Oleanolic Acid Glycosides from Scabiosa caucasica and Scabiosa ochroleuca: Structural Analysis and Cytotoxicity
Abstract
1. Introduction
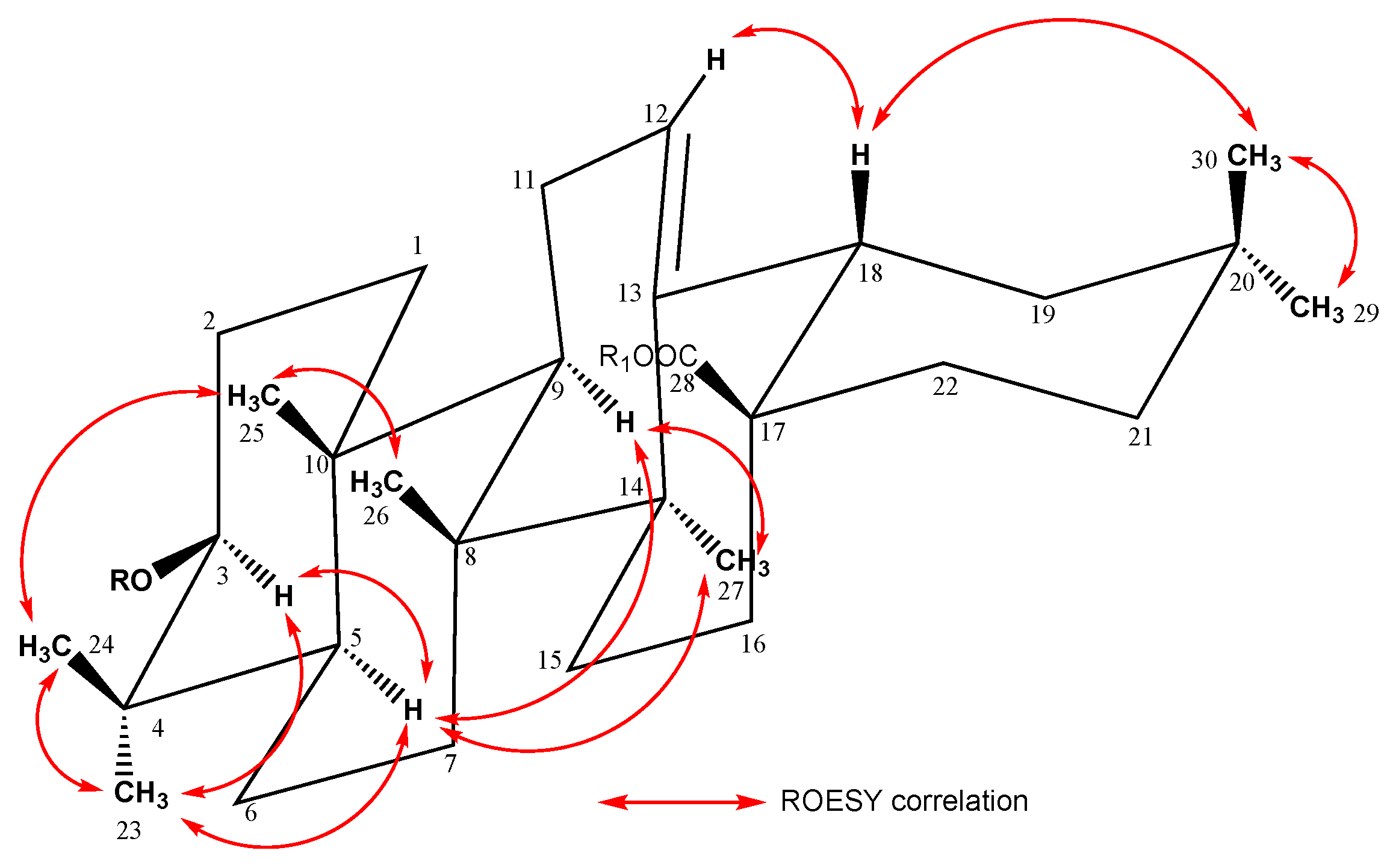
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
- 3-O-α-L-rhamnopyranosyl-(1→3)-β-D-glucopyranosyl-(1→4)-β-D-glucopyranosyl-(1→4)-β-D-xylopyranosyl-(1→3)-α-L-rhamnopyranosyl-(1→2)-α-L-arabinopyranosyloleanolic acid 28-O-β-D-glucopyranosyl-(1→6)-β-D-glucopyranosyl ester (1). White, amorphous powder. Melting point, 220°C. [α]25D-15 (c 0.2, MeOH). For 1H and 13C NMR data (600 MHz, CD3OD), see Table 1 and Table 2. ESIMS (positive-ion mode) m/z: 1683.7553 [M + Na]+ (calcd. for C76H124O39Na, 1683,7617).
- 3-O-β-D-xylopyranosyl-(1→2)-[α-L-rhamnopyranosyl-(1→4)]-β-D-glucopyranosyl-(1→4)-β-D-glucopyranosyl-(1→4)-β-D-xylopyranosyl-(1→3)-α-L-rhamnopyranosyl-(1→2)-α-L-arabinopyranosyloleanolic acid 28-O-β-D-glucopyranosyl-(1→6)-β-D-glucopyranosyl ester (2). White, amorphous powder. Melting point, 238°C. [α]25D-10 (c 0.2, MeOH). For 1H and 13C NMR data (600 MHz, CD3OD), see Table 1 and Table 2. ESIMS (positive-ion mode) m/z: 1815.8003 [M + Na]+ (calcd. for C81H132O43Na, 1815.8040).
- 3-O-β-D-xylopyranosyl-(1→2)-[α-L-rhamnopyranosyl-(1→4)]-β-D-glucopyranosyl-(1→4)-β-D-glucopyranosyl-(1→4)-β-D-xylopyranosyl-(1→3)-α-L-rhamnopyranosyl-(1→2)-α-L-arabinopyranosyloleanolic acid (3). White, amorphous powder. Melting point, 250 °C. [α]25D-17 (c 0.2, MeOH). For 1H and 13C NMR data (600 MHz, CD3OD), see Table 1 and Table 2. ESIMS (positive-ion mode) m/z: 1491.6949 [M + Na]+ (calcd. for C69H112O33Na, 1491.6984).
- 3-O-β-D-xylopyranosyl-(1→2)-[α-L-rhamnopyranosyl-(1→4)]-β-D-xylopyranosyl-(1→4)-β-D-glucopyranosyl-(1→4)-β-D-xylopyranosyl-(1→3)-α-L-rhamnopyranosyl-(1→2)-α-L-arabinopyranosyloleanolic acid 28-O-β-D-glucopyranosyl-(1→6)-β-D-glucopyranosyl ester (4). White, amorphous powder. [α]25D-20 (c 0.2, MeOH). For 1H and 13C NMR data (600 MHz, CD3OD), see Table 1 and Table 2. ESIMS (positive-ion mode) m/z: 1785.7940 [M + Na]+ (calcd. for C80H130O42Na, 1785.7934).
- 3-O-α-L-rhamnopyranosyl-(1→4)-β-D-glucopyranosyl-(1→4)-β-D-glucopyranosyl-(1→4)-β-D-xylopyranosyl-(1→3)-α-L-rhamnopyranosyl-(1→2)-α-L-arabinopyranosyloleanolic acid 28-O-β-D-glucopyranosyl-(1→6)-β-D-glucopyranosyl ester (5). White, amorphous powder. Melting point, 226°C. [α]25D-12 (c 0.2, MeOH). For 1H and 13C NMR data (600 MHz, CD3OD), see Table 1 and Table 2. ESIMS (positive-ion mode) m/z: 1683.7553 [M + Na]+ (calcd. for C76H124O39Na, 1683,7617).
3.4. Acid Hydrolysis and Absolute Configuration Determination
3.5. Bioactivity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Sample Availability
References
- International Agency for Research on Cancer (IARC), World Health Organization. Cancer Today: GLOBOCAN 2020 Estimates. 2021. Available online: https://gco.iarc.fr/today/home (accessed on 28 March 2023).
- Bedirian, K.; Aghabekyan, T.; Mesrobian, A.; Shekherdimian, S.; Zohrabyan, D.; Safaryan, L.; Sargsyan, L.; Avagyan, A.; Harutyunyan, L.; Voskanyan, A.; et al. Overview of Cancer Control in Armenia and Policy Implications. Front. Oncol. 2022, 11, 782581. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, H.; Zhang, L.; Guo, T.; Wang, P.; Geng, M.; Li, Y. Synthesis and antitumor activities of naturally occurring oleanolic acid triterpenoid saponins and their derivatives. Eur. J. Med. Chem. 2013, 64, 1–15. [Google Scholar] [CrossRef]
- Rezgui, A.; Mitaine-Offer, A.-C.; Miyamoto, T.; Tanaka, C.; Delemasure, S.; Dutartre, P.; Lacaille-Dubois, M.-A. Oleanolic acid and hederagenin glycosides from Weigela stelzneri. Phytochemistry 2016, 123, 40–47. [Google Scholar] [CrossRef]
- Nguyen, D.H.; Mitaine-Offer, A.-C.; Maroso, S.; Papini, A.-M.; Paululat, T.; Bellaye, P.-S.; Collin, B.; Chambin, O.; Lacaille-Dubois, M.-A. Cytotoxic glycosides from Weigela × “Bristol Ruby”. Fitoterapia 2019, 137, 104242. [Google Scholar] [CrossRef]
- Podolak, I.; Galanty, A.; Sobolewska, D. Saponins as cytotoxic agents: A review. Phytochem. Rev. 2010, 9, 425–474. [Google Scholar] [CrossRef]
- Medicinal Plants and Their Uses and Benefits with Reference to Small Farmers in Armenia. Available online: https://armeniandinner.com/wp-content/uploads/2020/09/Armen-Medicinal-plants5.pdf (accessed on 28 March 2023).
- Carlson, S.E.; Linder, H.P.; Donoghue, M.J. The historical biogeography of Scabiosa (Dipsacaceae): Implications for Old World plant disjunctions: Historical biogeography of Scabiosa. J. Biogeogr. 2012, 39, 1086–1100. [Google Scholar] [CrossRef]
- Lack, H.W. The discovery and naming of Lomelosia caucasica (Dipsacaceae) with notes on its nomenclature and its early cultivation. Willdenowia 2018, 48, 185–194. [Google Scholar] [CrossRef]
- Bendamene, S.; Boutaghane, N.; Bellik, Y.; Sayagh, C.; Alabdul Magid, A.; Harakat, D.; Kabouche, Z.; Voutquenne-Nazabadioko, L. Semipapposides A-M, triterpenoid bidesmosides saponins from the roots of Scabiosa semipapposa. Phytochemistry 2020, 180, 112526. [Google Scholar] [CrossRef] [PubMed]
- Lehbili, M.; Alabdul Magid, A.; Kabouche, A.; Voutquenne-Nazabadioko, L.; Morjani, H.; Harakat, D.; Kabouche, Z. Triterpenoid saponins from Scabiosa stellata collected in North-eastern Algeria. Phytochemistry 2018, 150, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Koike, K.; Han, L.-K.; Okuda, H.; Nikaido, T. New biologically active triterpenoid saponins from Scabiosa tschiliensis. J. Nat. Prod. 2004, 67, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Baykal, T.; Panayir, T.; Tasdemir, D.; Sticher, O.; Çalis, I. Triterpene saponins from Scabiosa rotate. Phytochemistry 1998, 48, 867–873. [Google Scholar] [CrossRef] [PubMed]



| Position | 1 | 2 | 3 | 4 | 5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | δC | δH | δC | δH | |
| 1(CH2) | 38.5 | α 1.01 m β 1.64 m | 38.6 | α 1.01 m β 1.64 m | 38.5 | α 0.98 m β 1.63 m | 38.9 | α 0.98 m β 1.62 m | 38.5 | α 0.98 m β 1.63 m |
| 2(CH2) | 25.8 | β 1.75 m α 1.86 m | 25.4 | β 1.75 m α 1.87 m | 22.4 | β 1.72 m α 1.85 m | 25.8 | β 1.70 m α 1.83 m | 26.1 | β 1.70 m α 1.82 m |
| 3(CH) | 89.2 | α 3.14 dd (11.5, 4.0) | 89.2 | α 3.14 | 89.2 | α 3.13 | 89.2 | α 3.11 dd (11.7, 4.1) | 91.0 | α 3.11 dd (11.8, 4.0) |
| 4(C) | 39.0 | - | 38.9 | - | 38.8 | - | 38.7 | - | 38.7 | - |
| 5(CH) | 55.8 | α 0.80 | 55.8 | α 0.80 | 55.7 | α 0.80 br d (11.2) | 55.8 | α 0.77 | 55.8 | α 0.77 |
| 6(CH2) | 18.0 | β 1.42 α 1.56 | 18.1 | β 1.43 α 1.56 | 18.2 | β 1.43 α 1.56 | 18.0 | β 1.42 α 1.54 | 18.0 | β 1.49 α 1.54 |
| 7(CH2) | 32.4 | β 1.34 m α 1.49 m | 32.6 | β 1.35 m α 1.50 m | 32.7 | β 1.33 m α 1.53 m | 32.5 | β 1.32 m α 1.48 m | 32.7 | β 1.32 m α 1.48 m |
| 8(C) | 39.5 | - | 39.4 | - | 39.2 | - | 39.4 | - | 39.4 | - |
| 9(CH) | 47.8 | α 1.60 | 47.9 | α 1.61 | 47.8 | α 1.60 | 47.8 | α 1.58 | 47.8 | α 1.58 |
| 10(C) | 36.7 | - | 36.6 | - | 36.5 | - | 36.5 | - | - | |
| 11(CH2) | 23.2 | 1.92 1.95 | 23.1 | 1.90 1.92 | 23.1 | 1.86 1.91 | 23.1 | 1.88 1.90 | 23.1 | 1.88 1.90 |
| 12(CH) | 122.4 | 5.27 t (3.6) | 122.4 | 5.28 t (3.6) | 122.0 | 5.25 t (3.6) | 122.4 | 5.24 t (3.4) | 122.4 | 5.24 t (3.5) |
| 13(C) | 143.5 | - | 143.5 | - | 144.2 | - | 143.5 | - | 143.5 | - |
| 14(C) | 41.4 | - | 41.5 | - | 41.6 | - | 41.5 | - | 41.4 | - |
| 15(CH2) | 27.6 | α 1.09 m β 1.81 m | 27.5 | α 1.10 m β 1.81 m | 27.5 | α 1.05 m β 1.79 m | 27.6 | α 1.08 m β 1.78 m | 27.6 | α 1.08 m β 1.78 m |
| 16(CH2) | 22.7 | β 1.73 α 2.07 m | 22.7 | β 1.74 α 2.07 m | 22.9 | β 1.60 α 1.97 m | 22.6 | β 1.72 α 2.04 m | 22.6 | β 1.70 α 2.04 m |
| 17(C) | 46.7 | - | 46.7 | - | - | 46.5 | - | 46.4 | - | |
| 18(CH) | 41.2 | β 2.89 dd (14.5, 3.3) | 41.2 | β 2.89 dd (12.7, 3.5) | 41.5 | β 2.87 dd (12.8, 3.4) | 41.2 | β 2.85 dd (13.3, 3.4) | 41.2 | β 2.85 dd (12.7, 3.3) |
| 19(CH2) | 45.9 | β 1.16 α 1.74 | 45.9 | β 1.18 α 1.74 | 46.2 | β 1.13 α 1.68 | 45.8 | β 1.14 α 1.71 | 45.8 | β 1.13 α 1.70 |
| 20(C) | 30.2 | - | 30.2 | - | 30.2 | - | 30.2 | - | 30.2 | - |
| 21(CH2) | 33.6 | β 1.24 α 1.42 m | 33.5 | β 1.25 α 1.43 m | 33.7 | β 1.20 α 1.40 m | 33.5 | β 1.23 α 1.40 m | 33.8 | β 1.21 α 1.39 m |
| 22(CH2) | 31.8 | α 1.63 β 1.76 | 31.8 | α 1.61 β 1.75 | 32.6 | α 1.53 β 1.74 | 32.0 | α 1.60 β 1.72 | 32.0 | α 1.60 β 1.73 |
| 23(CH3) | 27.3 | α 1.05 s | 27.3 | α 1.05 s | 27.3 | α 1.05 s | 27.1 | α 1.02 s | 27.1 | α 1.02 s |
| 24(CH3) | 15.8 | β 0.88 s | 15.8 | β 0.88 s | 15.8 | β 0.87 s | 15.8 | β 0.84 s | 15.8 | β 0.85 s |
| 25(CH3) | 14.8 | β 0.99 s | 14.8 | β 0.99 s | 14.7 | β 0.97 s | 14.9 | β 0.96 s | 14.8 | β 0.96 s |
| 26(CH3) | 16.6 | β 0.83 s | 16.4 | β 0.83 s | 16.6 | β 0.85 s | 16.6 | β 0.80 s | 16.7 | β 0.80 s |
| 27(CH3) | 24.9 | α 1.18 s | 24.9 | α 1.18 s | 25.1 | α 1.18 s | 24.9 | α 1.15 s | 24.9 | α 1.15 s |
| 28(CH3) | 178.0 | - | 176.7 | - | 180.2 | - | 176.7 | - | 176.6 | - |
| 29(CH3) | 32.1 | α 0.93 s | 32.1 | α 0.94 s | 32.2 | α 0.92 s | 32.0 | α 0.90 s | 32.0 | α 0.90 s |
| 30(CH3) | 22.7 | β 0.96 s | 22.7 | β 0.97 s | 22.7 | β 0.96 s | 22.8 | β 0.93 s | 22.7 | β 0.93 s |
| Position | 1 | 2 | 3 | 4 | 5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | δC | δH | δC | δH | |
| 3-O- | ||||||||||
| Ara-1 | 103.8 | 4.52 d (5.0) | 103.8 | 4.52 d (5.3) | 103.8 | 4.52 d (5.3) | 103.8 | 4.48 d (5.5) | 103.8 | 4.48 d (5.5) |
| 2 | 74.9 | 3.79 | 75.2 | 3.78 | 75.0 | 3.78 | 74.8 | 3.76 | 75.5 | 3.76 |
| 3 | 72.3 | 3.73 | 72.3 | 3.75 | 72.3 | 3.74 | 72.3 | 3.75 | 72.7 | 3.72 |
| 4 | 67.6 | 3.79 | 67.6 | 3.80 | 67.6 | 3.79 | 68.1 | 4.11 | 68.6 | 4.12 |
| 5 | 63.2 | 3.53 3.87 | 63.2 | 3.52 3.87 | 63.2 | 3.52 3.86 | 63.1 | 3.49 3.83 | 63.1 | 3.50 3.83 |
| Rha I-1 | 100.1 | 5.21 s | 100.1 | 5.22 s | 100.1 | 5.21 s | 100.0 | 5.18 s | 100.3 | 5.18 s |
| 2 | 70.4 | 4.09 s | 70.4 | 4.09 s | 70.4 | 4.09 s | 70.4 | 4.05 s | 70.8 | 4.06 s |
| 3 | 80.7 | 3.87 | 80.7 | 3.86 | 80.7 | 3.85 | 80.6 | 3.84 | 81.2 | 3.83 |
| 4 | 71.4 | 3.59 | 71.4 | 3.60 | 71.4 | 3.60 | 71.3 | 3.56 t (9.4) | 72.1 | 3.57 |
| 5 | 68.6 | 3.92 | 68.6 | 3.93 | 68.6 | 3.92 | 68.6 | 3.88 | 68.9 | 3.89 |
| 6 | 16.4 | 1.26 d (6.2) | 16.5 | 1.26 d (6.1) | 16.6 | 1.26 d (5.9) | 16.6 | 1.22 d (5.9) | 16.5 | 1.23 d (6.5) |
| Rha II-1 | 101.4 | 5.19 s | 100.3 | 5.37 s | 100.3 | 5.37 s | 100.9 | 5.17 s | 100.0 | 5.40 s |
| 2 | 70.9 | 3.97 s | 71.2 | 3.93 s | 71.2 | 3.92 s | 71.2 | 3.90 s | 70.5 | 3.92 s |
| 3 | 70.8 | 3.71 | 70.5 | 3.86 | 70.5 | 3.85 | 70.5 | 3.74 | 70.9 | 3.88 |
| 4 | 72.3 | 3.42 | 72.7 | 3.40 | 72.3 | 3.42 | 72.6 | 3.36 | 73.8 | 3.31 |
| 5 | 68.7 | 4.02 dq (9.6, 6.2) | 68.1 | 4.39 | 68.0 | 4.37 | 68.1 | 4.22 | 68.0 | 4.46 |
| 6 | 16.4 | 1.27 d (6.2) | 16.4 | 1.24 d (6.2) | 16.5 | 1.24 d (6.2) | 16.6 | 1.21 d (6.5) | 16.5 | 1.22 d (5.9) |
| Xyl I-1 | 104.9 | 4.55 d (7.6) | 104.9 | 4.53 d (7.4) | 104.9 | 4.53 d (7.8) | 104.9 | 4.49 d (7.6) | 104.9 | 4.49 d (7.6) |
| 2 | 73.9 | 3.36 | 74.0 | 3.37 | 73.7 | 3.35 | 73.8 | 3.34 | 73.8 | 3.34 |
| 3 | 74.4 | 3.52 | 74.4 | 3.53 | 74.4 | 3.52 | 74.0 | 3.48 | 75.1 | 3.50 |
| 4 | 76.9 | 3.72 | 76.8 | 3.73 | 76.9 | 3.71 | 76.9 | 3.69 | 77.8 | 3.69 |
| 5 | 63.2 | 3.34 4.06 dd (11.9, 5.4) | 63.1 | 3.32 4.06 dd (11.7, 5.2) | 63.1 | 3.32 4.06 dd (11.8, 5.3) | 62.9 | 3.34 4.02 dd (11.9, 5.4) | 63.1 | 3.31 4.02 dd (11.7, 5.3) |
| Xyl II-1 | 103.0 | 4.38 d (7.8) | 103.0 | 4.38 d (7.7) | 103.2 | 4.34 d (7.6) | ||||
| 2 | 73.7 | 3.22 | 73.7 | 3.22 | 77.0 | 3.40 | ||||
| 3 | 76.6 | 3.28 | 76.7 | 3.30 | 75.0 | 3.76 | ||||
| 4 | 69.5 | 3.60 | 69.5 | 3.60 | 78.6 | 3.62 | ||||
| 5 | 65.4 | 3.15 3.91 | 65.4 | 3.14 3.90 | 63.1 | 3.32 4.11 | ||||
| Xyl III-1 | 101.9 | 4.25 d (7.6) | ||||||||
| 2 | 73.9 | 3.16 | ||||||||
| 3 | 76.6 | 3.27 | ||||||||
| 4 | 69.6 | 3.49 | ||||||||
| 5 | 65.6 | 3.14 3.88 | ||||||||
| Glc I-1 | 101.7 | 4.42 d (7.7) | 101.7 | 4.42 d (7.8) | 101.7 | 4.41 d (7.7) | 101.7 | 4.38 d (7.6) | 102.5 | 4.38 d (7.6) |
| 2 | 73.1 | 3.30 | 72.9 | 3.30 | 72.7 | 3.29 | 73.0 | 3.27 | 73.5 | 3.27 |
| 3 | 74.2 | 3.50 | 74.9 | 3.42 | 74.9 | 3.50 | 75.0 | 3.44 | 75.5 | 3.44 |
| 4 | 78.9 | 3.58 | 78.7 | 3.58 | 78.7 | 3.57 | 78.8 | 3.50 | 80.0 | 3.58 |
| 5 | 75.0 | 3.47 | 75.7 | 3.46 | 75.7 | 3.45 | 75.7 | 3.45 | 76.1 | 3.52 |
| 6 | 60.2 | 3.87 3.93 | 60.2 | 3.87 3.92 | 60.1 | 3.87 3.92 | 60.1 | 3.81 3.90 | 61.3 | 3.79 3.86 |
| Glc II-1 | 103.0 | 4.43 d (7.8) | 103.1 | 4.43 d (7.7) | 103.1 | 4.42 d (7.7) | 103,8 | 4.41 d (8.2) | ||
| 2 | 73.9 | 3.35 | 76.7 | 3.44 | 75.8 | 3.47 | 73.5 | 3.43 | ||
| 3 | 82.7 | 3.53 | 76.4 | 3.54 | 76.4 | 3.54 | 74.5 | 3.45 | ||
| 4 | 68.6 | 3.38 | 76.9 | 3.74 | 76.9 | 3.73 | 77.0 | 3.78 | ||
| 5 | 76.5 | 3.32 | 75.8 | 3.48 | 75.8 | 3.44 | 75.5 | 3.52 | ||
| 6 | 61.0 | 3.92, nd | 59.8 | 3.93, nd | 59.6 | 3.93, nd | 60.3 | 3.91, nd | ||
| 28-O- | ||||||||||
| Glc III-1 | 94.4 | 5.38 d (8.1) | 94.4 | 5.38 d (8.1) | 94.3 | 5.34 d (8.2) | 94.8 | 5.34 d (8.2) | ||
| 2 | 72.4 | 3.36 | 72.5 | 3.36 | 72.5 | 3.36 | 73.0 | 3.35 | ||
| 3 | 76.7 | 3.44 | 74.7 | 3.55 | 74.9 | 3.50 | 74.9 | 3.54 | ||
| 4 | 69.5 | 3.46 | 69.5 | 3.45 | 69.6. | 3.42 | 70.5 | 3.44 | ||
| 5 | 76.2 | 3.53 | 76.7 | 3.43 | 76.6 | 3.41 | 76.8 | 3.42 | ||
| 6 | 68.1 | 3.79 4.15 dd (11.9, 1.5) | 68.1 | 3.80 4.14 dd (11.8, 1.9) | 68.1 | 3.89 4.11 | 68.6 | 3.83 4.12 dd (10.9, 1.5) | ||
| Glc IV-1 | 103.2 | 4.37 d (7.7) | 103.2 | 4.37 d (7.4) | 103.2 | 4.33 d (7.6) | 103.2 | 4.33 d (7.6) | ||
| 2 | 73.7 | 3.23 dd (9.0, 8.0) | 73.6 | 3.23 | 73.0 | 3.20 dd (8.8, 7.6) | 74.4 | 3.20 dd (9.4, 7.6) | ||
| 3 | 76.7 | 3.39 | 76.7 | 3.38 | 76.8 | 3.35 | 77.1 | 3.37 | ||
| 4 | 70.1 | 3.32 | 70.1 | 3.32 | 71.0 | 3.28 | 71.0 | 3.29 | ||
| 5 | 76.7 | 3.27 | 76.6 | 3.30 | 76.8 | 3.25 | 77.1 | 3.25 | ||
| 6 | 61.3 | 3.70 3.87 | 61.3 | 3.69 dd (12.0, 5.6) 3.88 | 61.3 | 3.67 3.87 | 62.0 | 3.68 dd (11.7, 5.3) 3.86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nazaryan, S.; Bruguière, A.; Hovhannisyan, N.; Miyamoto, T.; Dias, A.M.M.; Bellaye, P.-S.; Collin, B.; Briand, L.; Mitaine-Offer, A.-C. Oleanolic Acid Glycosides from Scabiosa caucasica and Scabiosa ochroleuca: Structural Analysis and Cytotoxicity. Molecules 2023, 28, 4329. https://doi.org/10.3390/molecules28114329
Nazaryan S, Bruguière A, Hovhannisyan N, Miyamoto T, Dias AMM, Bellaye P-S, Collin B, Briand L, Mitaine-Offer A-C. Oleanolic Acid Glycosides from Scabiosa caucasica and Scabiosa ochroleuca: Structural Analysis and Cytotoxicity. Molecules. 2023; 28(11):4329. https://doi.org/10.3390/molecules28114329
Chicago/Turabian StyleNazaryan, Samvel, Antoine Bruguière, Nelli Hovhannisyan, Tomofumi Miyamoto, Alexandre M. M. Dias, Pierre-Simon Bellaye, Bertrand Collin, Loïc Briand, and Anne-Claire Mitaine-Offer. 2023. "Oleanolic Acid Glycosides from Scabiosa caucasica and Scabiosa ochroleuca: Structural Analysis and Cytotoxicity" Molecules 28, no. 11: 4329. https://doi.org/10.3390/molecules28114329
APA StyleNazaryan, S., Bruguière, A., Hovhannisyan, N., Miyamoto, T., Dias, A. M. M., Bellaye, P.-S., Collin, B., Briand, L., & Mitaine-Offer, A.-C. (2023). Oleanolic Acid Glycosides from Scabiosa caucasica and Scabiosa ochroleuca: Structural Analysis and Cytotoxicity. Molecules, 28(11), 4329. https://doi.org/10.3390/molecules28114329







