Aggregation of Amyloidogenic Peptide Uperin—Molecular Dynamics Simulations
Abstract
1. Introduction
2. Results
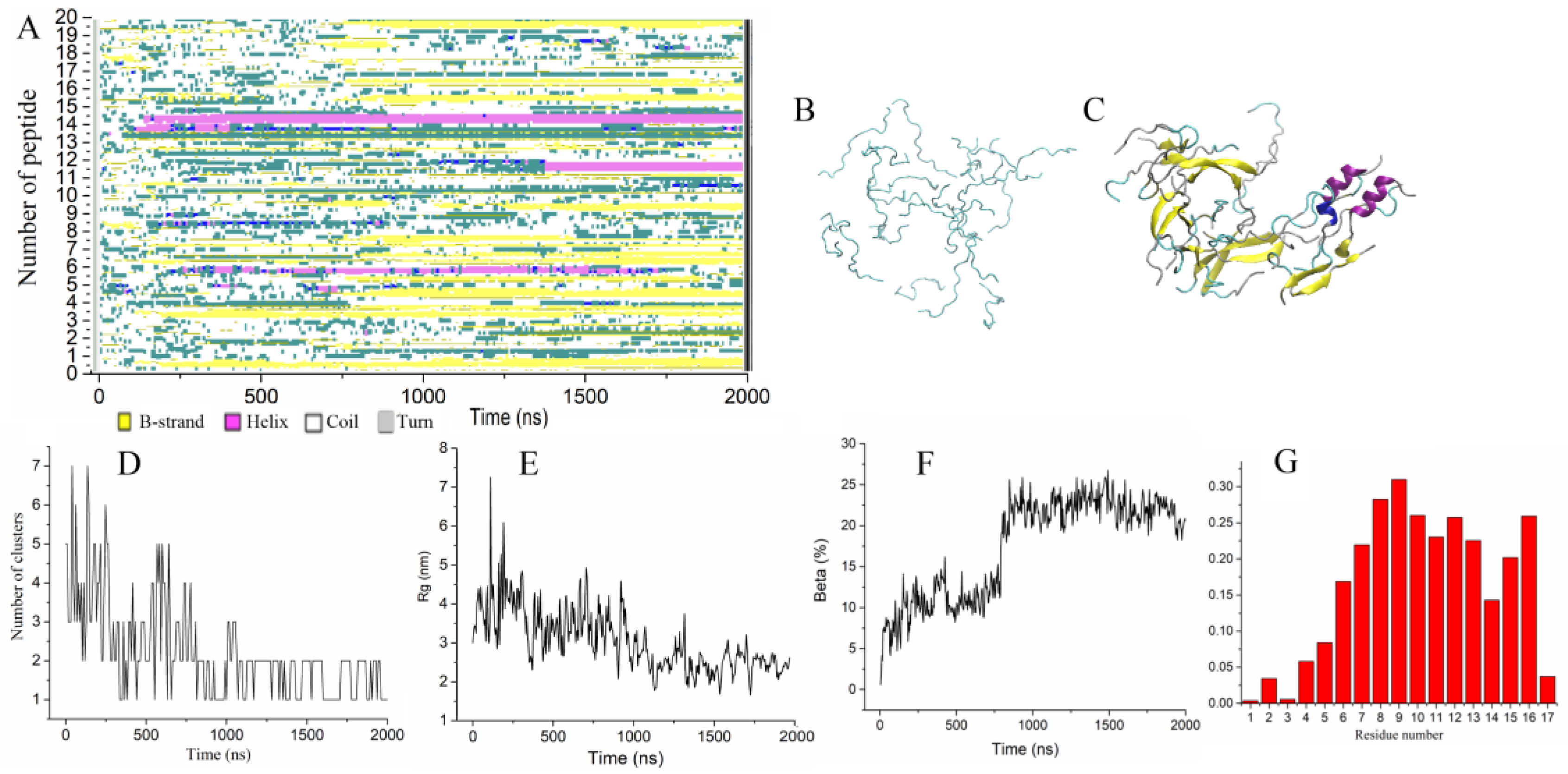
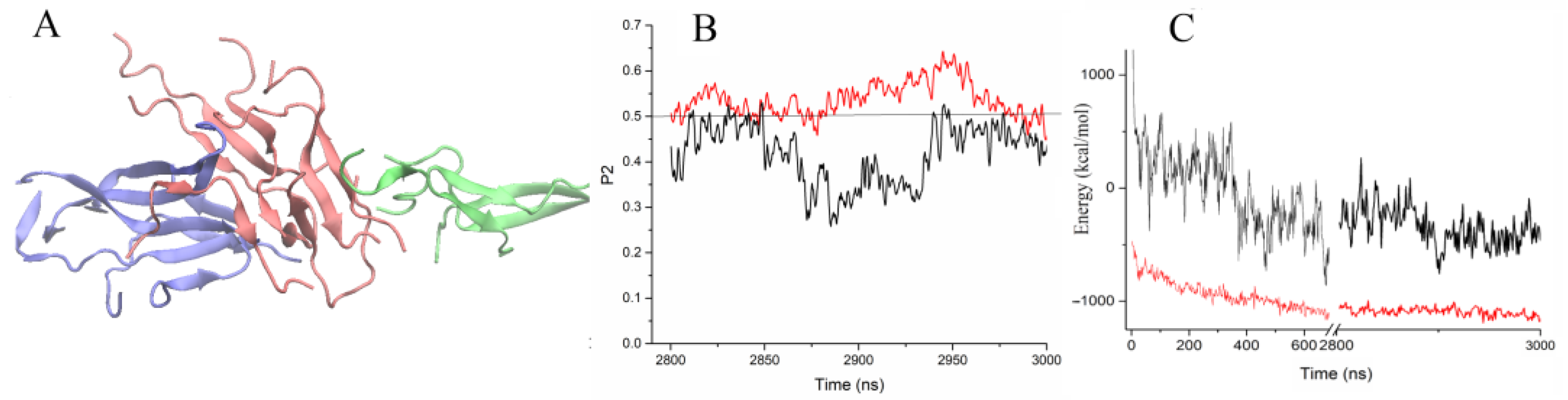
2.1. Formation of Dimers and Beta-Strands Is the First Step of Peptide Aggregation
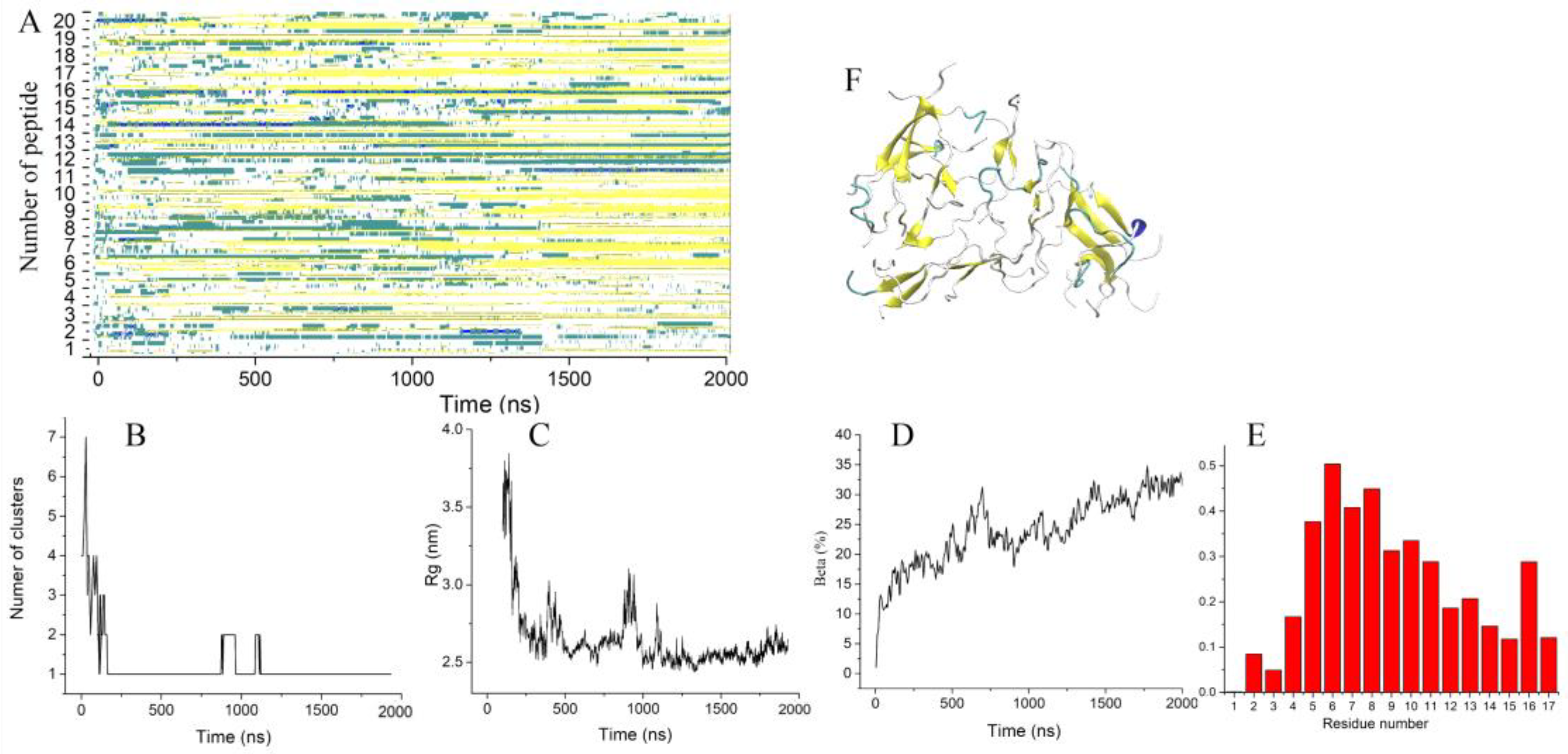
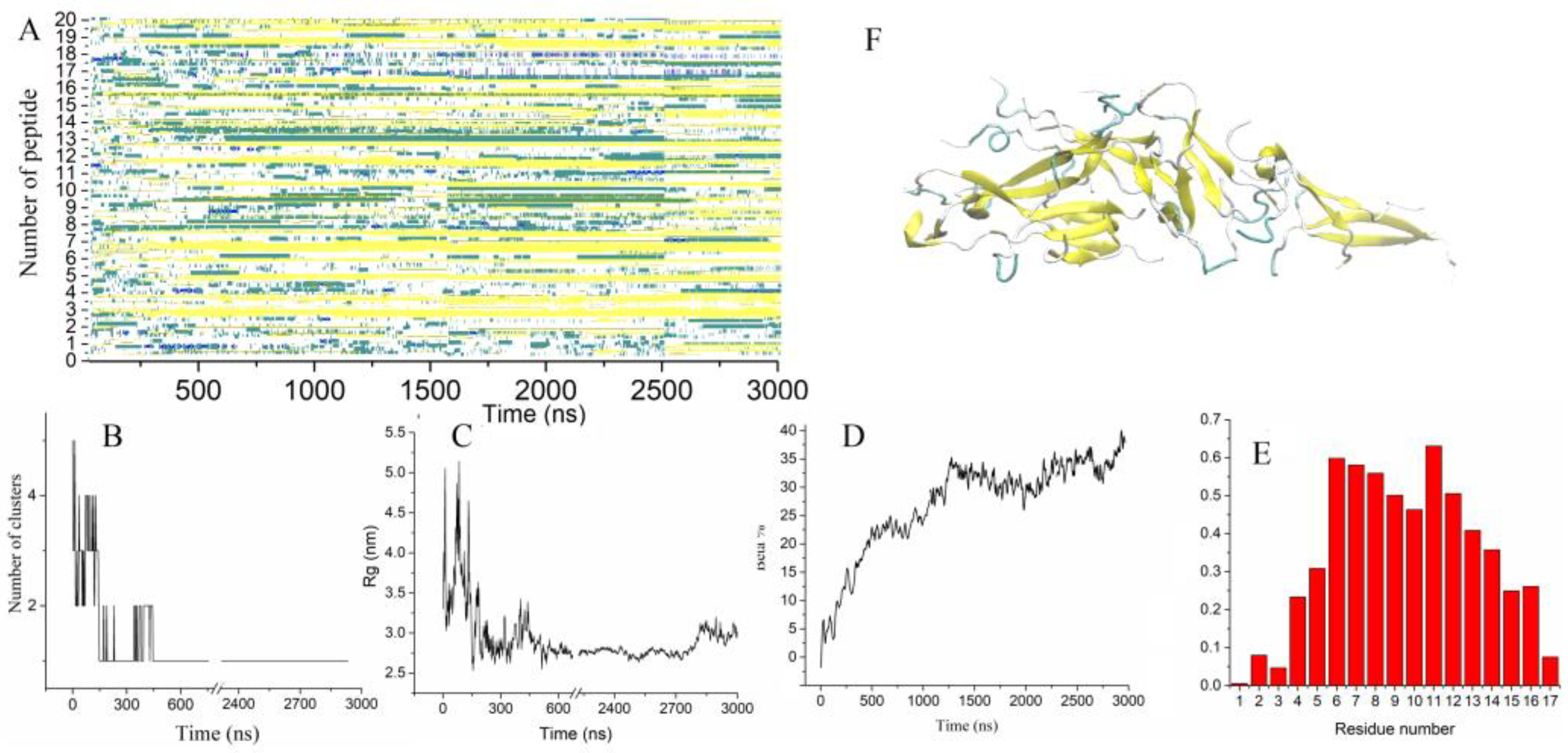
2.2. Influence of a Single Mutation on the Aggregation Properties of Uperins
2.2.1. Aggregation of Wild-Type Uperin
2.2.2. Aggregation of Uperin Mutants
3. Discussion
4. Methods
Molecular Dynamics Simulations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Samdin, T.D.; Kreutzer, A.G.; Nowick, J.S. Exploring Amyloid Oligomers with Peptide Model Systems. Curr. Opin. Chem. Biol. 2021, 64, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.K.; Tiwari, C.; Ray, S.; Holden, S.; Armstrong, D.A.; Rosengren, K.J.; Rodger, A.; Panwar, A.S.; Martin, L.L. Secondary Structure Transitions for a Family of Amyloidogenic, Antimicrobial Uperin 3 Peptides in Contact with Sodium Dodecyl Sulfate. ChemPlusChem 2022, 87, e202100408. [Google Scholar] [CrossRef] [PubMed]
- Yadav, J.K. Structural and Functional Swapping of Amyloidogenic and Antimicrobial Peptides: Redefining the Role of Amyloidogenic Propensity in Disease and Host Defense. J. Pept. Sci. 2022, 28, e3378. [Google Scholar] [CrossRef]
- van Gils, J.H.M.; van Dijk, E.; Peduzzo, A.; Hofmann, A.; Vettore, N.; Schützmann, M.P.; Groth, G.; Mouhib, H.; Otzen, D.E.; Buell, A.K.; et al. The Hydrophobic Effect Characterises the Thermodynamic Signature of Amyloid Fibril Growth. PLoS Comput. Biol. 2020, 16, e1007767. [Google Scholar] [CrossRef] [PubMed]
- Saurabh, A.; Prabhu, N.P. Concerted Enhanced-Sampling Simulations to Elucidate the Helix-Fibril Transition Pathway of Intrinsically Disordered α-Synuclein. Int. J. Biol. Macromol. 2022, 223, 1024–1041. [Google Scholar] [CrossRef]
- Katyal, N.; Deep, S. Inhibition of GNNQQNY Prion Peptide Aggregation by Trehalose: A Mechanistic View. Phys. Chem. Chem. Phys. 2017, 19, 19120–19138. [Google Scholar] [CrossRef] [PubMed]
- Li, D.-W.; Mohanty, S.; Irbäck, A.; Huo, S. Formation and Growth of Oligomers: A Monte Carlo Study of an Amyloid Tau Fragment. PLoS Comput. Biol. 2008, 4, e1000238. [Google Scholar] [CrossRef] [PubMed]
- Matthes, D.; Gapsys, V.; Daebel, V.; de Groot, B.L. Mapping the Conformational Dynamics and Pathways of Spontaneous Steric Zipper Peptide Oligomerization. PLoS ONE 2011, 6, e19129. [Google Scholar] [CrossRef]
- Sun, X.; Lai, L. Protein Fibrils Formed by Rationally Designed α-Helical Peptides. Langmuir 2020, 36, 6126–6131. [Google Scholar] [CrossRef]
- Szała-Mendyk, B.; Molski, A. Clustering and Fibril Formation during GNNQQNY Aggregation: A Molecular Dynamics Study. Biomolecules 2020, 10, 1362. [Google Scholar] [CrossRef]
- Jalali, S.; Yang, Y.; Mahmoudinobar, F.; Singh, S.M.; Nilsson, B.L.; Dias, C. Using All-Atom Simulations in Explicit Solvent to Study Aggregation of Amphipathic Peptides into Amyloid-like Fibrils. J. Mol. Liq. 2022, 347, 118283. [Google Scholar] [CrossRef]
- Bradford, A.; Bowie, J.; Tyler, M.; Wallace, J. New Antibiotic Uperin Peptides From the Dorsal Glands of the Australian Toadlet Uperoleia Mjobergii. Aust. J. Chem. 1996, 49, 1325. [Google Scholar] [CrossRef]
- Chia, B.C.S.; Bowie, J.H.; Carver, J.A.; Mulhern, T.D. The Solution Structure of Uperin 3.6, an Antibiotic Peptide from the Granular Dorsal Glands of the Australian Toadlet, Uperoleia mjobergii: Solution Structure of Uperin 3.6. J. Pept. Res. 1999, 54, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, A.N.; Liu, Y.; Wang, T.; Musgrave, I.F.; Pukala, T.L.; Tabor, R.F.; Martin, L.L.; Carver, J.A.; Bowie, J.H. The Amyloid Fibril-Forming Properties of the Amphibian Antimicrobial Peptide Uperin 3.5. ChemBioChem 2016, 17, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Bücker, R.; Seuring, C.; Cazey, C.; Veith, K.; García-Alai, M.; Grünewald, K.; Landau, M. The Cryo-EM Structures of Two Amphibian Antimicrobial Cross-β Amyloid Fibrils. Nat. Commun. 2022, 13, 4356. [Google Scholar] [CrossRef]
- Ray, S.; Holden, S.; Martin, L.L.; Panwar, A.S. Mechanistic Insight into the Early Stages of Amyloid Formation Using an Anuran Peptide. Pept. Sci. 2019, 111, e24120. [Google Scholar] [CrossRef]
- Ray, S.; Holden, S.; Prasad, A.K.; Martin, L.L.; Panwar, A.S. Exploring the Role of Peptide Helical Stability in the Propensity of Uperin 3.x Peptides toward Beta-Aggregation. J. Phys. Chem. B 2020, 124, 11659–11670. [Google Scholar] [CrossRef]
- Salinas, N.; Tayeb-Fligelman, E.; Sammito, M.D.; Bloch, D.; Jelinek, R.; Noy, D.; Usón, I.; Landau, M. The Amphibian Antimicrobial Peptide Uperin 3.5 Is a Cross-α/Cross-β Chameleon Functional Amyloid. Proc. Natl. Acad. Sci. USA 2021, 118, e2014442118. [Google Scholar] [CrossRef]
- Chatani, E.; Yamamoto, N. Recent Progress on Understanding the Mechanisms of Amyloid Nucleation. Biophys. Rev. 2018, 10, 527–534. [Google Scholar] [CrossRef]
- Cecchini, M.; Rao, F.; Seeber, M.; Caflisch, A. Replica Exchange Molecular Dynamics Simulations of Amyloid Peptide Aggregation. J. Chem. Phys. 2004, 121, 10748–10756. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Hamelberg, D.; Mongan, J.; McCammon, J.A. Accelerated Molecular Dynamics: A Promising and Efficient Simulation Method for Biomolecules. J. Chem. Phys. 2004, 120, 11919–11929. [Google Scholar] [CrossRef] [PubMed]
- Martínez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. PACKMOL: A Package for Building Initial Configurations for Molecular Dynamics Simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Frishman, D.; Argos, P. Knowledge-Based Protein Secondary Structure Assignment. Proteins 1995, 23, 566–579. [Google Scholar] [CrossRef]
- Seeber, M.; Cecchini, M.; Rao, F.; Settanni, G.; Caflisch, A. Wordom: A Program for Efficient Analysis of Molecular Dynamics Simulations. Bioinformatics 2007, 23, 2625–2627. [Google Scholar] [CrossRef]

| Trajectory | Peptides | Concentration, mM | Ion Strength, M | Number of Peptides | Number of Water Molecules | Length of Trajectory, μs |
|---|---|---|---|---|---|---|
| Tr1 | Up | 7.5 | 0.3 | 10 | 38,243 | 1.1 |
| Tr2 | Up7a | 7.5 | 0.3 | 10 | 38,310 | 1.2 |
| Tr3 | Up | 15 | 0.3 | 20 | 41,108 | 2 |
| Tr4 | Up8a | 15 | 0.3 | 20 | 41,274 | 2 |
| Tr5 | Up7a | 15 | 0.3 | 20 | 41,300 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ermakova, E.; Makshakova, O.; Kurbanov, R.; Ibraev, I.; Zuev, Y.; Sedov, I. Aggregation of Amyloidogenic Peptide Uperin—Molecular Dynamics Simulations. Molecules 2023, 28, 4070. https://doi.org/10.3390/molecules28104070
Ermakova E, Makshakova O, Kurbanov R, Ibraev I, Zuev Y, Sedov I. Aggregation of Amyloidogenic Peptide Uperin—Molecular Dynamics Simulations. Molecules. 2023; 28(10):4070. https://doi.org/10.3390/molecules28104070
Chicago/Turabian StyleErmakova, Elena, Olga Makshakova, Rauf Kurbanov, Ilya Ibraev, Yuriy Zuev, and Igor Sedov. 2023. "Aggregation of Amyloidogenic Peptide Uperin—Molecular Dynamics Simulations" Molecules 28, no. 10: 4070. https://doi.org/10.3390/molecules28104070
APA StyleErmakova, E., Makshakova, O., Kurbanov, R., Ibraev, I., Zuev, Y., & Sedov, I. (2023). Aggregation of Amyloidogenic Peptide Uperin—Molecular Dynamics Simulations. Molecules, 28(10), 4070. https://doi.org/10.3390/molecules28104070









