Docking-Based Evidence for the Potential of ImmunoDefender: A Novel Formulated Essential Oil Blend Incorporating Synergistic Antiviral Bioactive Compounds as Promising Mpro Inhibitors against SARS-CoV-2
Abstract
1. Introduction
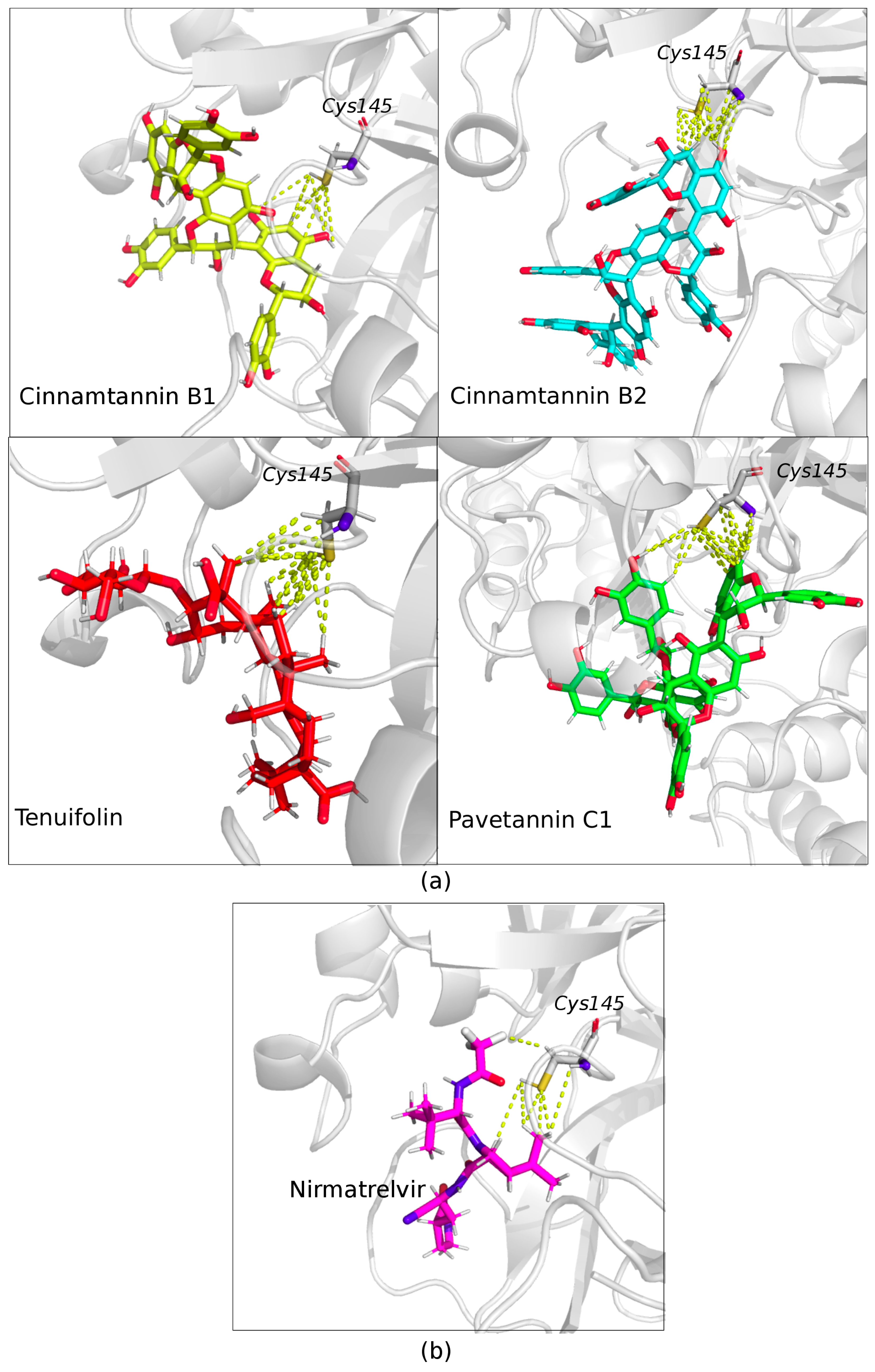
2. Results
3. Materials and Methods
3.1. Essential Oil Quantification
3.2. Data Source: Preparation or Protein and Ligands Molecule Files
3.3. Receptor-Ligand Docking Process
3.4. Drug-Likeness Prediction for the Major EO Bioactive Compound
4. Discussion
5. Conclusions
- -
- ImmunoDefender is a novel formulated bioactive antiviral compound based on a well-established mixture of essential oils and is considered highly effective in the treatment of SARS-CoV-2 infections.
- -
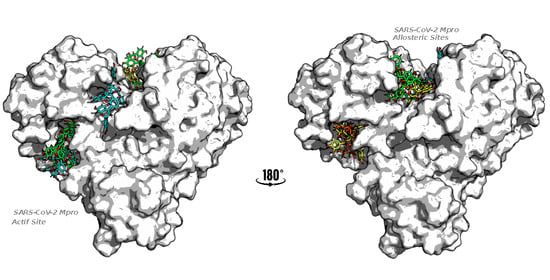
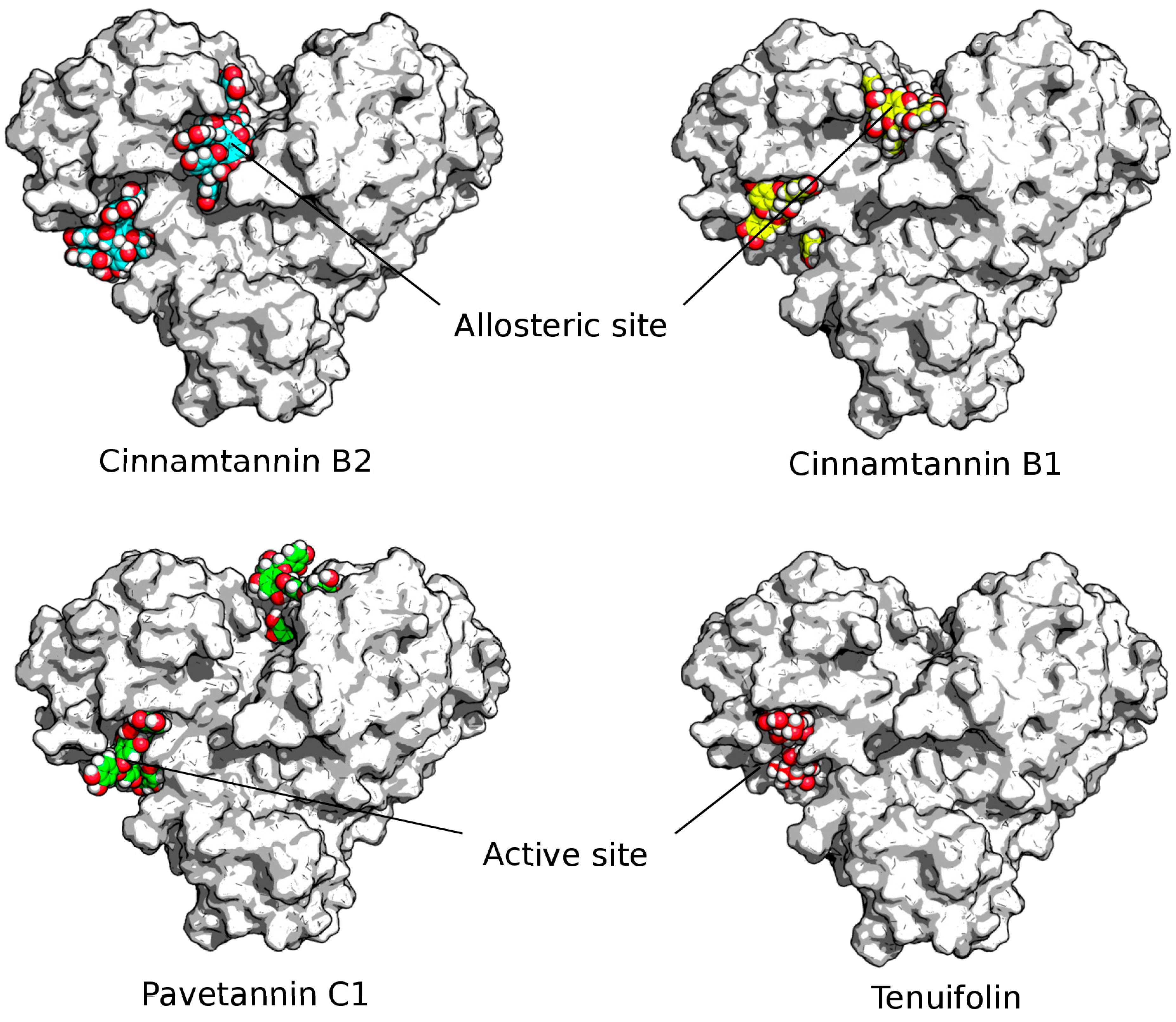
- High affinity and capacity of blocking the main protease (Mpro) catalytic and allosteric sites are considered critical in SARS-CoV-2 pathogenesis and virus transmission pathways.
- -
- Identification of lead compounds was mainly focused on molecule linkage associations and the binding intensity demonstrated by the free binding energy scores.
- -
- The study followed a rigorous methodology to ensure the accuracy, safety, and efficacy of the antiviral herbal medicinal extract, including careful selection and dosing of essential oils based on their purity and potency and a consideration of toxicity data and previous studies.
- -
- The use of an inverse problem of mathematical expectation to determine the appropriate weighting for each compound based on their kinetic effects of temporarily irreversible inhibition of the key enzymes in the replication of the SARS-CoV-2 virus increased the reliability and validity of the study’s findings.
- -
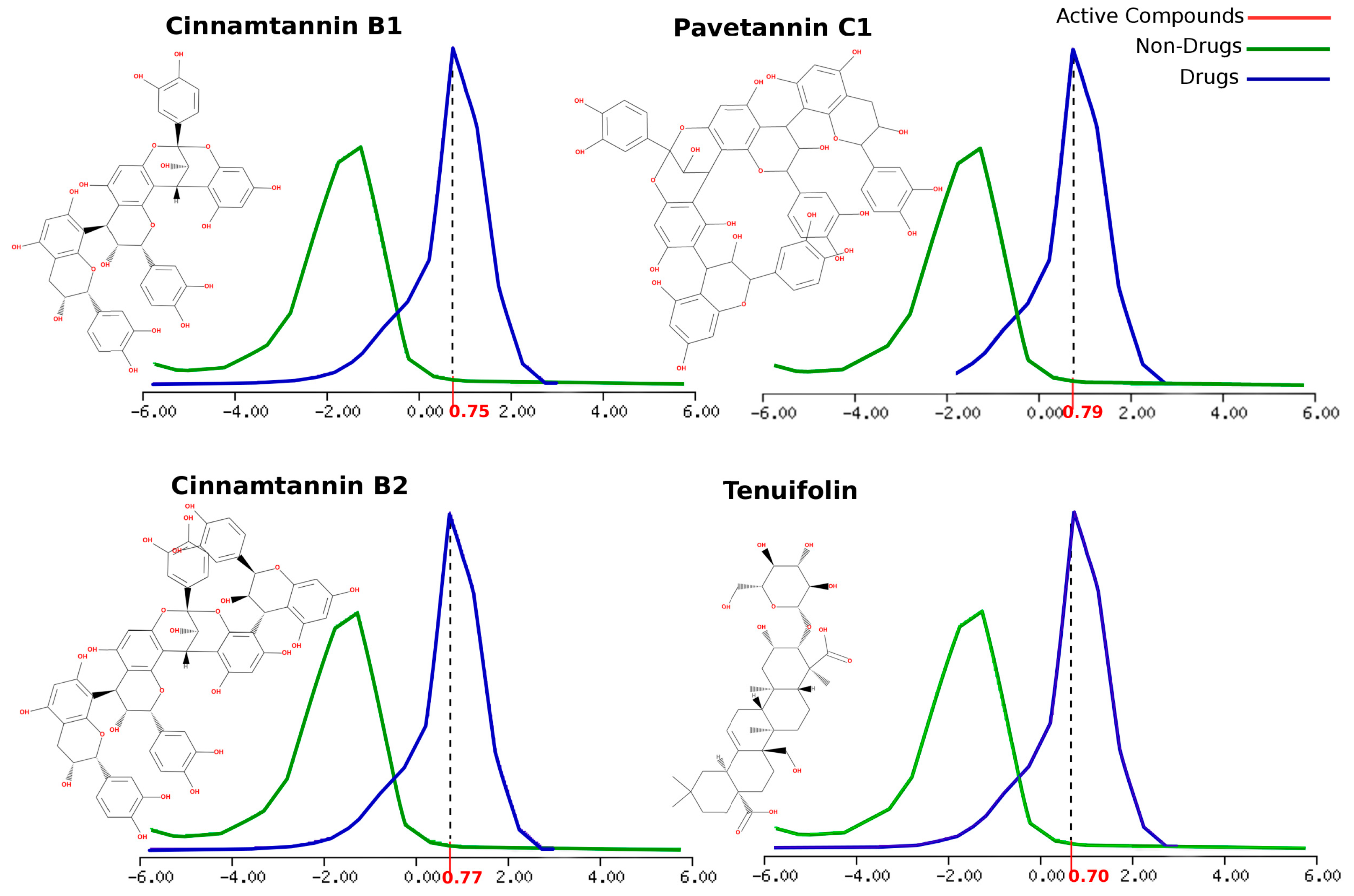
- The predicted overall drug-likeness score chemical fingerprints for Cinnamtannin B1, Cinnamtannin B2, Pavetannin C1, Syzyginin B, and Tenuifolin showed very good agreement, with perfect approval for tolerability and efficacy.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Sample Availability
References
- World Health Organization. Clinical Management of COVID-19: Interim Guidance, 27 May 2020; World Health Organization: Geneva, Switzerland, 2020; p. 1445.
- Public Health England. Investigation of Novel SARS-CoV-2 Variant, Variant of Concern 202012/01 Technical Briefing 2–28 December 2020; PHE: London, UK, 2020.
- Wang, Z.; Wang, N.; Yang, L.; Song, X.Q. Bioactive natural products in COVID-19 therapy. Front. Pharmacol. 2022, 13, 926507. [Google Scholar] [CrossRef] [PubMed]
- Narkhede, R.R.; Pise, A.V.; Cheke, R.S.; Shinde, S.D. Recognition of natural products as potential inhibitors of COVID-19 main protease (Mpro): In-silico evidences. Nat. Prod. Bioprospect. 2020, 10, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Ying, T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef] [PubMed]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Coronaviruses 2015, 1282, 1–23. [Google Scholar]
- Oudshoorn, D.; Rijs, K.; Limpens, R.W.; Groen, K.; Koster, A.J.; Snijder, E.J.; Bárcena, M. Expression and cleavage of middle east respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. MBio 2017, 8, e01658-17. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef]
- Sivropoulou, A.; Nikolaou, C.; Papanikolaou, E.; Kokkini, S.; Lanaras, T.; Arsenakis, M. Antimicrobial, cytotoxic, and antiviral activities of Salvia fructicosa essential oil. J. Agric. Food Chem. 1997, 45, 3197–3201. [Google Scholar] [CrossRef]
- Benencia, F.; Courreges, M.C. In vitro and in vivo activity of eugenol on human herpesvirus. Phytother. Res. Int. J. Devoted Pharmacol. Toxicol. Eval. Nat. Prod. Deriv. 2000, 14, 495–500. [Google Scholar] [CrossRef]
- Hayashi, K.; Imanishi, N.; Kashiwayama, Y.; Kawano, A.; Terasawa, K.; Shimada, Y.; Ochiai, H. Inhibitory effect of cinnamaldehyde, derived from Cinnamomi cortex, on the growth of influenza A/PR/8 virus in vitro and in vivo. Antivir. Res. 2007, 74, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Buchbauer, G.; Jirovetz, L.; Jager, W. Aromatherapy: Evidence for sedative effects of the essential oil of lavender after inhalation. Z. Fur Nat. C 1993, 48, 796–801. [Google Scholar] [CrossRef]
- He, W.; Han, H.; Wang, W.; Gao, B. Antiviral Activity of Essential Oils and Their Major Components against Herpes Simplex Virus Type 1 (HSV-1) In Vitro. Molecules 2017, 22, 1068. [Google Scholar]
- Tisserand, R.; Young, R. Essential Oil Safety: A Guide for Health Care Professionals; Elsevier Health Sciences: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Edwards, J. The Practice of Aromatherapy; Churchill Livingstone: London, UK, 2000. [Google Scholar]
- Gordienko, A.I.; Kazmirchuk, V.V.; Dmytrenko, O.P. Essential oils as effective tools for viral diseases’ control. Phytochem. Rev. 2020, 19, 1539–1555. [Google Scholar]
- Sun, S.; Li, W.; Deng, W.; Mo, X. Antiviral properties and mechanisms of natural polyphenols. Nutrients 2021, 13, 2561. [Google Scholar]
- Zhang, H.; Wang, W.; Li, C.; Yang, J.; Xie, J.; Zhang, Y.; Yuan, J. Antibacterial mechanism of cinnamtannin B-2 isolated from Rhododendron formosanum in vitro and in vivo. Sci. Rep. 2018, 8, 1–13. [Google Scholar]
- Prasanth, D.S.N.B.K.; Murahari, M.; Chandramohan, V.; Panda, S.P.; Atmakuri, L.R.; Guntupalli, C. In silico identification of potential inhibitors from Cinnamon against main protease and spike glycoprotein of SARS CoV-2. J. Biomol. Struct. Dyn. 2021, 39, 4618–4632. [Google Scholar] [CrossRef]
- Galeotti, N.; Mannelli, L.D.C.; Mazzanti, G.; Bartolini, A.; Ghelardini, C. Menthol: A natural analgesic compound. Neurosci. Lett. 2002, 322, 145–148. [Google Scholar] [CrossRef]
- World Health Organization. Evaluations of the Joint FAO/WHO Expert Committee on Food Additives (JECFA); WHO: Geneva, Switzerland, 2019.
- Vijayasteltar, L.; Nair, G.G.; Maliakel, B.; Kuttan, R.; Krishnakumar, I.M. Safety assessment of a standardized polyphenolic extract of clove buds: Subchronic toxicity and mutagenicity studies. Toxicol. Rep. 2016, 3, 439–449. [Google Scholar] [CrossRef]
- Nair, B. Final report on the safety assessment of Mentha Piperita (Peppermint) Oil, Mentha Piperita (Peppermint) Leaf Extract, Mentha Piperita (Peppermint) Leaf, and Mentha Piperita (Peppermint) Leaf Water. Int. J. Toxicol. 2001, 20, 61. [Google Scholar] [PubMed]
- Bhowal, M.; Gopal, M.M. Eucalyptol: Safety and pharmacological profile. J. Pharm. Sci. 2015, 5, 125–131. [Google Scholar] [CrossRef]
- Laekeman, G. Assessment Report on Cinnamomum Verum JS Presl, Cortex and Corticisaetheroleum; European Medicines Agency: London, UK, 2011.
- Cravotto, G.; Cintas, P. Extraction of flavourings from natural sources. In Modifying Flavour in Food; Woodhead Publishing: Sawston, UK, 2007; pp. 41–63. [Google Scholar]
- European Food Safety Authority (EFSA). Camphor in flavourings and other food ingredients with flavouring properties-Opinion of the Scientific Panel on Food Additives, Flavourings, Processing Aids and Materials in Contact with Food on a request from the Commission. EFSA J. 2008, 6, 729. [Google Scholar]
- FAO; WHO. Toxicological Evaluation of Some Flavouring Substances and Non-Nutritive Sweetening Agents; FAO Nutrition Meetings Report, Series No. 44a; WHO/Food Add./68.33; FAO: Rome, Italy; WHO: Geneva, Switzerland, 1967.
- Zebib, H.; Bultosa, G.; Abera, S. Physico-chemical properties of sesame (Sesamum indicum L.) varieties grown in Northern Area, Ethiopia. Agric. Sci. 2015, 6, 238. [Google Scholar]
- PyMOL Molecular Graphics System. Version 1.3r1; Schrödinger, LLC: New York, NY, USA, 2010; Available online: https://pymol.org/ (accessed on 12 March 2023).
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Bolton, E.E. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminf. 2011, 3, 1–14. [Google Scholar] [CrossRef]
- Rose, P.W.; Prlić, A.; Altunkaya, A.; Bi, C.; Bradley, A.R.; Christie, C.H.; Burley, S.K. The RCSB protein data bank: Integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2016, 45, D271–D281. [Google Scholar]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Yang, H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Bowman, G.R.; Pande, V.S. Simulated tempering yields insight into the low-resolution Rosetta scoring functions. Proteins Struct. Funct. Bioinform. 2009, 74, 777–788. [Google Scholar] [CrossRef]
- Unni, S.; Huang, Y.; Hanson, R.M.; Tobias, M.; Krishnan, S.; Li, W.W.; Baker, N.A. Web servers and services for electrostatics calculations with APBS and PDB2PQR. J. Comput. Chem. 2011, 32, 1488–1491. [Google Scholar] [CrossRef]
- Sanner, M. Python: A Programming Language for Software Integration and Development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Bitencourt-Ferreira, G.; Azevedo, W.F.D. Docking with SwissDock. In Docking Screens for Drug Discovery; Humana: New York, NY, USA, 2019; pp. 189–202. [Google Scholar]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39 (Suppl. 2), W270–W277. [Google Scholar] [CrossRef] [PubMed]
- Molsoft L.L.C. ICM Molecular Modeling Software, Version 3.9-3c; Molsoft L.L.C.: San Diego, CA, USA, 2021. [Google Scholar]
- Astani, A.; Reichling, J.; Schnitzler, P. Comparative study on the antiviral activity of selected monoterpenes derived from essential oils. Phytother. Res. 2010, 24, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Abebe, W.; Sousa, S.M.; Duarte, V.G.; Machado, M.I.; Matos, F.J. Analgesic and anti-inflammatory effects of essential oils of Eucalyptus. J. Ethnopharmacol. 2003, 89, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Ranasinghe, P.; Pigera, S.; Premakumara, G.S.; Galappaththy, P.; Constantine, G.R.; Katulanda, P. Medicinal Properties of ’True’ Cinnamon (Cinnamomum zeylanicum): A Systematic Review. Molecules 2013, 18, 10066–10098. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Dong, H.; Hou, X.; Zhang, X.; Zeng, H. Antiviral Activity of Cinnamon Bark Extracts and Its Effect on Viral Morphology. Molecules 2020, 25, 904. [Google Scholar]
- Borkotoky, S.; Banerjee, M.; Choudhury, B. In-silico screening of potential inhibitors of SARS-CoV-2 main protease (Mpro) from tea (Camellia sinensis) bioactive molecules. J. Biomol. Struct. Dyn. 2021, 39, 4392–4404. [Google Scholar]
- Aloufi, B.H.; Snoussi, M.; Sulieman, A.M.E. Antiviral Efficacy of Selected Natural Phytochemicals against SARS-CoV-2 Spike Glycoprotein Using Structure-Based Drug Designing. Molecules 2022, 27, 2401. [Google Scholar] [CrossRef]
- Mouhieddine, T.H.; Darwish, R.M.; Soukieddine, M.A.; Ghanem, A.A.; Kobeissy, F.H. Antiviral strategies against coronaviruses and other emerging viral respiratory infections. Front. Pharmacol. 2020, 11, 1791. [Google Scholar]
- Shi, Q.; Hu, Y.; Tan, X.; Hu, Y.; Zhou, P. Inhibition of SARS-CoV-2 entry by quercetin-3-rhamnoside through binding to viral spike protein. Phytomedicine 2021, 85, 153429. [Google Scholar]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Zhang, B.; Jiang, X.M.; Su, H.; Li, J.; Zhao, Y.; Xie, X.; Jin, Z.; Peng, J.; Liu, F.; et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 2020, 368, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, H.; Kim, S.; Shin, D.H.; Kim, M.S. Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors. Chem. Biol. Drug Des. 2020, 95, 876–884. [Google Scholar] [CrossRef]
- Kumar, B.K.; Sekhar, K.V.G.C.; Kunjiappan, S.; Jamalis, J.; Balaña-Fouce, R.; Tekwani, B.L.; Sankaranarayanan, M. Druggable targets of SARS-CoV-2 and treatment opportunities for COVID-19. Bioorg. Chem. 2020, 104, 104269. [Google Scholar]
- Alzyoud, L.; Ghattas, M.A.; Atatreh, N. Allosteric binding sites of the SARS-CoV-2 main protease: Potential targets for broad-spectrum anti-coronavirus agents. Drug Des. Dev. Ther. 2022, 16, 2463–2478. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cui, R.; Zhang, Y.; Yang, W.; Shi, J.; Guo, Y.; Yang, L. Quercetin, a flavonoid, inhibits the entry of SARS-CoV-2 into host cells and suppresses pro-inflammatory cytokines production. J. Clin. Virol. 2021, 139, 104812. [Google Scholar]
- Gendrot, M.; Andreani, J.; Boxberger, M.; Jardot, P.; Fonta, I.; Le Bideau, M.; Duflot, I.; Mosnier, J.; Rolland, C.; Bogreau, H.; et al. Antimalarial drugs inhibit the replication of SARS-CoV-2: An in vitro evaluation. Travel Med. Infect. Dis. 2021, 37, 101873. [Google Scholar] [CrossRef]
- Das, S.; Sarmah, S.; Lyndem, S.; Singha Roy, A.; Bajpai, V.K. In-silico investigation of phytochemicals from Asparagus racemosus as plausible antiviral agent in COVID-19. J. Biomol. Struct. Dyn. 2020, 39, 5033–5047. [Google Scholar]
- Mahmud, S.; Uddin, M.A.; Paul, G.K.; Shimu, M.S.; Islam, S.; Rahman, E.; Islam, A.; Islam, M.S.; Promi, M.M.; Emran, T.B.; et al. Virtual screening and molecular dynamics simulation study of plant-derived compounds to identify potential inhibitors of main protease from SARS-CoV-2. Brief. Bioinform. 2021, 22, 1402–1414. [Google Scholar] [CrossRef]
- Heilmann, E.; Costacurta, F.; Moghadasi, S.A.; Ye, C.; Pavan, M.; Bassani, D.; von Laer, D. SARS-CoV-2 3CLpro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376. Sci. Transl. Med. 2022, 15, eabq7360. [Google Scholar] [CrossRef] [PubMed]
- Cosar, B.; Karagulleoglu, Z.Y.; Unal, S.; Ince, A.T.; Uncuoglu, D.B.; Tuncer, G.; Demir-Dora, D. SARS-CoV-2 mutations and their viral variants. Cytokine Growth Factor Rev. 2022, 63, 10–22. [Google Scholar] [CrossRef] [PubMed]
| Plants | Essential Oils (Eos) | Acceptable Daily Intake (mg/kg Body Weight/Day) | References |
|---|---|---|---|
| Mentha spicata | Spearmint Eo | 40 | [23] |
| Mentha | Menthol (crystals) | 4 | [24] |
| Melaleuca cajuputii | Cajeput Eo | 0.17 | [24] |
| Mentha aquatica | Watermint Eo | 5 | [25] |
| Syzygium aromaticum | Cloves Eo | 2.5 | [26] |
| Mentha piperita | peppermint Eo | 200 | [27] |
| Mentha poulegium | Pennyroyal Eo | 2.3 | [28] |
| Eucalyptus globulus | Eucalyptus Eo | 4.28 | [29] |
| Cinnamomum camphora | Camphor | 50 | [30] |
| Cinnamomum zeylenicum | Cinnamon Eo | 0.1 | [31] |
| Sesamum indicum | Sesame Oil | 15,000 | [32] |
| No | Ligands | Ligand Topological Polar Surface Area (Å2) | Binding Affinity (kcal/mol) “Active Site” | Binding Affinity (kcal/mol) “Allosteric Site” |
|---|---|---|---|---|
| 1 | Cinnamtannin B1 | 320 | −9.56 | −11.12 |
| 2 | Cinnamtannin B2 | 431 | −9.40 | −10.74 |
| 3 | Pavetannin C1 | 431 | −10.30 | −10.79 |
| 4 | Syzyginin B | 349 | −10.10 | - |
| 5 | Procyanidin C1 | 331 | −9.05 | - |
| 6 | Tenuifolin | 214 | −8.75 | - |
| 7 | Nirmatrelvir | 131 | −9.24 | - |
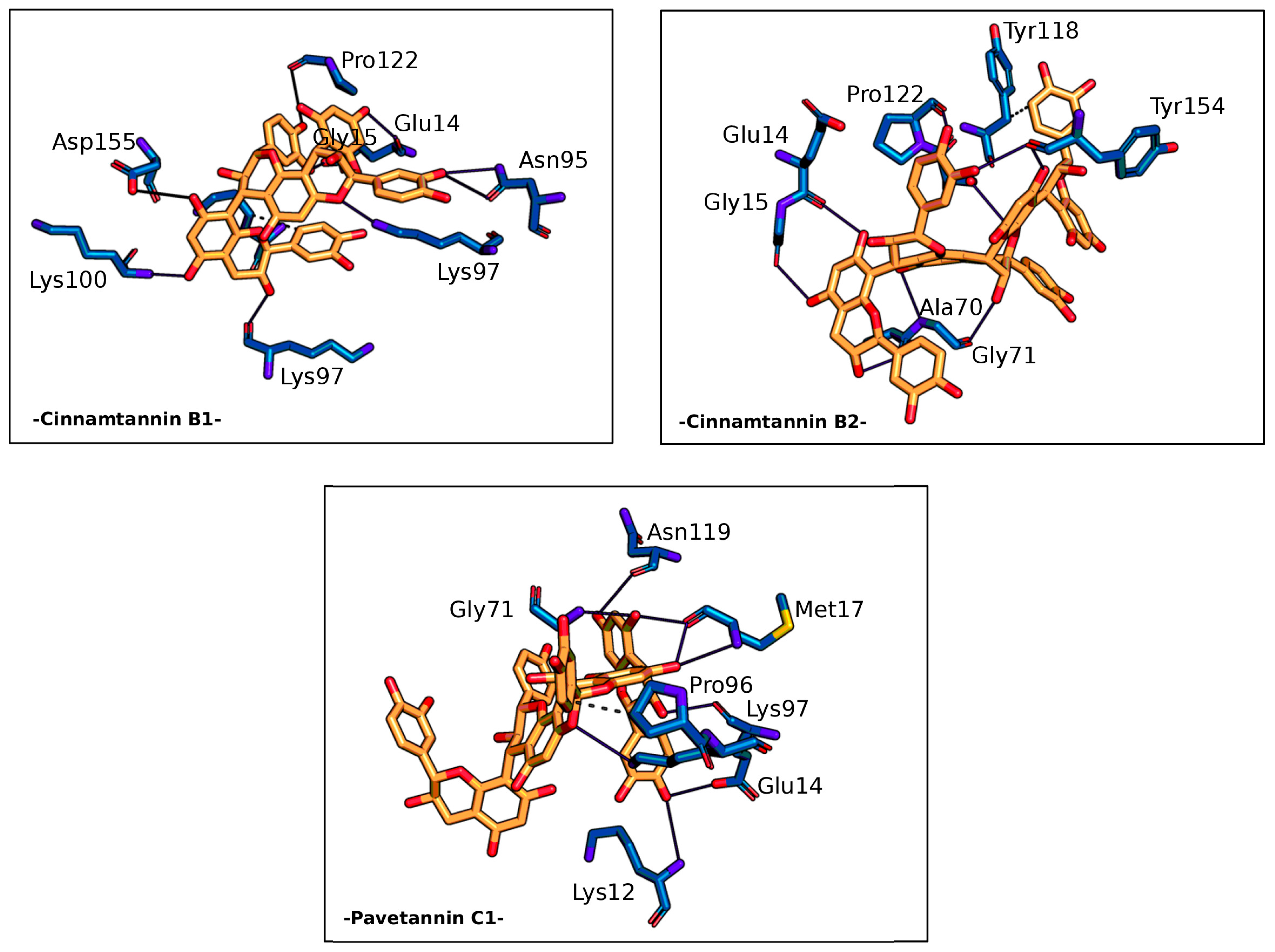
| Residue | AA | Distance H-A | Distance D-A | Donor Angle | Donor Atom | Acceptor Atom |
|---|---|---|---|---|---|---|
| -Cinnamtannin B1- | ||||||
| 14A | GLU | 2.18 | 3.07 | 150.97 | 28 [O2] | 311 [O−] |
| 14A | GLU | 1.74 | 2.69 | 163.65 | 20 [O2] | 306 [O2] |
| 95A | ASN | 3.53 | 3.94 | 106.70 | 1568 [Nam] | 24 [O2] |
| 95A | ASN | 2.51 | 3.10 | 118.88 | 24 [O2] | 1569 [O2] |
| 97A | LYS | 2.40 | 3.13 | 131.54 | 18 [O3] | 6275 [O2] |
| 97A | LYS | 2.37 | 3.31 | 150.71 | 1598 [N3+] | 63 [O2] |
| 100A | LYS | 3.22 | 3.75 | 115.47 | 6322 [Nam] | 22 [O2] |
| 122A | PRO | 2.91 | 3.86 | 162.78 | 30 [O2] | 1996 [O2] |
| 155A | ASP | 3.13 | 3.82 | 129.57 | 16 [O2] | 7160 [O−] |
| -Cinnamtannin B2- | ||||||
| 14A | GLU | 2.04 | 2.80 | 132.84 | 9387 [O2] | 4889 [O2] |
| 15A | GLY | 2.00 | 2.83 | 141.70 | 9395 [O2] | 4904 [O2] |
| 70A | ALA | 2.11 | 3.04 | 159.71 | 9389 [O3] | 5762 [O2] |
| 71A | GLY | 2.65 | 3.20 | 114.54 | 5769 [Nam] | 9381 [O2] |
| 71A | GLY | 3.19 | 3.64 | 110.53 | 9371 [O3] | 5772 [O2] |
| 121A | SER | 3.38 | 3.89 | 115.24 | 6570 [O3] | 9449 [O2] |
| 122A | PRO | 2.86 | 3.58 | 131.51 | 9403 [O2] | 6579 [O2] |
| 154A | TYR | 2.74 | 3.15 | 105.73 | 9401 [O2] | 2354 [O2] |
| 154A | TYR | 2.86 | 3.81 | 165.39 | 9385 [O2] | 2354 [O2] |
| -Pavetannin C1- | ||||||
| 14A | GLU | 1.90 | 2.82 | 156.43 | 9409 [O2] | 212 [O−] |
| 14A | GLU | 1.87 | 2.80 | 159.59 | 9385 [O3] | 207 [O2] |
| 17A | MET | 2.13 | 2.77 | 122.36 | 9397 [O2] | 240 [O2] |
| 17A | MET | 3.17 | 3.48 | 100.00 | 237 [Nam] | 9387 [O2] |
| 17A | MET | 2.07 | 2.82 | 131.63 | 9387 [O2] | 240 [O2] |
| 71A | GLY | 2.29 | 3.04 | 130.92 | 1087 [Nam] | 9397 [O2] |
| 97A | LYS | 3.14 | 3.87 | 128.25 | 1499 [N3+] | 9367 [O2] |
| 119A | ASN | 2.57 | 3.19 | 121.41 | 9403 [O2] | 1865 [O2] |
| Active Compounds | Molecular Formula | Molecular Weight (KDa) | Number of HBA | Number of HBD | MolLogP | MolLogS Log(moles/L) | MolPSA (Å2) | MolVol (Å3) | pKa | BBB Score | Number of Stereo Centers | Drug-Likeness Model Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pavetannin C1 | C60H48O24 | 1152.25 | 24 | 19 | 3.46 | −3.04 | 344.95 | 1039.90 | <0./9.52 | 0 | 11 | 0.79 |
| Tenuifolin | C36H56O12 | 680.38 | 12 | 8 | 1.02 | −1.30 | 168.92 | 737.65 | <0./5.17 | 0.34 | 15 | 0.70 |
| Cinnamtannin B1 | C45H36 O18 | 864.19 | 18 | 14 | 2.40 | −2.60 | 257.26 | 782.13 | <0./9.52 | 0 | 8 | 0.75 |
| Cinnamtannin B2 | C60H48O24 | 1152.25 | 14 | 19 | 3.12 | −3.06 | 345.71 | 1039.96 | <0./9.52 | 0 | 11 | 0.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ksouri, A.; Klouz, A.; Bouhaouala-Zahar, B.; Moussa, F.; Bezzarga, M. Docking-Based Evidence for the Potential of ImmunoDefender: A Novel Formulated Essential Oil Blend Incorporating Synergistic Antiviral Bioactive Compounds as Promising Mpro Inhibitors against SARS-CoV-2. Molecules 2023, 28, 4296. https://doi.org/10.3390/molecules28114296
Ksouri A, Klouz A, Bouhaouala-Zahar B, Moussa F, Bezzarga M. Docking-Based Evidence for the Potential of ImmunoDefender: A Novel Formulated Essential Oil Blend Incorporating Synergistic Antiviral Bioactive Compounds as Promising Mpro Inhibitors against SARS-CoV-2. Molecules. 2023; 28(11):4296. https://doi.org/10.3390/molecules28114296
Chicago/Turabian StyleKsouri, Ayoub, Anis Klouz, Balkiss Bouhaouala-Zahar, Fathi Moussa, and Mounir Bezzarga. 2023. "Docking-Based Evidence for the Potential of ImmunoDefender: A Novel Formulated Essential Oil Blend Incorporating Synergistic Antiviral Bioactive Compounds as Promising Mpro Inhibitors against SARS-CoV-2" Molecules 28, no. 11: 4296. https://doi.org/10.3390/molecules28114296
APA StyleKsouri, A., Klouz, A., Bouhaouala-Zahar, B., Moussa, F., & Bezzarga, M. (2023). Docking-Based Evidence for the Potential of ImmunoDefender: A Novel Formulated Essential Oil Blend Incorporating Synergistic Antiviral Bioactive Compounds as Promising Mpro Inhibitors against SARS-CoV-2. Molecules, 28(11), 4296. https://doi.org/10.3390/molecules28114296