1. Introduction
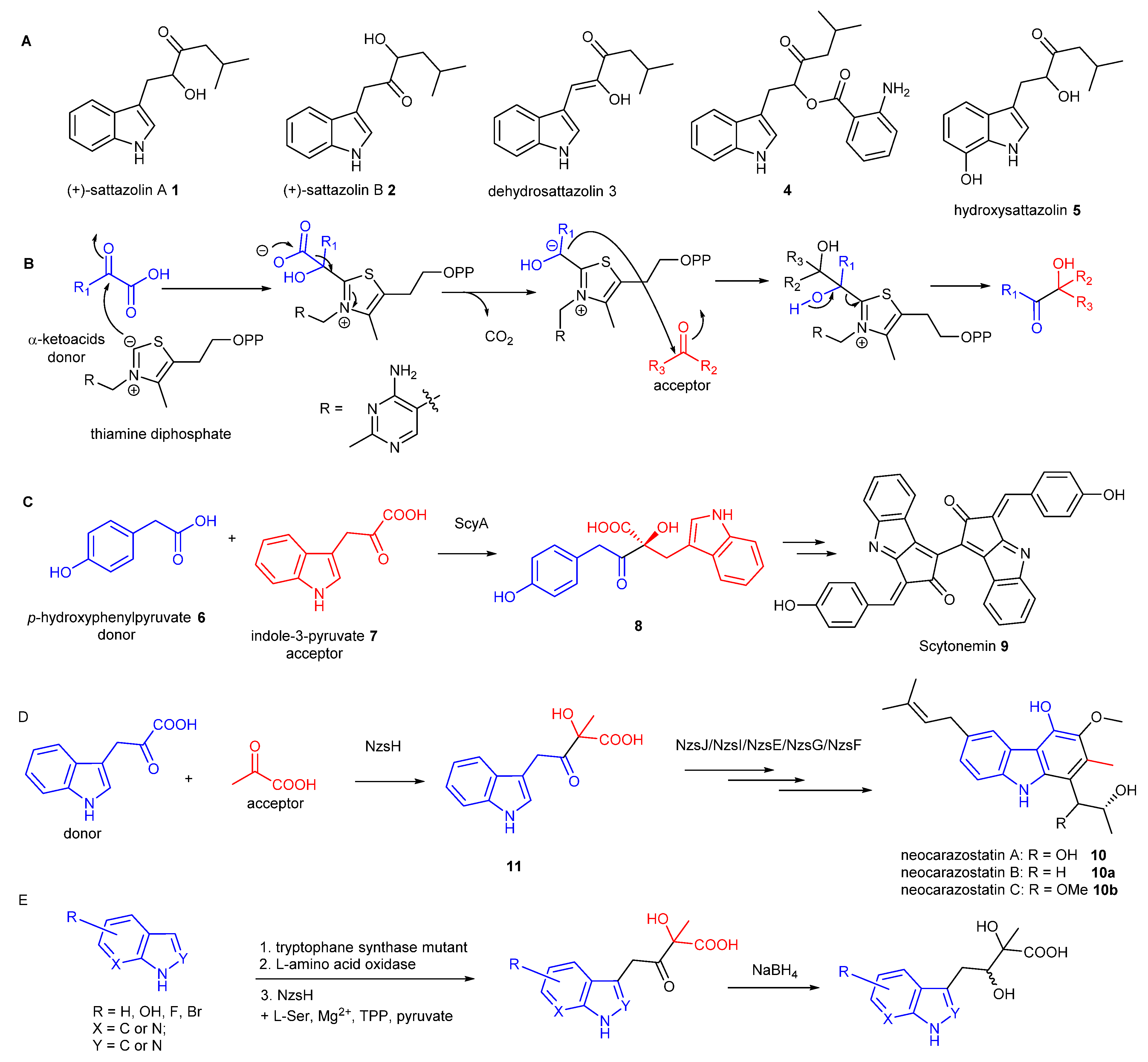
Indole-containing acyloins are key precursor of antimicrobial/antiviral agents, such as sattazolins
1–
5 (
Figure 1A) [
1,
2,
3]. They are also useful intermediates in synthesis of biologically active molecules such as carbazoles, β-carbolines and other indole-containing molecules [
4]. However, access to structurally diverse indole-containing acyloins has presented a synthetic challenge.
Thiamine diphosphate (ThDP)-dependent enzymes are widely found in the biological systems and have been demonstrated to be involved in diverse biotransformation, including C–C, C–N, C–S, and C–O bond cleavage and formation [
5]. The general mechanism underlying all these reactions is that the enzyme’s active site-mediated dissociation of the C2–H proton from the thiazolium ring of ThDP will generate the C2 anion ylid which is covalently bound to the donor substrate (
Figure 1B). Subsequent decarboxylation results in the ThDP-bound enolate intermediate with nucleophilic reactivity which readily react with electrophilic acceptors, such as aldehydes, ketones or α-ketoacids to form the α-hydroxyl ketone (an acyloin) [
6].
In many cases, pyruvate is the donor in the ThDP-dependent reactions. However, some of ThDP-dependent enzymes utilise a range of α-ketoacids to initiate carboligation reactions. For example, the ThDP dependent enzyme, ScyA, was found to couple
p-hydroxyphenylpyruvate
6 as the donor and indole-3-pyruvate
7 as the acceptor to afford the β-ketoacid product
8, the key intermediate during the biosynthesis of scytonemin
9, a pigment produced by cyanobacteria [
7]. During our biosynthetic studies of the bacterial carbazole alkaloid, neocarazostatin A
10 [
8,
9,
10,
11], we found that the ThDP-dependent enzyme NzsH catalyses an unusual carboligation between indole-3-pyruvate
7 as the donor and pyruvate as the acceptor to generate an indole-containing acyloin intermediate
11 [
9], followed by further modifications to finally decorate
10 [
10,
11], consistent with a parallel study in the biosynthetic pathway of the bacterial carbazole analogue, carbazomycin [
12]. NzsH only accepts 2-oxobutyrate, an analogue of pyruvate, as the acceptor, unlike other ThDP enzymes that normally display a considerable range of aldehyde and ketones [
9]. Phylogenetic analysis demonstrated that NzsH formed a distinct clade with other ThDP-dependent enzymes, suggesting that NzsH represents a unique group of ThDP-dependent enzymes that utilises indole-3-pyruvate as the donor for the carboligation reaction [
9]. However, the donor substrate plasticity of NzsH has remain elusive. More recently, two NzsH homologues, CbeiHKI805_0381 and Cbei2730, were shown to be responsible to produce the indole-containing acyloin core of antimicrobial/antiviral agents, sattazolins
1-
5 and its derivatives, isolated from various strains of
Clostridium beijerinckii, a bacterium used for industrial solvent production [
13]. Interestingly, the enzymes can accept branched chain α-ketoacids as acceptor substrates and both pyruvate and indole-3-pyruvate as donor substrates [
13].
Here, we report a pilot study of new three-enzyme coupled biotransformation to access indole-containing acyloin derivatives with structural diversity. The reaction mixture includes an engineered tryptophane synthase β-subunit, a commercially available L-amino acid oxidase (LAAO), and NzsH. This coupled reaction is initiated from commercially available indole or indole derivatives with L-Ser catalysed by the engineered tryptophane synthetase to provide tryptophane derivatives, followed by addition of LAAO for oxidation reactions to give the corresponding indole-3-pyruvate analogues. Inclusion of NzsH in the reactions finally allow access of a new range of indole-containing acyloin derivatives. This newly introduced functionalised indole-3-pyruvate analogues substantially broaden the donor substrate range of the ThDP-dependent NzsH enzyme, thus holding potential to generate new antiviral/antimicrobial acyloins with different indole ring systems.
2. Results and Discussion
To generate structurally diverse indole-containing acyloin derivatives, indole-3-pyruvate derivatives with structural diversity to evaluate the substrate plasticity of NzsH is key. However, most of indole-3-pyruvate derivatives are not commercially available. In nature, indole-3-pyruvate is directly descended from L-tryptophane via deamination reactions. There has been a considerable interest in the application of tryptophane synthase to generate structurally diverse tryptophane derivatives due to its importance as a key building block for many bioactive molecules [
14]. Tryptophan synthase is a heterodimeric complex consisting of two subunits, TrpA (α-subunit) catalysing the cleavage of indole glycerol phosphate to indole, and TrpB (β-subunit) mediating the coupling between indole and L-Ser to provide L-Tryptophan using pyridoxal phosphate (PLP) as cofactors (
Supplementary Scheme S1A). The activities of both subunits are required for wild type tryptophan synthases, although only the reactivity of TrpB, coupling L-Ser and indole is necessary and desirable for L-tryptophan synthesis. Recent studies reported an engineered β-subunit of tryptophan synthase (
PfTrpB) from
Pyrococcus furiosus as standalone function that restore the catalytic efficiency and surpass the activity of the native complex (
Supplementary Scheme S1B) [
15,
16,
17]. One variant (
PfTrpB
6) with six amino acid site mutations displayed the best kinetics towards L-Ser and indole and a wide range of indole derivatives. To this end, we synthesized the gene coding
PfTrpB
6 for overexpression in
E. coli.
PfTrpB
6 was expressed as an N-terminal pHis
6 recombinant protein and was purified to near homogenesis by Ni-NTA chromatography, giving estimated molecular weight of ~45 kDa as previously reported (
Supplementary Figure S1A). Incubation of
PfTrpB
6 with indole, L-Ser and PLP resulted in the accumulation of L-tryptophan
13 as evidenced in our LC-MS and tandem MS analyses (
Figure 2 and
Supplementary Figure S2).
To further in situ generate indole-3-pyruvate from L-tryptophan, we decided to use commercially available L-amino acid oxidase (LAAO) from snake venom (Sigma Aldrich catalogue number: A5147) because this enzyme displays broad substrate tolerance toward amino acids and does not require any additional cofactors (
Supplementary Scheme S1C). It also can efficiently shift the reaction equilibrium during the coupled reaction as observed in our previous report when the fluorination enzyme, FlA, from
Streptomyces cattleya, was investigated for a chlorination reaction [
18]. Addition of LAAO into the aforementioned enzyme system led to the formation of indole-3-pyruvate
14 as evidenced in our MS and tandem MS analysis (
Figure 2 and
Supplementary Figure S3). We next overexpressed the synthetic construct containing the gene
nzsH in
E coli BL-21 (DE3). NzsH was expressed as an N-terminal pHis
6 recombinant protein and was purified to near homogenesis by Ni-NTA chromatography, giving estimated molecular weight of ~64 kDa as previously reported [
9] (
Supplementary Figure S1B). Inclusion of NzsH into the coupled reaction of
PfTrpB
6 and LAAO together with TPP and Mg
2+ resulted in the formation of the indole-containing acyloin scaffold. However, this molecule is not stable and is subjected to facile decarboxylation in the LC-MS analysis [
9]. We used NaBH
4 to reduce the β-ketone of the molecule to generate two diastereomeric diols,
15 and
15′, as evidenced in our LC-MS and tandem MS analyses (
Figure 2 and
Supplementary Figure S4).
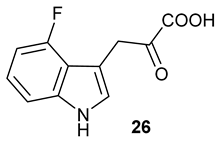
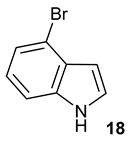
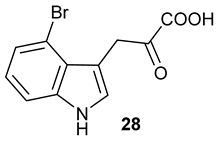
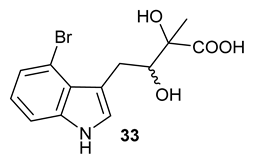
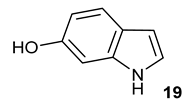
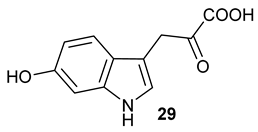
Encouraged by the results of this three-enzyme coupling system, we then acquired four commercially available indole derivatives including 4-fluoroindole
16, 5-fluoroindole
17, 4-bromoindole
18 and 6-hydroxyl-indole
19 (
Table 1). Our LC-MS and tandem MS analysis demonstrated that all of these indole derivatives can be efficiently transformed into the corresponding indole-3-pyruvate derivatives,
26–
29 as well as the corresponding diols,
31–
35, (
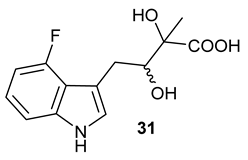
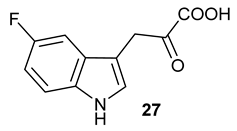
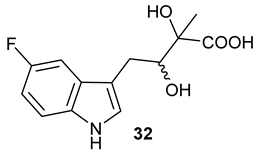
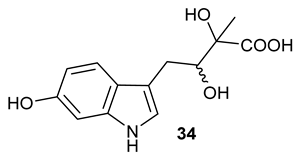
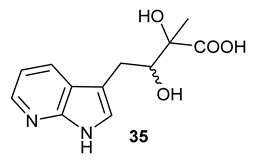
Table 1), in the corresponding coupled chemoenzymatic systems.
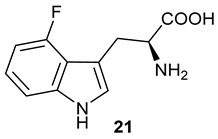
We also monitored the biotransformation of the formation of fluorinated indole-containing acyloins in
19F NMR. As shown in
Figure 3A,E, the reaction was initiated by addition of 4-fluoroindole
16 (−122.8 ppm) and 5-fluoroindole
17 (−125.2 ppm), respectively, in the
PfTrpB
6 -mediated systems to provide 4-fluoro-tryptophan
21 (−124.3 ppm) and 5-fluoro-tryptophan
22 (−125.4 ppm), respectively, in
19FNMR spectrum (
Figure 3B,F). When LAAO was added to the reactions, 4-fluoro-indole-3-pyruvate
22 (−124.3 ppm) and 5-fluoroindole-3-pyruvate
27 (−124.8 ppm), respectively, appeared (
Figure 3C,G). Finally, after inclusion of NzsH, the corresponding 4-fluoro-acyloin
31 (−124.4 ppm) and 5-fluoro-acyloin
32 (−125.6 ppm) was formed (
Figure 3D,H), respectively.
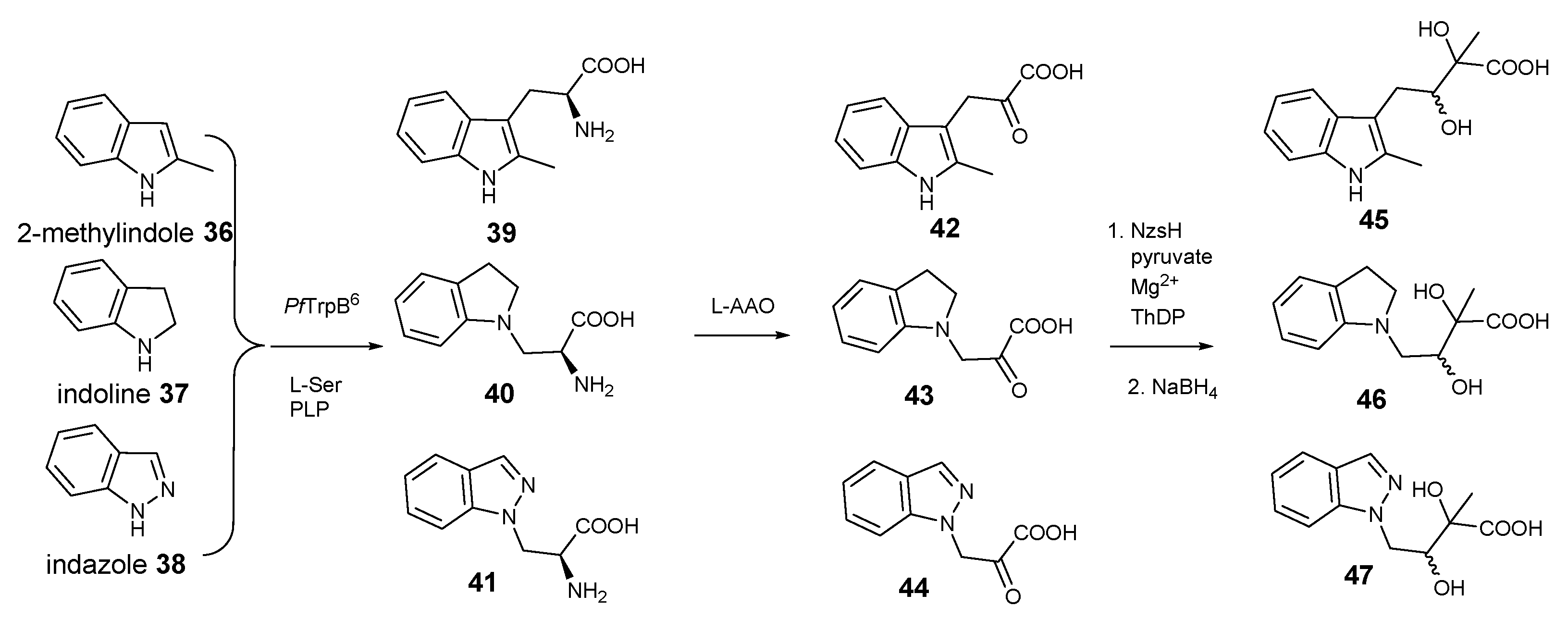
Next, we acquired three commercially available indole derivatives, 2-methyl-indole
36, indoline
37, and indazole
38 that contain modified indole rings (
Figure 4). Consistent with the previous report [
15,
16,
17], the biotransformation catalysed by
PfTrpB
6 gave the corresponding tryptophan derivatives,
39–
41, respectively. In all three cases, addition of LAAO provided the corresponding indole-pyruvate derivatives,
42–
44, respectively, further demonstrating the considerable substrate promiscuity of LAAO. To our surprise, these indole-pyruvate derivatives can be further utilized as the donor substrates in the NzsH-mediated enzymatic reaction to provide the final indole-containing acyloins,
45–
47, respectively, as observed in our LC-MS and tandem MS analyses of the corresponding reduced forms (
Supplementary Figure S20–S28). Taken together, our studies demonstrated that NzsH, unlike its activities towards acceptor substrates, displays considerable substrate tolerances toward its donor substrates. However, further studies are required to improve the reactivities of NzsH enzyme toward the unnatural indole-3-pyruvate derivatives. This can be achieved via either identification of new NzsH homologues through comparative genomics or directed evolution to provide an engineered NzsH with the aim of finding a suitable biocatalyst with better kinetics in order to efficiently generate structurally diverse indole-containing acyloin derivatives.
In conclusion, we developed a coupled biotransformation system including a genetically modified β-subunit of tryptophan synthase (PfTrpB6) from Pyrococcus furiosus, and a commercially available L-amino acid oxidase (LAAO) from snake venom and the ThDP-dependent NzsH enzyme from the biosynthetic pathway of neocarazostatins to access structurally diverse (aza)indole-containing acyloin derivatives. The combination of PfTrpB6 and LAAO allowed generation of (aza)indole-3-pyruvate derivatives, starting from commercially available (aza)indole derivatives, which were subsequently accessed by NzsH-mediated reaction. Our results demonstrated that NzsH displays considerably good substrate tolerance, suggesting that this three-enzyme coupled system holds potential as a means of new biotransformation to produce indole-containing acyloins, important precursors of many biologically relevant molecules.
3. Methods and Materials
3.1. General Chemicals, Reagents, and Analytical Methods
All starting materials and reagents were bought from commercial sources and used as received. All biochemical reactions apart from where noted were carried out in triplicate. Before every set of measurements, triplicate control reactions were performed to ensure that the assay were functioning correctly. All flash column chromatography was carried out using silica purchased from Sigma Aldrich using the solvent system noted. 19F NMR spectra were recorded at 298 K on Bruker Avance III 400 using CFCl3 as an external reference. Chemical shifts are reported in parts per million (ppm) and coupling constants (J) are reported in Hertz (Hz).
Enzymatic assays were analyzed on a Bruker MaXis II ESI-Q-TOF-MS connected to an Agilent 1290 Infinity II UHPLC fitted with a Phenomenex Kinetex XB-C18 (2.6 μM, 100 × 2.1 mm) column. The column was eluted with a linear gradient of 5–100% MeCN containing 0.1% formic acid over 15 min. The mass spectrometer was operated in positive ion mode with a scan range of 200–3000 m/z. Source conditions were: end plate offset at −500 V; capillary at −4500 V; nebulizer gas (N2) at 4.0 bar; dry gas (N2) at 9.0 L min−1; dry temperature at 200 °C. Ion transfer conditions were: ion funnel RF at 400 Vpp; multiple RF at 200 Vpp; quadrupole low mass at 200 m/z; collision energy at 8.0 eV; collision RF at 2000 Vpp; transfer time at 110.0 μs; pre-pulse storage time at 10.0 μs. MS data were analysed using Bruker DataAnalysis or Thermo Xcalibur.
3.2. General Methods of Protein Expression and Purification
The synthetic constructs encoding PfTrpB6 and NzsH proteins were purchased from Genscript Ltd. and were individually transformed into E. coli BL21 (DE3). Single colonies from each transformation were grown overnight in LB media (5 mL) containing kanamycin (50 µg/mL) and chloramphenicol (25 µg/mL). The overnight culture was transferred to fresh LB medium (500 mL) supplemented with kanamycin (50 µg/mL) and cultivated at 37 °C until the cell density reached an OD600 of 0.6. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM to induce protein expression. Cells were grown for 16–20 h at 16 °C and then harvested by centrifugation at 4 °C. The cells pellets were resuspended in ice-cold lysis buffer (20 mM Tris-HCl, 300 mM NaCl, 10 mM imidazole, pH 8.0), and further disrupted by Ultrasonic Homogenizer JY92-IIN. Then, the supernatant of cell debris was loaded onto Ni-NTA-affinity column. Bound proteins were eluted with the same Tris-HCl buffer containing different concentrations of imidazole. The desired elution fractions were combined and concentrated using a Centrifugal Filter Unit (Millipore). The final yields of PfTrpB6 and NzsH were estimated to be 10 mg/100 mL culture and 5 mg/100 mL culture, respectively.
3.3. Biochemical Reactions
A sample of PfTrpB6 (20 µM) was incubated with indole or indole derivatives (1 mM), L-Ser (1 mM) and PLP (1 mM) in phosphate buffer (50 mM, pH 7.5) to the final volume of 100 µL at 28 °C for 3 h and then quenched by addition of 100 µL of acetonitrile. The mixture was centrifuged at 13,000 rpm for 10 min to remove protein precipitates.
For the production of indole-3-pyruvate derivatives, a sample of PfTrpB6 (20 µM) was incubated with indole or indole derivatives (1 mM), L-Ser (1 mM) and PLP (1 mM) in phosphate buffer (50 mM, pH 7.5) to a final volume of 50 µL at 28 °C for 3 h. To this mixture were added LAAO (20 µM) to a final volume of 100 µL at 28 °C for another 1 h. The reaction mix was then quenched by addition of 100 µL of acetonitrile. The mixture was centrifuged at 13,000 rpm for 10 min to remove protein precipitates.
For the production of indole-containing acyloin derivatives, a sample of PfTrpB6 (20 µM) was incubated with indole or indole derivatives (1 mM), L-Ser (1 mM) and PLP (1 mM) in phosphate buffer (50 mM, pH 7.5) to a final volume of 100 µL at 28 °C for 3 h. To this mixture were added LAAO (20 µM) to a volume of 100 µL at 28 °C for another 1 h. Inclusion of NzsH (25 µM), TPP (1 mM) and Mg2+ (1 mM) into the mixture was perform to the final volume of 100 µL at 28 °C for 3 h. Because of the instability of acyloins, overdose NaBH4 was treated to the three-enzyme systems. Finally, the reaction mix was quenched by addition of 100 µL of acetonitrile. The mixture was centrifuged at 13,000 rpm for 10 min to remove protein precipitates.
All the supernatants from the above reaction systems were then analyzed by Bruker MaXis II QTOF in tandem with an Agilent 1290 Infinity UHPLC. Samples were separated on a Phenomenex Kinetex XB-C18 (2.6 μM, 100 × 2.1 mm) column with a mobile phase of 5% ACN + 0.1% formic acid to 100% ACN + 0.1% formic acid in 15 min.