Studies on Chemical Composition of Pueraria lobata and Its Anti-Tumor Mechanism
Abstract
1. Introduction
2. Results
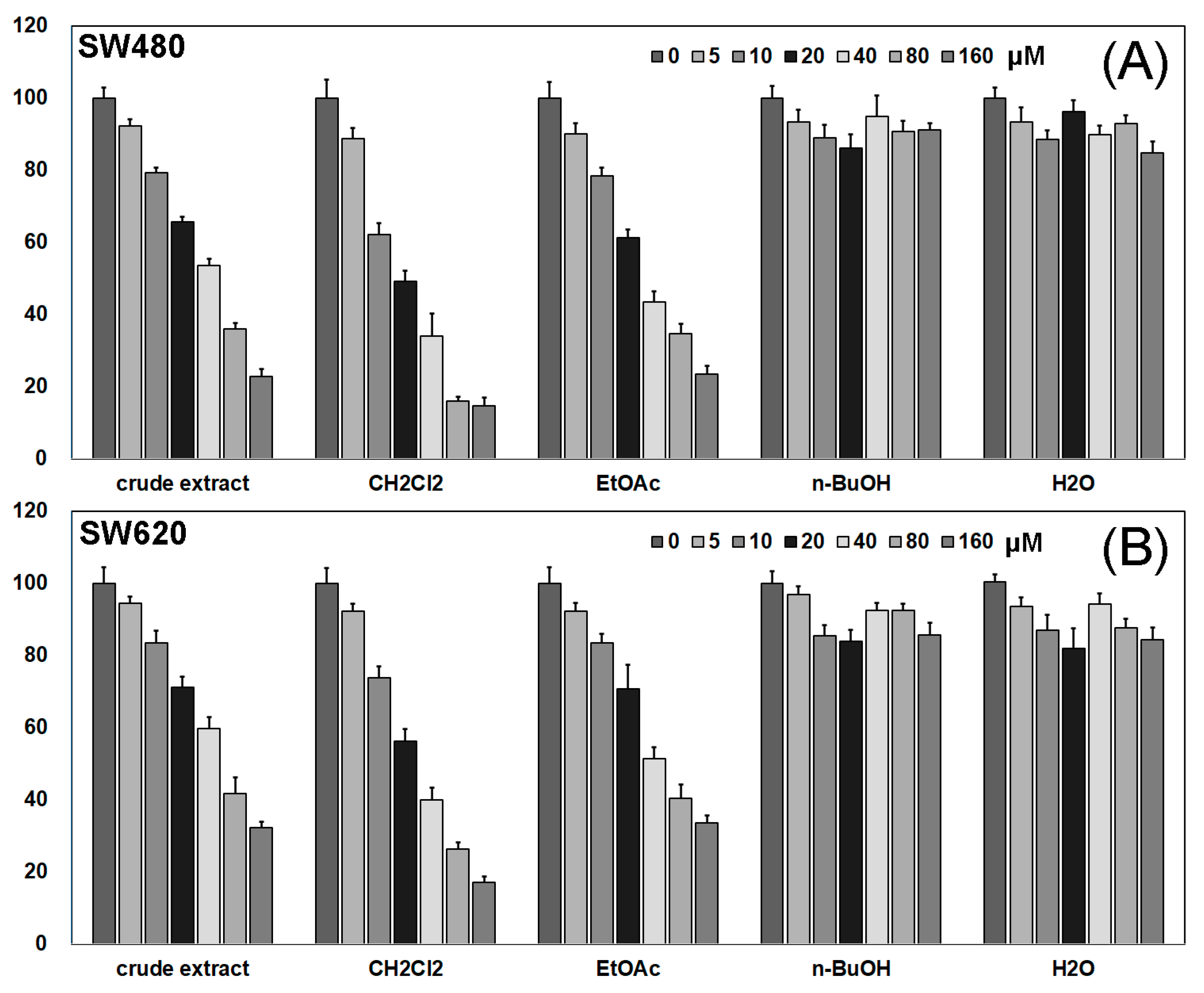
2.1. Activity Screening of Different Fractions of Pueraria lobata
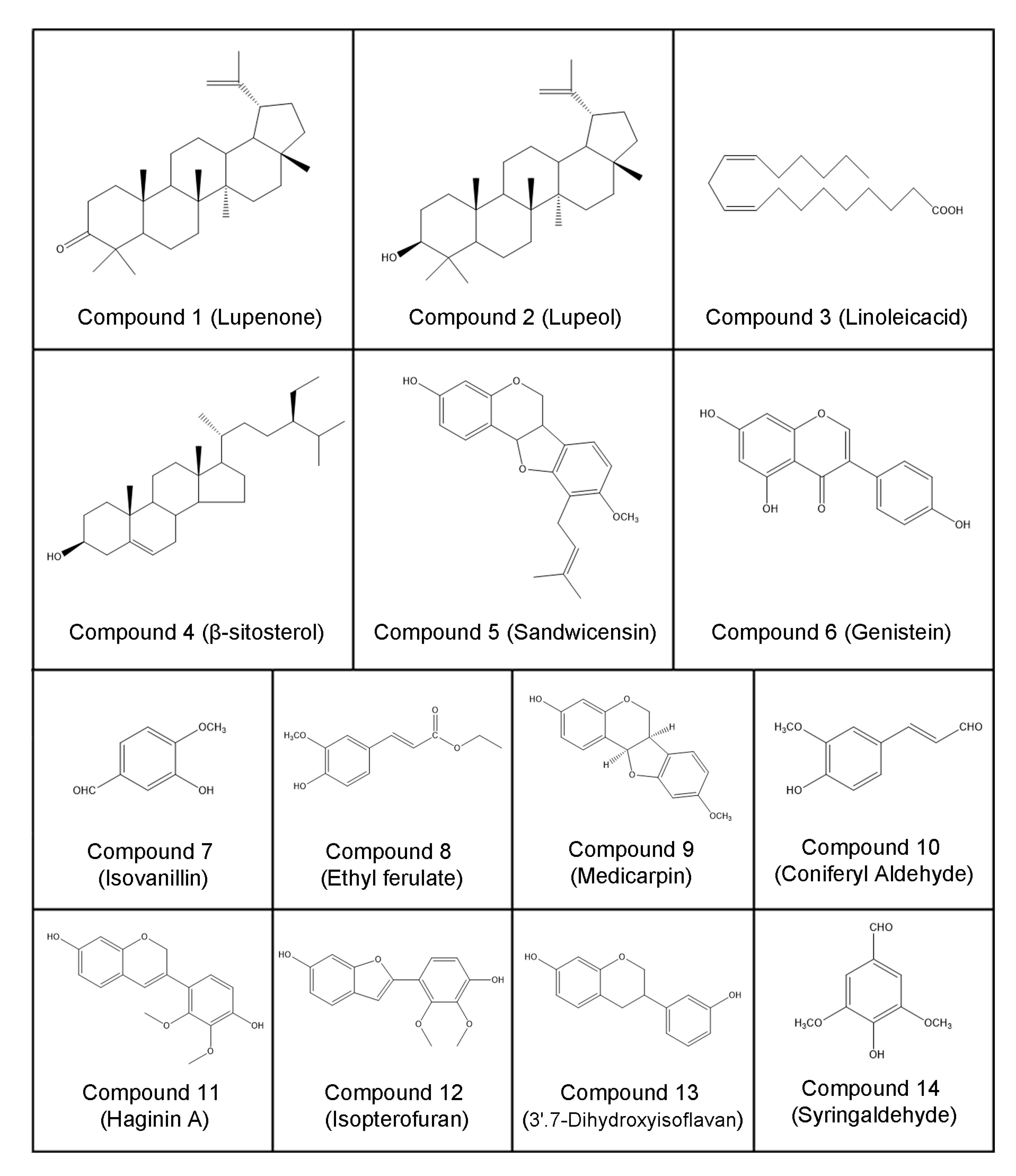
2.2. Structural Elucidation
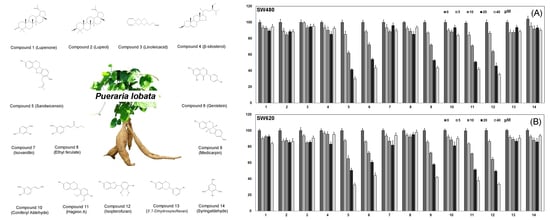
2.3. Activity Screening of Compounds 1–14
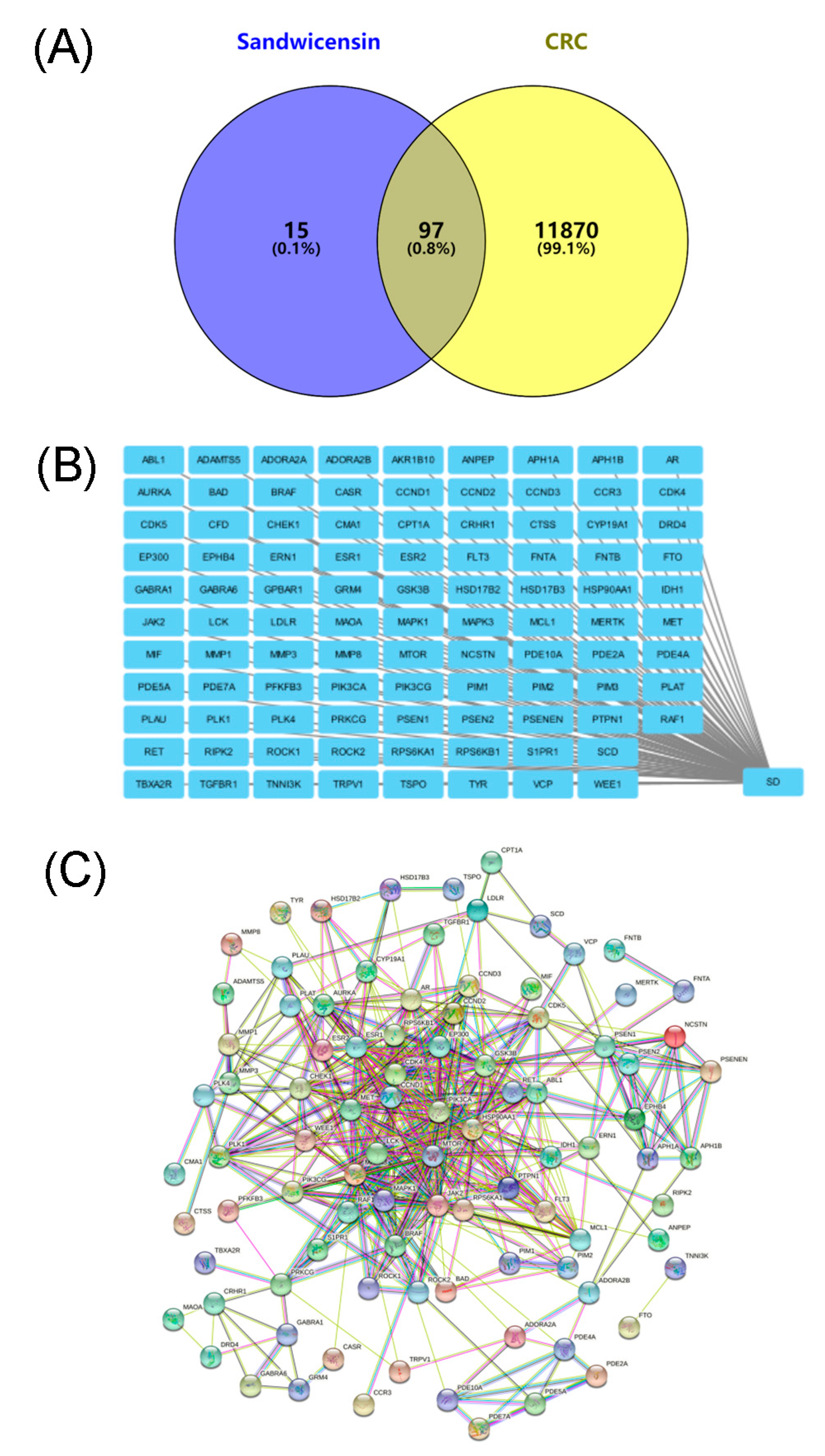
2.4. Target Screening of Sandwicensin in the Treatment of CRC and Construction of Its Network Relationship
2.5. PPI Network Analysis of Target Proteins of Sandwicensin in CRC
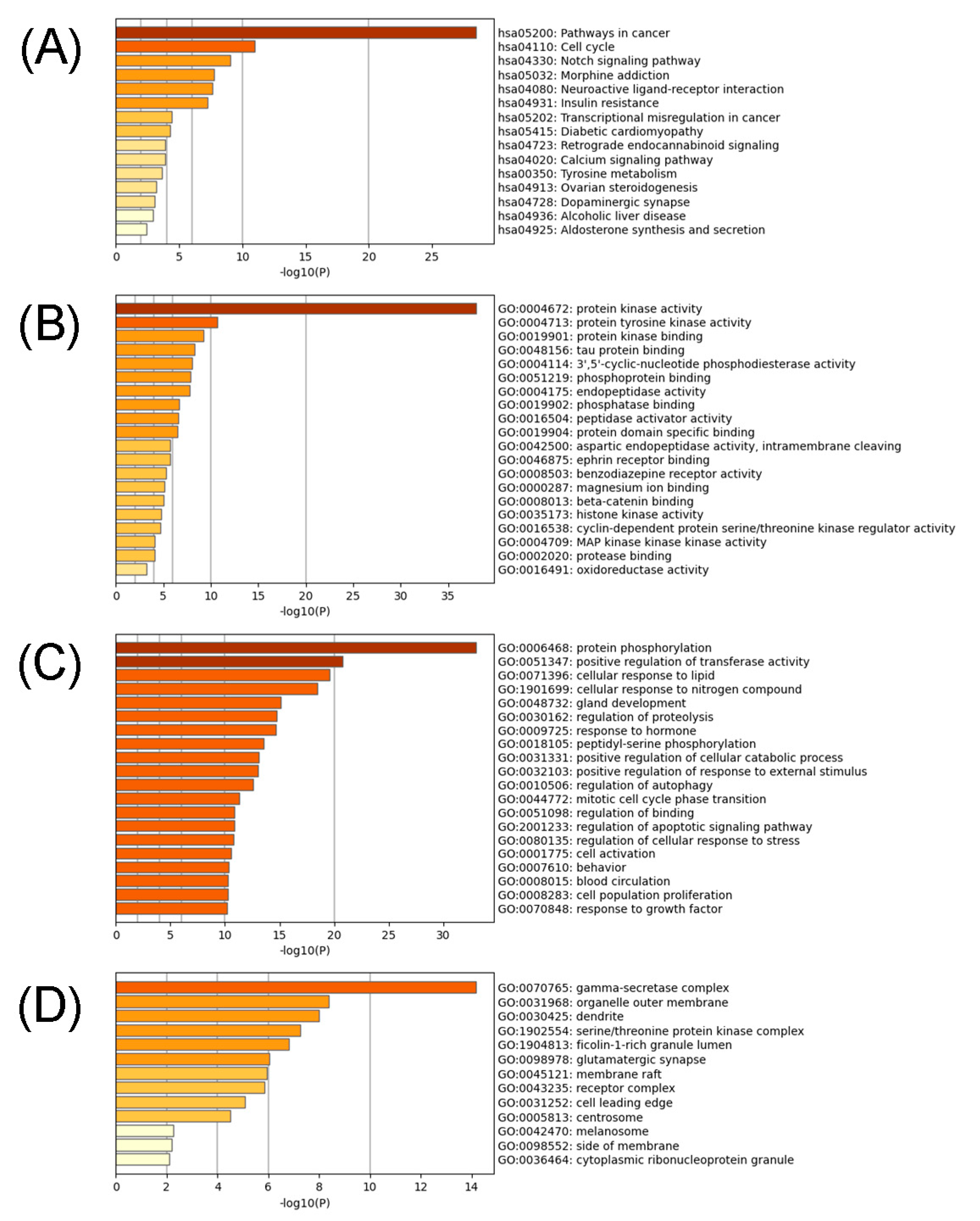
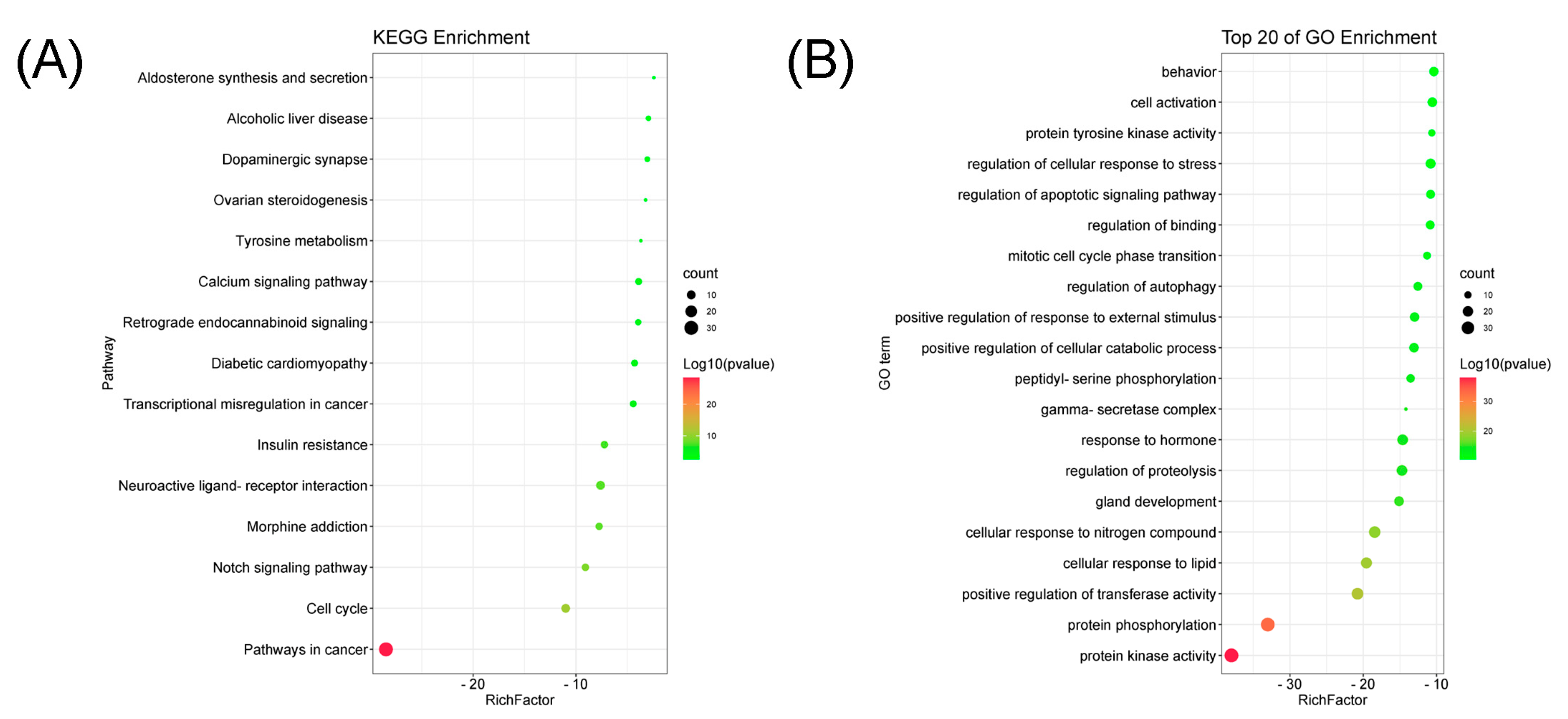
2.6. GO Annotation and KEGG Pathway Enrichment Analysis
3. Discussion
4. Materials and Methods
4.1. General Information
4.2. Plant Material
4.3. Cell Materials
4.4. Extraction and Isolation
4.5. Preparative Thin Layer Plates
4.6. Characterization
4.7. Cell Culture
4.8. Cytotoxicity Assessment
4.9. Network Pharmacology Technology
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Chang, K.F.; Chang, J.T.; Huang, X.F.; Lin, Y.L.; Liao, K.W.; Huang, C.W.; Tsai, N.M. Antitumor effects of N-butylidenephthalide encapsulated in lipopolyplexs in colorectal cancer cells. Molecules 2020, 25, 2394. [Google Scholar] [CrossRef] [PubMed]
- Uyemura, S.A.; Stopper, H.; Martin, F.L.; Kannen, V. A perspective discussion on rising pesticide levels and colon cancer burden in brazil. Front. Public Health 2017, 5, 273. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Xiao, X.; Zhang, C.; Yu, W.; Guo, W.; Zhang, Z.; Li, Z.; Feng, X.; Hao, J.; Zhang, K.; et al. Melatonin synergizes the chemotherapeutic effect of 5-fluorouracil in colon cancer by suppressing PI3K/AKT and NF-kappaB/iNOS signaling pathways. J. Pineal Res. 2017, 62, e12380. [Google Scholar] [CrossRef]
- Ma, S.; Lei, Y.; Zhang, L.; Wang, J. Research on the inhibiting effect of tanshinone IIA on colon cancer cell growth via COX-2-Wnt/beta-catenin signaling pathway. J. BUON 2018, 23, 1337–1342. [Google Scholar]
- Song, Q.; Sun, X.; Guo, H.; Yu, Q. Concomitant inhibition of receptor tyrosine kinases and downstream AKT synergistically inhibited growth of KRAS/BRAF mutant colorectal cancer cells. Oncotarget 2017, 8, 5003–5015. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Z.; Qi, F.; Cui, Y.; Zhao, L.; Sun, X.; Tang, W.; Cai, P. An update on Chinese herbal medicines as adjuvant treatment of anticancer therapeutics. Biosci. Trends 2018, 12, 220–239. [Google Scholar] [CrossRef]
- Zhang, L. Pharmacokinetics and drug delivery systems for puerarin, a bioactive flavone from traditional Chinese medicine. Drug Deliv. 2019, 26, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Yuan, R.; Chen, X.; Xin, Q.; Wang, Y.; Shang, X.; Cong, W.; Chen, K. Puerarin eeduces blood pressure in spontaneously hypertensive rats by targeting eNOS. Am. J. Chin. Med. 2019, 47, 19–38. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Kim, H.J.; Ok, H.M.; Jeong, H.Y.; Lee, W.J.; Weaver, C.; Kwon, O. Effect and interactions of Pueraria-Rehmannia and aerobic exercise on metabolic inflexibility and insulin resistance in ovariectomized rats fed with a high-fat diet. J. Funct. Foods 2018, 45, 146–154. [Google Scholar] [CrossRef]
- Jearapong, N.; Chatuphonprasert, W.; Jarukamjorn, K. Miroestrol, a phytoestrogen from Pueraria mirifica, improves the antioxidation state in the livers and uteri of beta-naphthoflavone-treated mice. J. Nat. Med. 2014, 68, 173–180. [Google Scholar] [CrossRef]
- Shukla, R.; Banerjee, S.; Tripathi, Y.B. Pueraria tuberosa extract inhibits iNOS and IL-6 through suppression of PKC-alpha and NF-kB pathway in diabetes-induced nephropathy. J. Pharm. Pharmacol. 2018, 70, 1102–1112. [Google Scholar] [CrossRef] [PubMed]
- Satpathy, S.; Patra, A.; Ahirwar, B.; Delwar Hussain, M. Antioxidant and anticancer activities of green synthesized silver nanoparticles using aqueous extract of tubers of Pueraria tuberosa. Artif. Cells Nanomed. Biotechnol. 2018, 46, S71–S85. [Google Scholar] [CrossRef]
- Zhang, X.L.; Wang, B.B.; Mo, J.S. Puerarin 6”-O-xyloside possesses significant antitumor activities on colon cancer through inducing apoptosis. Oncol. Lett. 2018, 16, 5557–5564. [Google Scholar] [CrossRef] [PubMed]
- Satpathy, S.; Patra, A.; Hussain, M.D.; Kazi, M.; Aldughaim, M.S.; Ahirwar, B. A fraction of Pueraria tuberosa extract, rich in antioxidant compounds, alleviates ovariectomized-induced osteoporosis in rats and inhibits growth of breast and ovarian cancer cells. PLoS ONE 2021, 16, e0240068. [Google Scholar] [CrossRef]
- Ahn, E.K.; Oh, J.S. Lupenone isolated from Adenophora triphylla var. japonica extract inhibits adipogenic differentiation through the downregulation of PPARgamma in 3T3-L1 cells. Phytother. Res. 2013, 27, 761–766. [Google Scholar] [CrossRef]
- Huang, H.M.; Ho, C.Y.; Chang, G.R.; Shia, W.Y.; Lai, C.H.; Chao, C.H.; Wang, C.M. HPLC/ESI-MS and NMR analysis of chemical constitutes in bioactive extract from the root nodule of vaccinium emarginatum. Pharmaceuticals 2021, 14, 1098. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S. Preliminary Study on Chemical Composition and the Activity of Dracocephalum Heterophyllum. Ph.D. Thesis, Lanzhou University, Gansu, China, 2013. [Google Scholar]
- Ododo, M.M.; Choudhury, M.K.; Dekebo, A.H. Structure elucidation of beta-sitosterol with antibacterial activity from the root bark of Malva parviflora. SpringerPlus 2016, 5, 1210. [Google Scholar] [CrossRef]
- McKee, T.C.; Bokesch, H.R.; McCormick, J.L.; Rashid, M.A.; Spielvogel, D.; Gustafson, K.R.; Alavanja, M.M.; Cardelline, J.H., 2nd; Boyd, M.R. Isolation and characterization of new anti-HIV and cytotoxic leads from plants, marine, and microbial organisms. J. Nat. Prod. 1997, 60, 431–438. [Google Scholar] [CrossRef]
- Mitscher, L.A.; Gollapudi, S.R.; Gerlach, D.C.; Drake, S.D.; Eduardo, A.V.; Ward, J.A. Erycristin, a new antimicrobial petrocarpan from Erythrina crista-galli. Phytochemistry 1988, 27, 381–385. [Google Scholar] [CrossRef]
- He, J.; Fan, P.; Feng, S.; Shao, P.; Sun, P. Isolation and purification of two isoflavones from hericium erinaceum mycelium by high-speed counter-current chromatography. Molecules 2018, 23, 560. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Son, D.; Kang, J.; Kim, H.S.; Kim, B.K.; Lee, S. A benzenoid from the stem of Acanthopanax senticosus. Arch. Pharmacal Res. 2004, 27, 912–914. [Google Scholar] [CrossRef]
- Jin, Q.; Jia-hong, Z.; Lu, C.; Ji-jun, C.; Bo-yang, Y.; Sheng-xiang, Q. Study on chemical constituents in tuber of harpagophytum procumbens D.C. Chin. Pharm. J. 2006, 41, 1613–1615. [Google Scholar]
- Hasan, N.; Osman, H.; Mohamad, S.; Chong, W.K.; Awang, K.; Zahariluddin, A.S. The chemical components of sesbania grandiflora root and their antituberculosis activity. Pharmaceuticals 2012, 5, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Ghanadian, M.; Ali, Z.; Khan, I.A.; Balachandran, P.; Nikahd, M.; Aghaei, M.; Mirzaei, M.; Sajjadi, S.E. A new sesquiterpenoid from the shoots of Iranian Daphne mucronata Royle with selective inhibition of STAT3 and Smad3/4 cancer-related signaling pathways. Daru 2020, 28, 253–262. [Google Scholar] [CrossRef]
- Miyase, T.; Ueno, A.; Noro, T.; Fukushima, S. Studies on the Constituentes of Lespedeza cyrtobotrya MIQ. I. The structures of a new chalcone and two new isoflav-3-ens. Chem. Pharm. Bull. 1980, 28, 1172–1177. [Google Scholar] [CrossRef][Green Version]
- Dewick, P.M.; Ingham, J.L. Isopterofuran, a new 2-arylbenzofuran phytoalexin from Coronilla emerus. Phytochemistry 1980, 19, 289–291. [Google Scholar] [CrossRef]
- Luk, K.C.; Stern, L.; Weigele, M.; O’Brien, R.A.; Spirt, N. Isolation and identification of “diazepam-like” compounds from bovine urine. J. Nat. Prod. 1983, 46, 852–861. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.R.; Selvaraj, S.; Jayaprakash, K.S.; Gunasekaran, S.; Kumaresan, S.; Devanathan, J.; Selvam, K.A.; Ramadass, L.; Mani, M.; Rajkumar, P. Multi-spectroscopic (FT-IR, FT-Raman, 1H NMR and 13C NMR) investigations on syringaldehyde. J. Mol. Struct. 2021, 1229, 129490. [Google Scholar] [CrossRef]
- Chen, S.; Li, L. Degradation strategy of cyclin D1 in cancer cells and the potential clinical application. Front. Oncol. 2022, 12, 949688. [Google Scholar] [CrossRef]
- Montalto, F.I.; De Amicis, F. Cyclin D1 in Cancer: A molecular connection for cell cycle control, adhesion and invasion in tumor and stroma. Cells 2020, 9, 2648. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Ruiz, L.; Gonzalez-Moles, M.A.; Gonzalez-Ruiz, I.; Ruiz-Avila, I.; Ramos-Garcia, P. Prognostic and clinicopathological significance of CCND1/Cyclin D1 upregulation in melanomas: A systematic review and comprehensive meta-analysis. Cancers 2021, 13, 1314. [Google Scholar] [CrossRef]
- Goel, B.; Tripathi, N.; Bhardwaj, N.; Jain, S.K. Small molecule CDK inhibitors for the therapeutic management of cancer. Curr. Top. Med. Chem. 2020, 20, 1535–1563. [Google Scholar] [CrossRef] [PubMed]
- Nardone, V.; Barbarino, M.; Angrisani, A.; Correale, P.; Pastina, P.; Cappabianca, S.; Reginelli, A.; Mutti, L.; Miracco, C.; Giannicola, R.; et al. CDK4, CDK6/cyclin-D1 complex inhibition and radiotherapy for cancer control: A Role for Autophagy. Int. J. Mol. Sci. 2021, 22, 8391. [Google Scholar] [CrossRef] [PubMed]
- Gan, C.P.; Sam, K.K.; Yee, P.S.; Zainal, N.S.; Lee, B.K.B.; Abdul Rahman, Z.A.; Patel, V.; Tan, A.C.; Zain, R.B.; Cheong, S.C. IFITM3 knockdown reduces the expression of CCND1 and CDK4 and suppresses the growth of oral squamous cell carcinoma cells. Cell. Oncol. 2019, 42, 477–490. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Liu, J.; Zhang, T. Knockdown of REG ialpha enhances the sensitivity to 5-fluorouracil of colorectal cancer cells via Cyclin D1/CDK4 pathway and BAX/BCL-2 pathways. Cancer Biother. Radiopharm. 2019, 34, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Sun, W.; Dong, X.; Yu, W.; Cai, J.; Yuan, Q.; Shan, L.; Efferth, T. Total ginsenosides of chinese ginseng induces cell cycle arrest and apoptosis in colorectal carcinoma HT-29 cells. Oncol. Lett. 2018, 16, 4640–4648. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, T.Y.; Hu, B.C.; Li, X.; Wu, Y.T.; Sun, X.T.; Jiang, X.W.; Wang, S.; Qin, X.C.; Ding, H.W.; et al. CK-3, A novel methsulfonyl pyridine derivative, suppresses hepatocellular carcinoma proliferation and invasion by blocking the PI3K/AKT/mTOR and MAPK/ERK pathways. Front. Oncol. 2021, 11, 717626. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cao, P.; Cao, F.; Wang, S.; He, Y.; Xu, Y.; Wang, Y. ANLN, Regulated by SP2, promotes colorectal carcinoma cell proliferation via PI3K/AKT and MAPK signaling pathway. J. Investig. Surg. 2022, 35, 268–277. [Google Scholar] [CrossRef]
- Li, F.; Zhou, Y.D.; Liu, J.; Cai, J.D.; Liao, Z.M.; Chen, G.Q. RBP-J promotes cell growth and metastasis through regulating miR-182-5p-mediated Tiam1/Rac1/p38 MAPK axis in colorectal cancer. Cell Signal 2021, 87, 110103. [Google Scholar] [CrossRef]
- Cheng, H.; Jiang, X.; Zhang, Q.; Ma, J.; Cheng, R.; Yong, H.; Shi, H.; Zhou, X.; Ge, L.; Gao, G. Naringin inhibits colorectal cancer cell growth by repressing the PI3K/AKT/mTOR signaling pathway. Exp. Ther. Med. 2020, 19, 3798–3804. [Google Scholar] [CrossRef] [PubMed]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef] [PubMed]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Amberger, J.S.; Hamosh, A. Searching Online Mendelian Inheritance in Man (OMIM): A Knowledgebase of human genes and genetic phenotypes. Curr. Protoc. Bioinform. 2017, 58, 1.2.1–1.2.12. [Google Scholar] [CrossRef] [PubMed]
- Pinero, J.; Ramirez-Anguita, J.M.; Sauch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The genecards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the drugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, X.; Zhang, Y.; Cao, Y.; Shan, M.; Song, D.; Ye, C.; Zhu, D. Studies on Chemical Composition of Pueraria lobata and Its Anti-Tumor Mechanism. Molecules 2022, 27, 7253. https://doi.org/10.3390/molecules27217253
Fang X, Zhang Y, Cao Y, Shan M, Song D, Ye C, Zhu D. Studies on Chemical Composition of Pueraria lobata and Its Anti-Tumor Mechanism. Molecules. 2022; 27(21):7253. https://doi.org/10.3390/molecules27217253
Chicago/Turabian StyleFang, Xiaoxue, Yegang Zhang, Yiming Cao, Mengyao Shan, Dimeng Song, Chao Ye, and Difu Zhu. 2022. "Studies on Chemical Composition of Pueraria lobata and Its Anti-Tumor Mechanism" Molecules 27, no. 21: 7253. https://doi.org/10.3390/molecules27217253
APA StyleFang, X., Zhang, Y., Cao, Y., Shan, M., Song, D., Ye, C., & Zhu, D. (2022). Studies on Chemical Composition of Pueraria lobata and Its Anti-Tumor Mechanism. Molecules, 27(21), 7253. https://doi.org/10.3390/molecules27217253