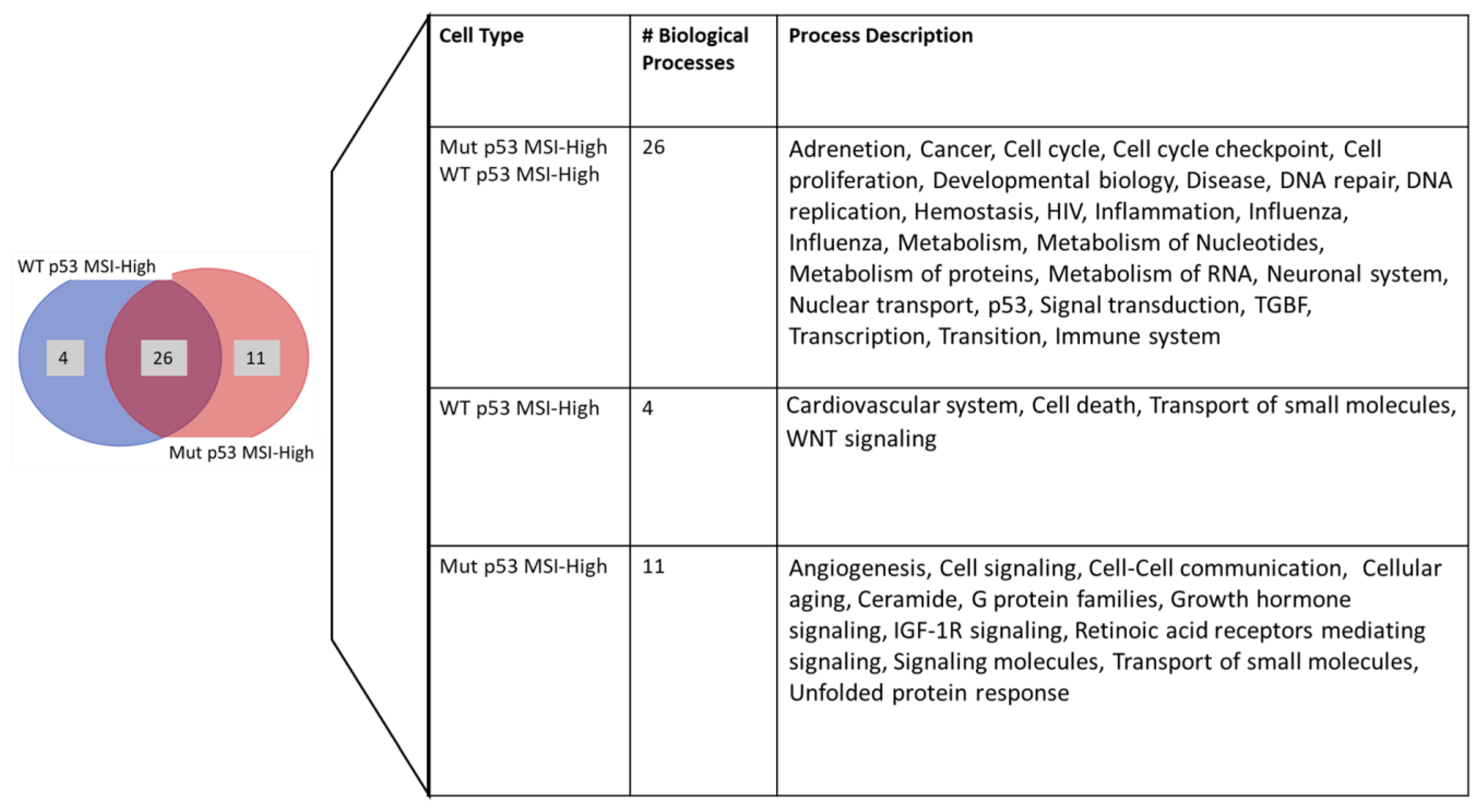
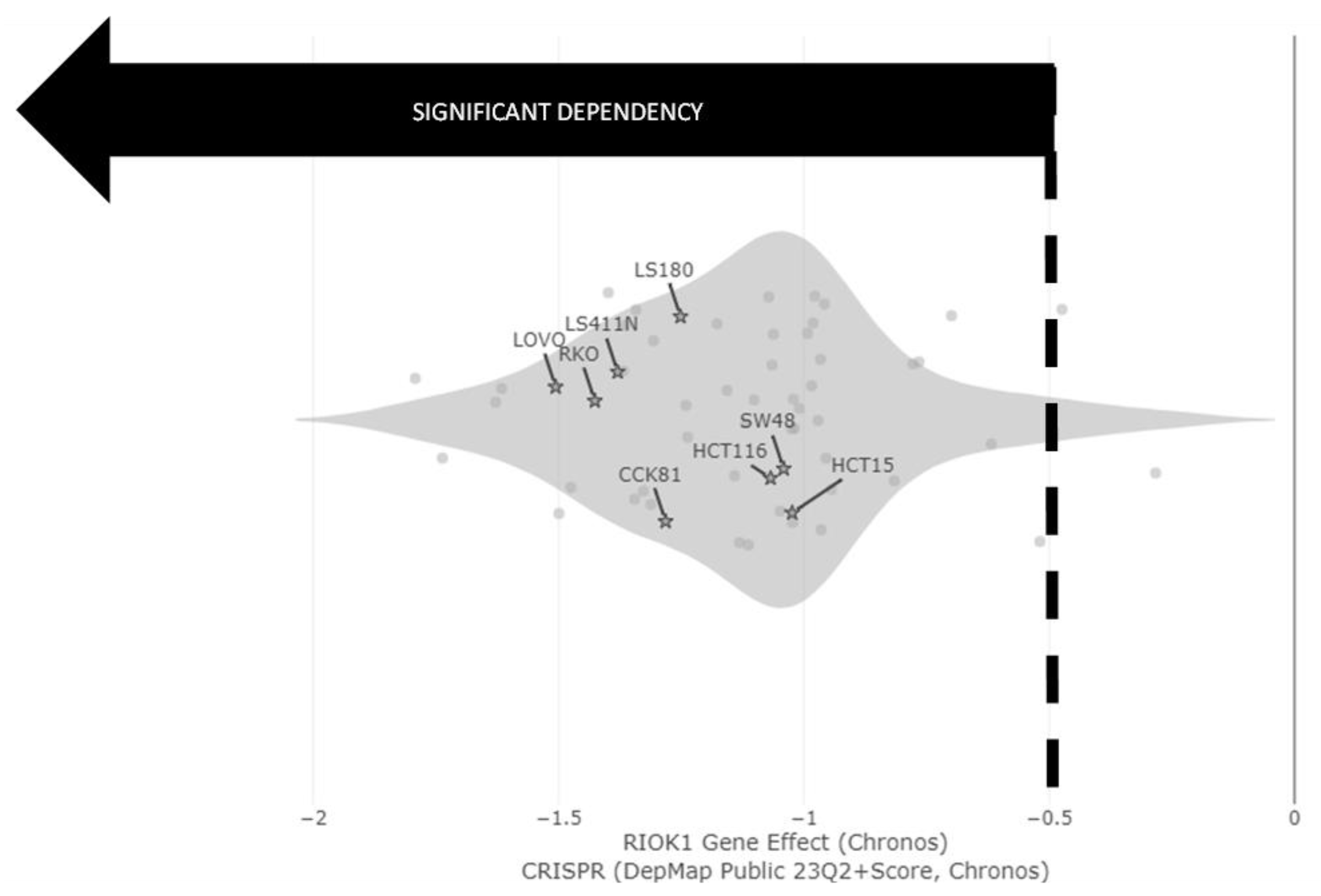
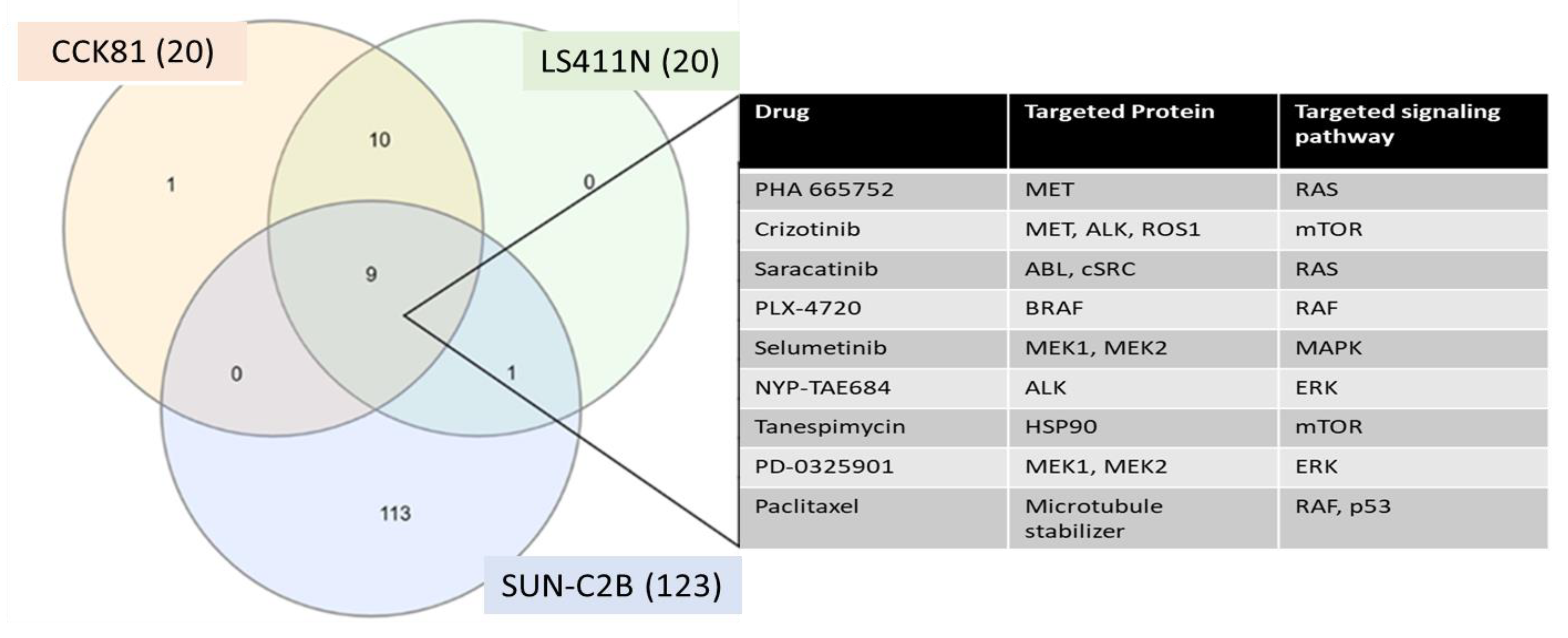
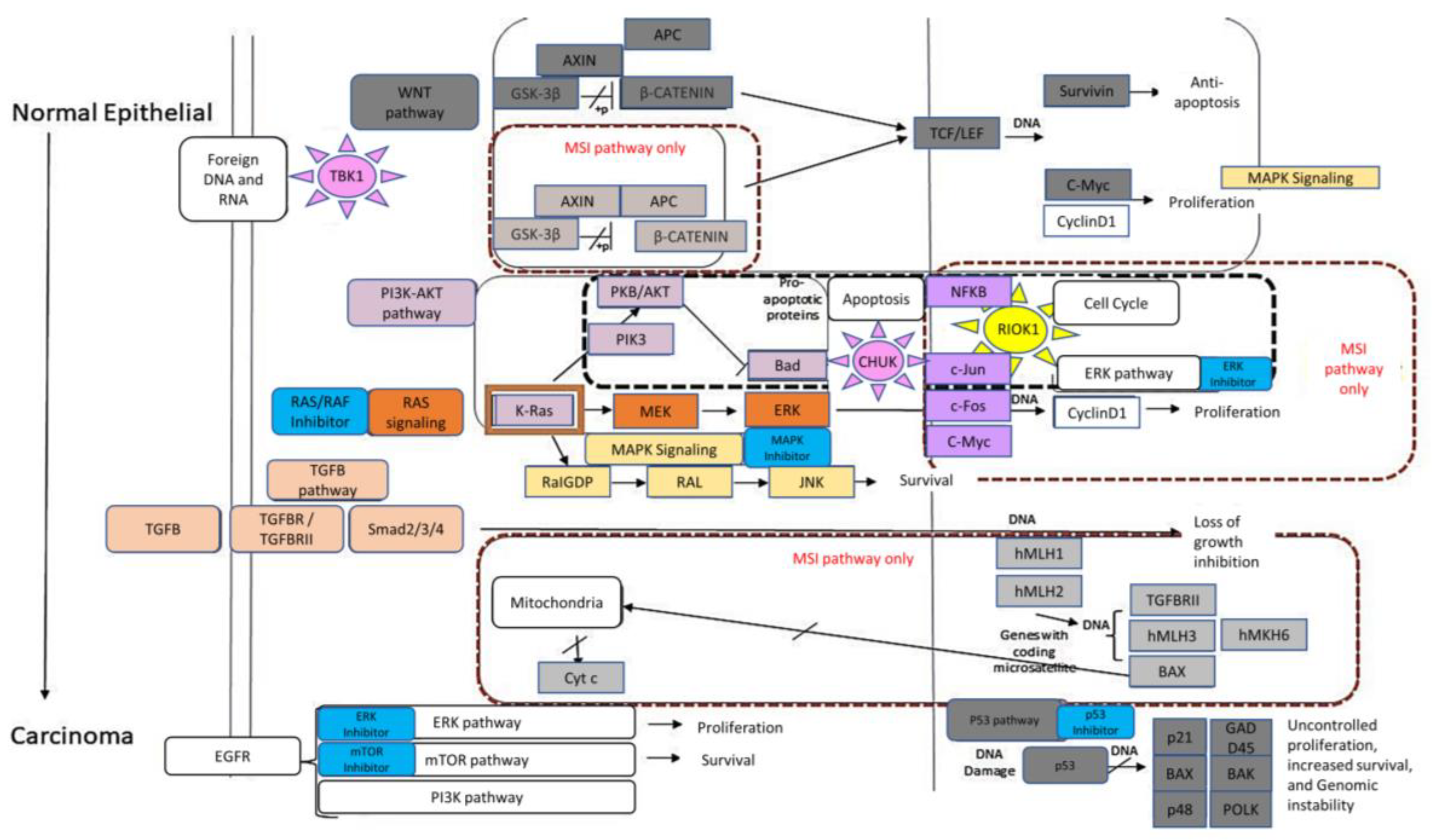
| Process Type | WT | MUT | χ2 | Significance |
|---|---|---|---|---|
| % Within WT | % Within MUT | |||
| (N = 485) | (N = 325) | |||
| Cell signaling | 0.0 (N = 0) | 1.5 (N = 5) | 7.51 | p < 0.01 |
| Cell-cycle checkpoint | 2.1 (N = 10) | 0.6 (N = 2) | 6.79 | p < 0.01 |
| DNA repair | 1.8 (N = 9) | 6.7 (N = 22) | 14.4 | p < 0.01 |
| Immune system | 13 (N = 63) | 21.8 (N = 71) | 11.06 | p < 0.01 |
| Metabolism of Proteins | 3.7 (N = 18) | 0.3 (N = 1) | 9.84 | p < 0.01 |
| Metabolism of RNA | 7.0 (N = 7) | 3.7 (N = 12) | 4 | p < 0.05 |
| Signal transduction | 5.2 (N = 25) | 1.5 (N = 5) | 7.14 | p < 0.01 |
| WNT signaling | 2.3 (N = 11) | 0.0 (N = 0) | 7.47 | p < 0.01 |
| Cell Type | TBK1 | RIOK1 | CHUK | CAMK1 |
|---|---|---|---|---|
| abundance (MSI-high-p53-Mut) | 0.81 | 0.81 | 0.78 | 0.57 |
| abundance (MSI-high-p53-WT) | 0.85 | 0.85 | 0.91 | 0.86 |
| abundance (MSS-p53-Mut) | 0.61 | 0.69 | 0.58 | 0.58 |
| abundance (MSS P53 WT) | 0.68 | 0.61 | 0.66 | 0.71 |
| Cell Line | p53 Status | KRAS Status | Toxicity |
|---|---|---|---|
| SW48 | WT | WT | - |
| LS411N | Mut | WT | 52% |
| HCT 116 | WT | Mut | 89% |
| HCT-15 | Mut | Mut | 37% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shechter, S.; Ya’ar Bar, S.; Khattib, H.; Gage, M.J.; Avni, D. Riok1, A Novel Potential Target in MSI-High p53 Mutant Colorectal Cancer Cells. Molecules 2023, 28, 4452. https://doi.org/10.3390/molecules28114452
Shechter S, Ya’ar Bar S, Khattib H, Gage MJ, Avni D. Riok1, A Novel Potential Target in MSI-High p53 Mutant Colorectal Cancer Cells. Molecules. 2023; 28(11):4452. https://doi.org/10.3390/molecules28114452
Chicago/Turabian StyleShechter, Sharon, Sapir Ya’ar Bar, Hamdan Khattib, Matthew J. Gage, and Dorit Avni. 2023. "Riok1, A Novel Potential Target in MSI-High p53 Mutant Colorectal Cancer Cells" Molecules 28, no. 11: 4452. https://doi.org/10.3390/molecules28114452
APA StyleShechter, S., Ya’ar Bar, S., Khattib, H., Gage, M. J., & Avni, D. (2023). Riok1, A Novel Potential Target in MSI-High p53 Mutant Colorectal Cancer Cells. Molecules, 28(11), 4452. https://doi.org/10.3390/molecules28114452






