How Theoretical Evaluations Can Generate Guidelines for Designing/Engineering Metalloproteins with Desired Metal Affinity and Selectivity
Abstract
1. Introduction
2. Intrinsic Physicochemical Properties of the Metal Cations Determine to a Great Extent the Metal Selectivity of the Binding Site
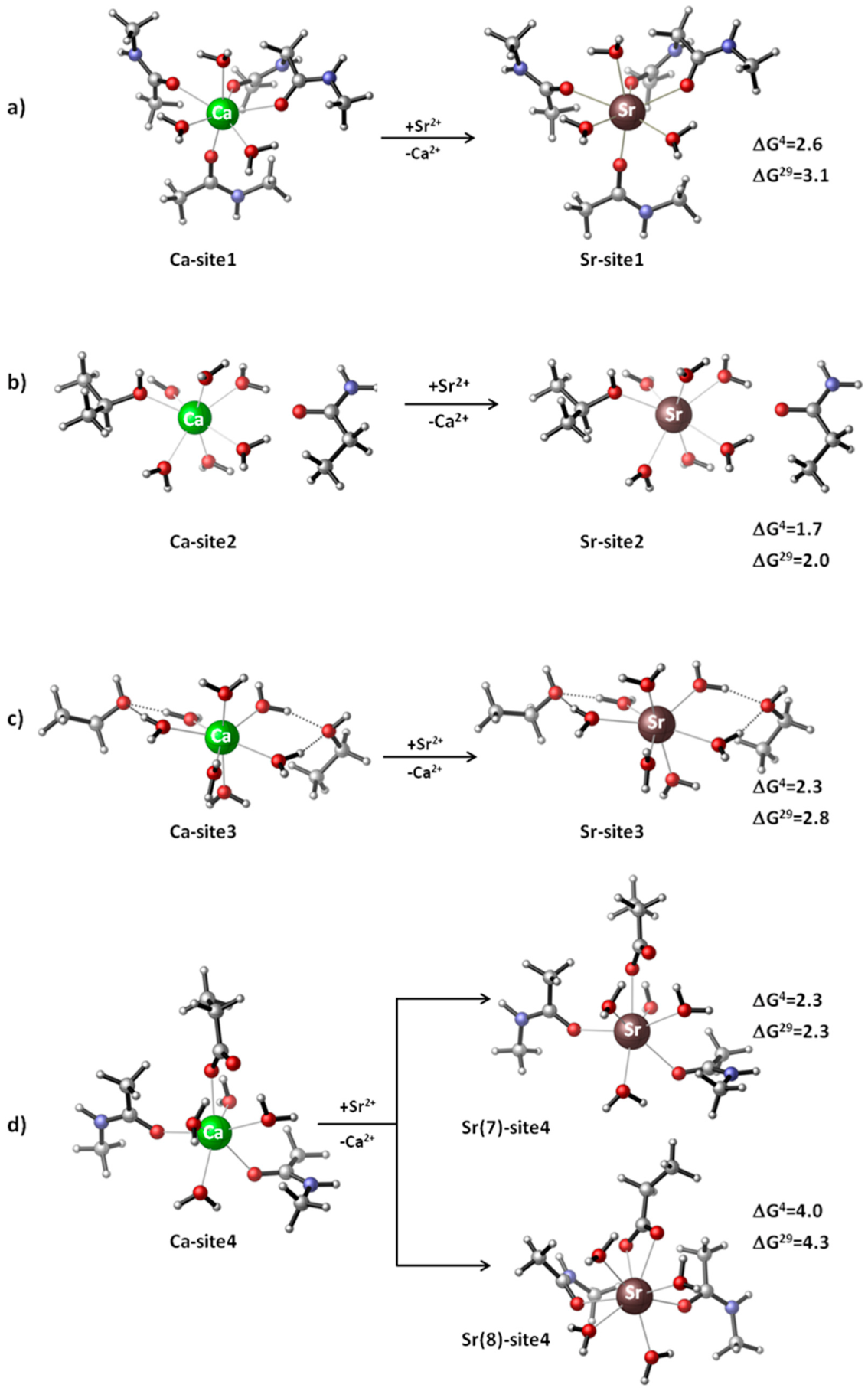
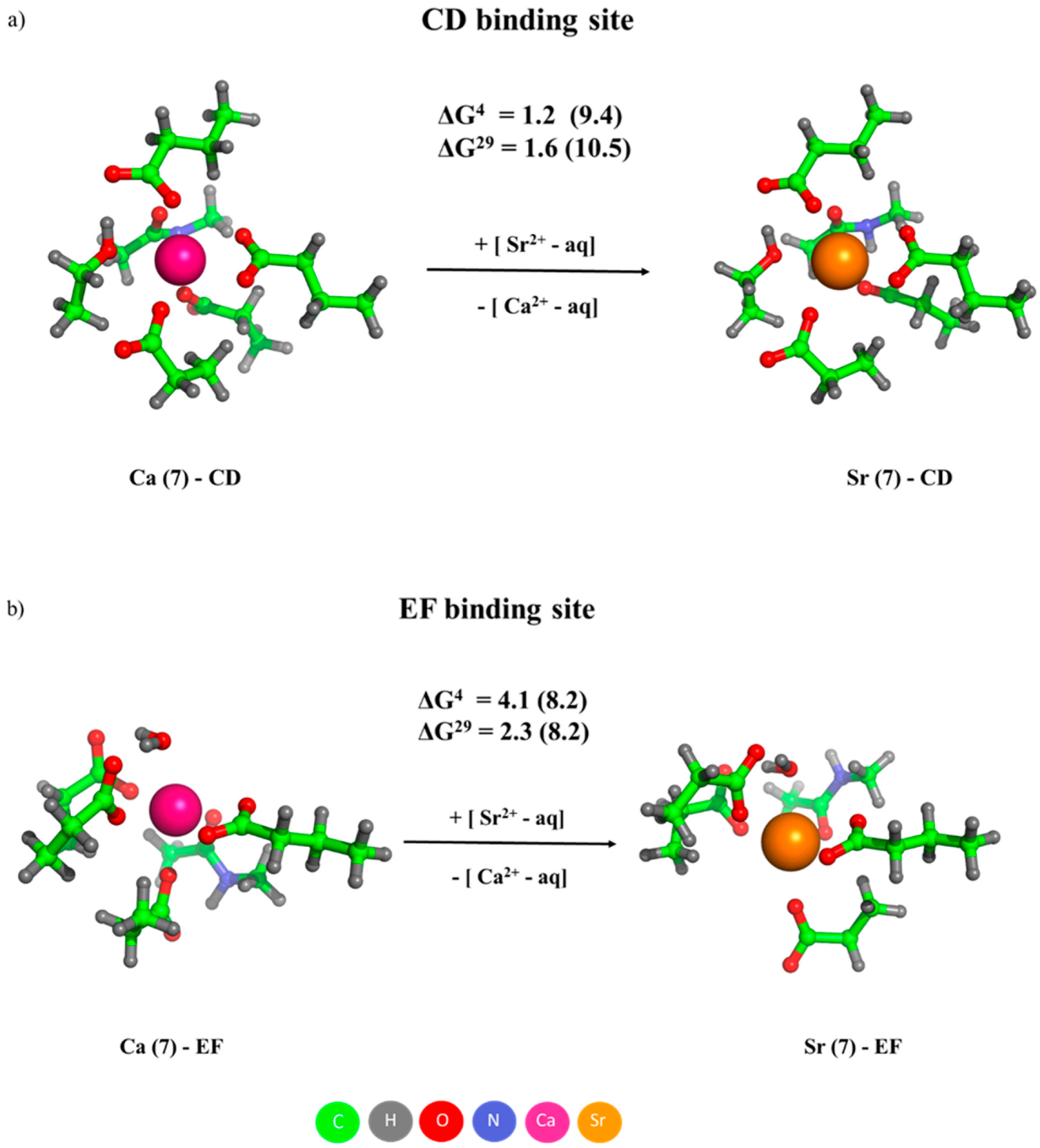
2.1. Competition between Ca2+ and Sr2+ in Calcium Receptors/Signaling Proteins
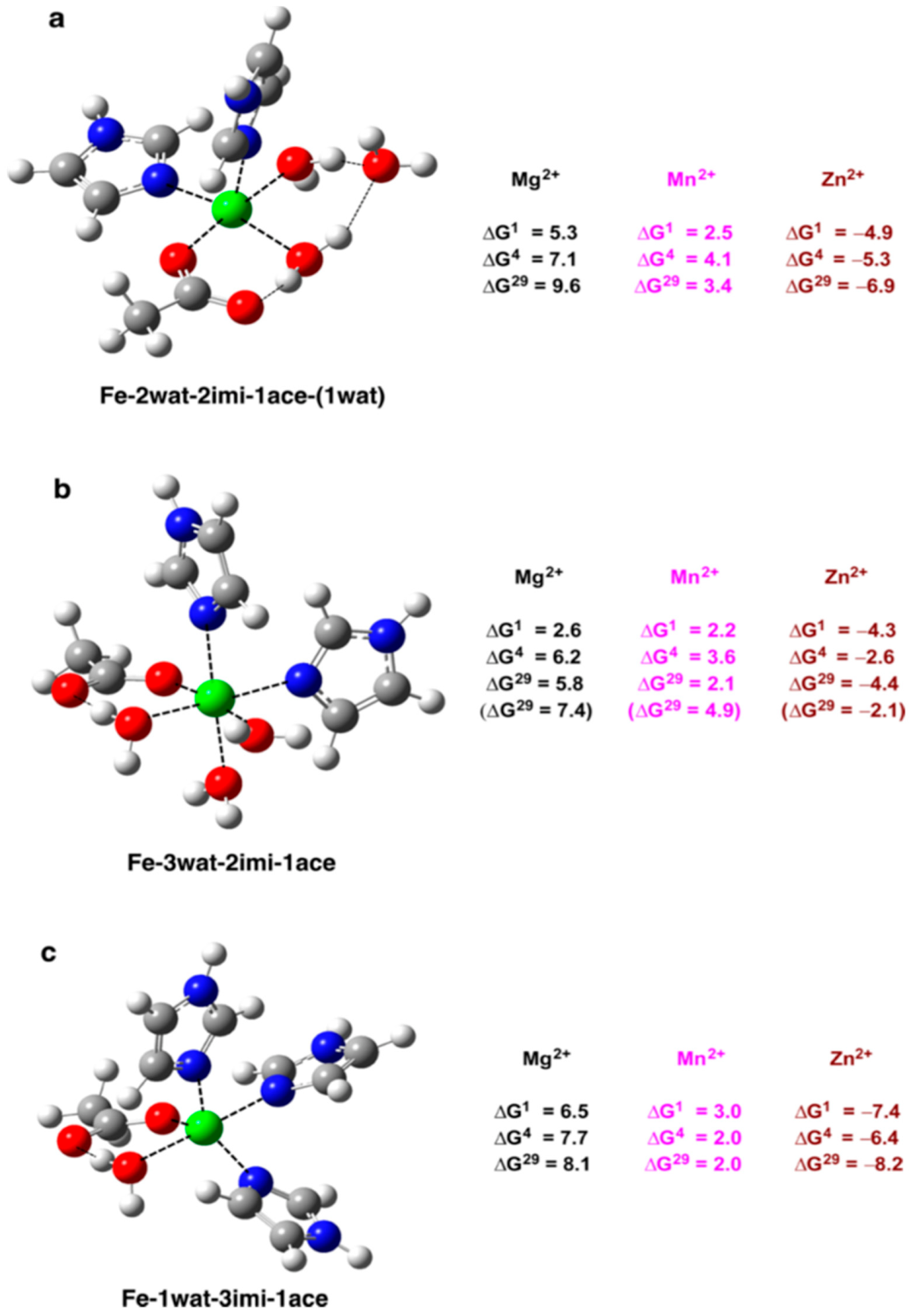
2.2. Fe2+ vs. Mg2+, Mn2+ and Zn2+ in Non-Heme Iron Proteins
- How does the Fe2+ binding site sequesters the “right” (native) cation from the cellular fluids and protect itself from attacks by other biogenic cations such as Mg2+, Mn2+, and Zn2+?
- What kind of selectivity strategies do iron binding sites employ toward metal cations having different ligand affinities and cytosolic concentrations?
- What are the key factors governing the metal selectivity in Fe2+ proteins?
2.3. Competition between Cr3+ and Fe3+, Fe2+, Mg2+ and Zn2+ in Chromodulin
3. Metal Coordination Number Is an Important Determinant of the Metal Selectivity
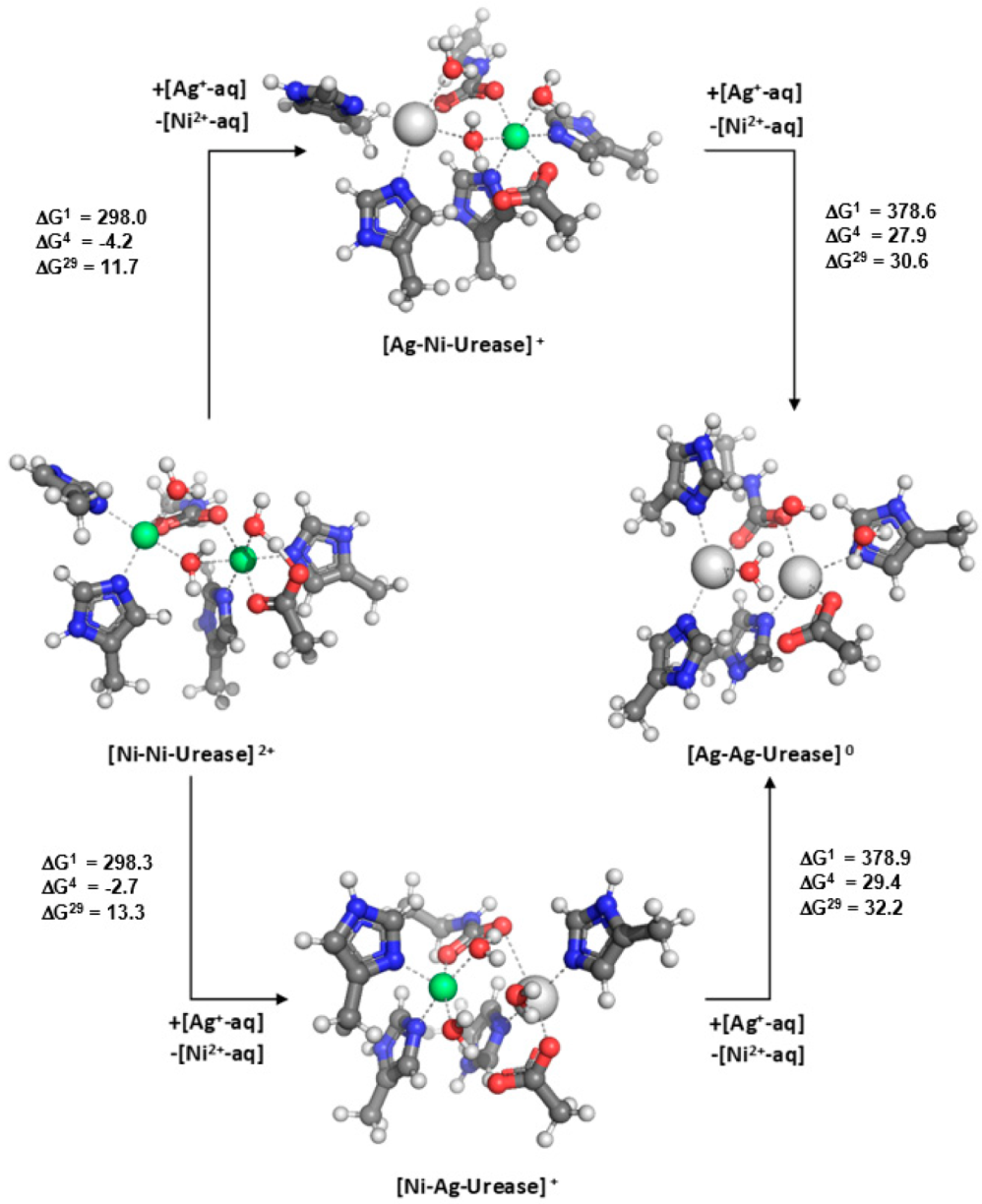
4. Adjacent Metal Cation May Reverse the Metal Selectivity in Binuclear/Trinuclear Metal Binding Sites
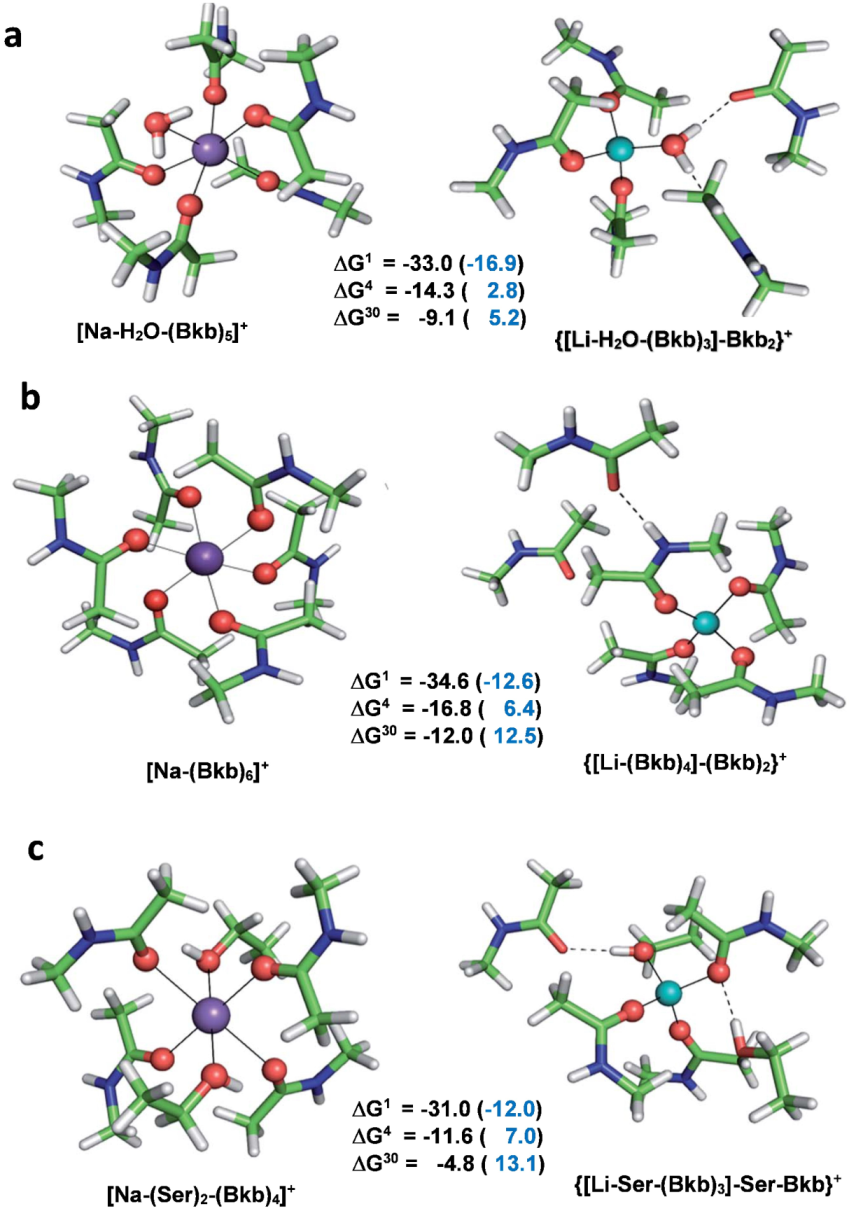
5. Rigid Binding Sites Adapted to the Specific Structural Requirements of the Native Metal Cation Enhance Its Competitiveness toward Other Metal Contenders
6. pH of the Medium Is a Key Factor Governing the Competition between the Native and Trivalent “Alien” Metal Species
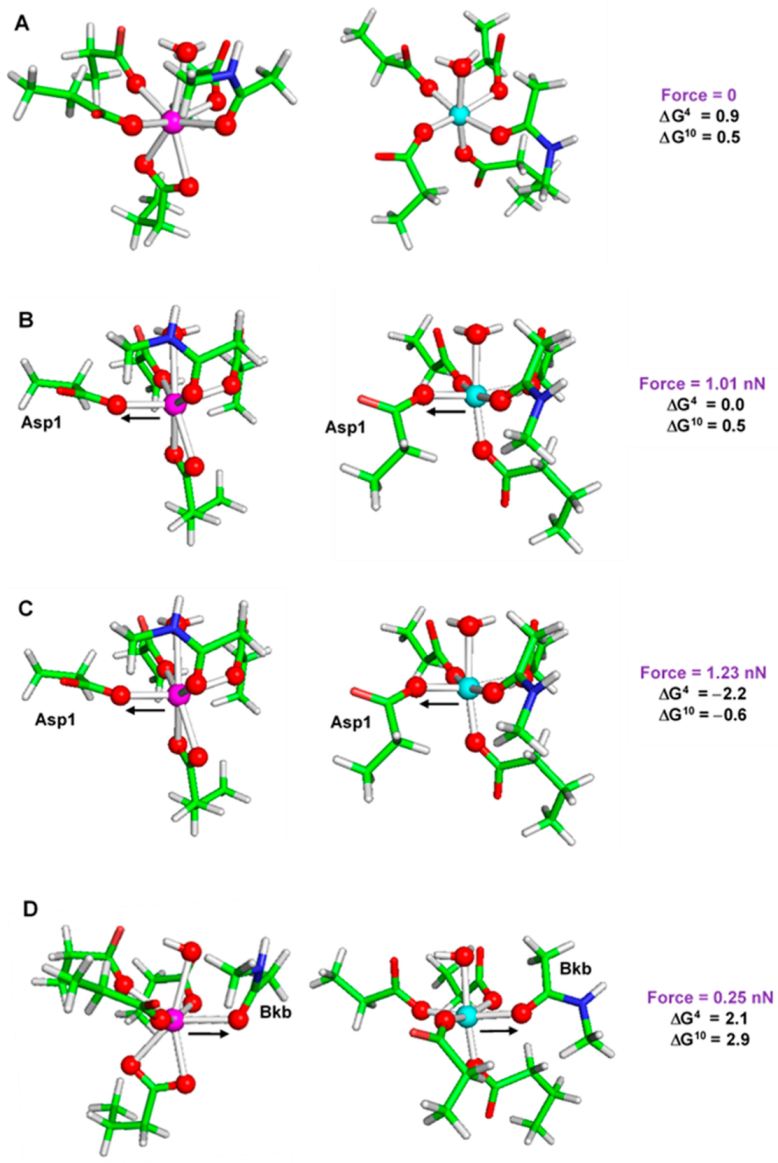
7. Mechanical Forces Can Modulate the Metal Selectivity in Metal Binding Sites in Proteins
8. Concluding Remarks
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dudev, T.; Lim, C. Competition among Metal Ions for Protein Binding Sites: Determinants of Metal Ion Selectivity in Proteins. Chem. Rev. 2013, 114, 538–556. [Google Scholar] [CrossRef] [PubMed]
- Dudev, T.; Lim, C. Principles Governing Mg, Ca and Zn Selectivity in Proteins. Chem. Rev. 2003, 103, 773–787. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A.; Merkle, D.L.; Berg, J.M. Ligand variation and metal ion binding specificity in zinc finger peptides. Inorg. Chem. 1993, 32, 937–940. [Google Scholar] [CrossRef]
- Payne, J.C.; Horst, M.A.T.; Godwin, H.A. Lead fingers: Pb2+ binding to structural zinc-binding domains determined directly by moni-toring lead-thiolate charge-transfer bands. J. Am. Chem. Soc. 1999, 121, 6850–6855. [Google Scholar] [CrossRef]
- Falke, J.J.; Snyder, E.E.; Thatcher, K.C.; Voertler, C.S. Quantitating and engineering the ion specificity of an EF-hand-like Ca2+ binding site. Biochemistry 1991, 30, 8690–8697. [Google Scholar] [CrossRef] [PubMed]
- Drake, S.K.; Zimmer, M.A.; Kundrot, C.; Falke, J.J. Molecular tuning of an EF-hand-like calcium binding loop—Contributions of the coordinating side chain at loop position 3. J. Gen. Physiol. 1997, 110, 173–184. [Google Scholar] [CrossRef]
- Rao, L.; Cui, Q.; Xu, X. Electronic Properties and Desolvation Penalties of Metal Ions Plus Protein Electrostatics Dictate the Metal Binding Affinity and Selectivity in the Copper Efflux Regulator. J. Am. Chem. Soc. 2010, 132, 18092–18102. [Google Scholar] [CrossRef]
- Rulisek, L.; Havlas, Z. Theoretical studies of metal ion selectivity. 1. DFT calculations of interaction energies of amino acid side chains with selected transition metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+). J. Am. Chem. Soc. 2000, 122, 10428–10439. [Google Scholar] [CrossRef]
- McCall, K.A.; Fierke, C.A. Probing Determinants of the Metal Ion Selectivity in Carbonic Anhydrase Using Mutagenesis. Biochemistry 2004, 43, 3979–3986. [Google Scholar] [CrossRef]
- Wang, S.C.; Dias, A.V.; Bloom, S.L.; Zamble, D.B. Selectivity of Metal Binding and Metal-Induced Stability of Escherichia coli NikR. Biochemistry 2004, 43, 10018–10028. [Google Scholar] [CrossRef]
- Tottey, S.; Waldron, K.J.; Firbank, S.J.; Reale, B.; Bessant, C.; Sato, K.; Cheek, T.R.; Gray, J.; Banfield, M.J.; Dennison, C.; et al. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature 2008, 455, 1138–1142. [Google Scholar] [CrossRef]
- Shamovsky, I.L.; Ross, G.M.; Riopelle, R.J.; Weaver, D.F. Theoretical and Biochemical Studies on the Selectivity of Nerve Growth Factor for Transition Metal Cations. J. Am. Chem. Soc. 1999, 121, 9797–9806. [Google Scholar] [CrossRef]
- Dudev, T.; Lim, C. Competition among Ca2+, Mg2+, and Na+ for Model Ion Channel Selectivity Filters: Determinants of Ion Selectivity. J. Phys. Chem. B 2012, 116, 10703–10714. [Google Scholar] [CrossRef]
- Nielsen, S.P. The biological role of strontium. Bone 2004, 35, 583–588. [Google Scholar] [CrossRef]
- Kraeber-Bodéré, F.; Campion, L.; Rousseau, C.; Bourdin, S.; Chatal, J.-F.; Resche, I. Treatment of bone metastases of prostate cancer with strontium-89 chloride: Efficacy in relation to the degree of bone involvement. Eur. J. Nucl. Med. 2000, 27, 1487–1493. [Google Scholar] [CrossRef]
- Cabrera, W.E.; Schrooten, I.; De Broe, M.E.; D’Haese, P.C. Strontium and bone. J. Bone Miner. Res. 1999, 14, 661–668. [Google Scholar] [CrossRef]
- Marie, P.J.; Ammann, P.; Boivin, G.; Rey, C. Mechanisms of Action and Therapeutic Potential of Strontium in Bone. Calcif. Tissue Int. 2001, 69, 121–129. [Google Scholar] [CrossRef]
- Cianferotti, L.; Gomes, A.R.; Fabbri, S.; Tanini, A.; Brandi, M.L. The calcium-sensing receptor in bone metabolism: From bench to bedside and back. Osteoporos. Int. 2015, 26, 2055–2071. [Google Scholar] [CrossRef]
- Geng, Y.; Mosyak, L.; Kurinov, I.; Zuo, H.; Sturchler, E.; Cheng, T.C.; Subramanyam, P.; Brown, A.P.; Brennan, S.C.; Mun, H.-C.; et al. Structural mechanism of ligand activation in human calcium-sensing receptor. eLife 2016, 5, e13662. [Google Scholar] [CrossRef]
- Shannon, R.D. Revised effective ionic radii and systematic studies of interatomic distances in halides and chalcogenides. Acta Cryst. 1976, A32, 751–766. [Google Scholar] [CrossRef]
- Marcus, Y. Thermodynamics of solvation of ions. Part 5—Gibbs free energy of hydration at 298.15 K. J. Chem. Soc. Faraday Trans. 1991, 87, 2995–2999. [Google Scholar] [CrossRef]
- Dudev, M.; Wang, J.; Dudev, T.; Lim, C. Factors Governing the Metal Coordination Number in Metal Complexes from Cambridge Structural Database Analyses. J. Phys. Chem. B 2006, 110, 1889–1895. [Google Scholar] [CrossRef] [PubMed]
- Hofer, T.S.; Randolf, B.R.; Rode, B.M. Sr(II) in Water: A Labile Hydrate with a Highly Mobile Structure. J. Phys. Chem. B 2006, 110, 20409–20417. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.M. Is the calcium receptor a molecular target for the actions of strontium on bone? Osteoporos. Int. 2003, 14, S25–S34. [Google Scholar] [CrossRef] [PubMed]
- Diepenhorst, N.A.; Leach, K.; Keller, A.N.; Rueda, P.; Cook, A.E.; Pierce, T.L.; Nowell, C.; Pastoureau, P.; Sabatini, M.; Summers, R.J.; et al. Divergent effects of strontium and calcium-sensing receptor positive allosteric modulators (calcimimetics) on human osteoclast activity. Br. J. Pharmacol. 2018, 175, 4095–4108. [Google Scholar] [CrossRef]
- Uversky, V.N. Strontium binding to proteins. In Encyclopedia of Metalloproteins; Uversky, V.N., Kretsinger, R.H., Permyakov, E.A., Eds.; Springer Science: New York, NY, USA, 2013; pp. 2133–2138. [Google Scholar]
- Vologzhannikova, A.A.; Shevelyova, M.P.; Kazakov, A.S.; Sokolov, A.S.; Borisova, N.I.; Permyakov, E.A.; Kircheva, N.; Nikolova, V.; Dudev, T.; Permyakov, S.E. Strontium Binding to α-Parvalbumin, a Canonical Calcium-Binding Protein of the “EF-Hand” Family. Biomolecules 2021, 11, 1158. [Google Scholar] [CrossRef]
- Cheshmedzhieva, D.; Ilieva, S.; Permyakov, E.A.; Permyakov, S.E.; Dudev, T. Ca2+/Sr2+ Selectivity in Calcium-Sensing Receptor (CaSR): Implications for Strontium’s Anti-Osteoporosis Effect. Biomolecules 2021, 11, 1576. [Google Scholar] [CrossRef]
- Coulombe, J.; Faure, H.; Robin, B.; Ruat, M. In vitro effects of strontium ranelate on the extracellular calcium-sensing receptor. Biochem. Biophys. Res. Commun. 2004, 323, 1184–1190. [Google Scholar] [CrossRef]
- Elizabeth, C.T.; Uversky, V.N. Encyclopedia of Metalloproteins; Uversky, V.N., Kretsinger, R.H., Permyakov, E.A., Eds.; Springer Science: New York, NY, USA, 2013; p. 1032. [Google Scholar]
- Crain, A.V.; Duschene, K.S.; Peters, J.W.; Broderick, J.B. Iron-Sulfur Cluster Proteins, Fe/SS-adenosylmethionine Enzymes and Hydrogenases. In Encyclopedia of Metalloproteins; Uversky, V.N., Kretsinger, R.H., Permyakov, E.A., Eds.; Springer Science: New York, NY, USA, 2013; pp. 1034–1044. [Google Scholar]
- Dudev, T.; Nikolova, V. Determinants of Fe2+over M2+ (M = Mg, Mn, Zn) Selectivity in Non-Heme Iron Proteins. Inorg. Chem. 2016, 55, 12644–12650. [Google Scholar] [CrossRef]
- Costas, M.; Mehn, M.P.; Jensen, M.P.; Que, J. Dioxygen Activation at Mononuclear Non-heme Iron Active Sites: Enzymes, Models and Intermediates. Chem. Rev. 2004, 104, 939–986. [Google Scholar] [CrossRef]
- Hausinger, R.P. Fell/alpha-ketoglutarate-dependent Hydroxylases and Related Enzymes. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 21–68. [Google Scholar] [CrossRef]
- Solomon, E.I.; Decker, A.; Lehnert, N. Non-heme iron enzymes: Contrasts to heme catalysis. Proc. Natl. Acad. Sci. USA 2003, 100, 3589–3594. [Google Scholar] [CrossRef]
- Bruijnincx, P.C.A.; van Koten, G.; Klein Gebbink, R.J.M. Mononuclear Non-heme Iron Enzymes with the 2-His-1 caboxylate Facial Triad: Recent Developments in Enzymology and Modeling Studies. Chem. Soc. Rev. 2008, 37, 2716–2744. [Google Scholar] [CrossRef]
- Kovaleva, E.G.; Lipscomb, J.D. Versatility of Biological Nonheme Fe(II) Centers in Oxyden Activation Reactions. Nat. Chem. Biol. 2008, 4, 186–193. [Google Scholar] [CrossRef]
- Mbughuni, M.M.; Lipscomb, J.D. Iron Proteins, Mononuclear (non-heme) Iron Oxygenases. In Encyclopedia of Metalloproteins; Uversky, V.N., Kretsinger, R.H., Permyakov, E.A., Eds.; Springer Science: New York, NY, USA, 2013; pp. 1006–1015. [Google Scholar]
- Irving, H.; Williams, R.J.P. Order of Stability of Metal Complexes. Nature 1948, 162, 746–747. [Google Scholar] [CrossRef]
- Anjem, A.; Varghese, S.; Imlay, J.A. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol. Microbiol. 2009, 72, 844–858. [Google Scholar] [CrossRef]
- Campos-Bermudez, V.A.; Leite, N.R.; Krog, R.; Costa-Filho, A.J.; Soncini, F.C.; Oliva, G.; Vila, A.J. Biochemical and Structural Char-acterization of Salmonella Typhimurium Glyoxalase II: New Insights into Metal Ion Selectivity. Biochemistry 2007, 46, 11069–11079. [Google Scholar] [CrossRef]
- Gattis, S.G.; Hernick, M.; Fierke, C.A. Active Site Metal Ion in UDP-3-O-((R)-3-Hydroxymyristoyl)-N-acetylglucosamine Deacetylase (LpxC) Switches between Fe(II) and Zn(II) Depending on Cellular Conditions. J. Biol. Chem. 2010, 285, 33788–33796. [Google Scholar] [CrossRef]
- Hernick, M.; Gattis, S.G.; Penner-Hahn, J.E.; Fierke, C.A. Activation of E. coli UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine Deacetylase by Fe2+ Yields a More Efficient Enzyme with Altered Ligand Affinity. Biochemistry 2010, 49, 2246–2255. [Google Scholar] [CrossRef]
- Kim, B.; Pithadia, A.S.; Fierke, C.A. Kinetics and thermodynamics of metal-binding to histone deacetylase 8. Protein Sci. 2015, 24, 354–365. [Google Scholar] [CrossRef]
- Dowling, D.; Gattis, S.G.; Fierke, C.A.; Christianson, D.W. Structures of Metal-Substituted Human Histone Deacetylase 8 Provide Mechanistic Inferences on Biological Function. Biochemistry 2010, 49, 5048–5056. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Schlichting, I.; Kabsch, W.; Groche, D.; Schultz, S.; Wagner, A.F.V. Iron center, substrate recognition and mechanism of peptide deformylase. Nat. Genet. 1998, 5, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Dizin, E.; Hu, X.; Wavreille, A.-S.; Park, J.; Pei, D. S-Ribosylhomocysteinase (LuxS) Is a Mononuclear Iron Protein. Biochemistry 2003, 42, 4717–4726. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Satpati, P. Divalent-Metal-Ion Selectivity of the CRISPR-Cas System-Associated Cas1 Protein: Insights from Classical Molecular Dynamics Simulations and Electronic Structure Calculations. J. Phys. Chem. B 2021, 125, 11943–11954. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.-H. Antioxidant Effects and Insulin Resistance Improvement of Chromium Combined with Vitamin C and E Supplementation for Type 2 Diabetes Mellitus. J. Clin. Biochem. Nutr. 2008, 43, 191–198. [Google Scholar] [CrossRef]
- Anderson, R.A.; Cheng, N.; Bryden, N.A.; Polansky, M.M.; Chi, J.; Feng, J. Elevated intakes of supplemental chromium improve glucose and insulin variables in individuals with type 2 diabetes. Diabetes 1997, 46, 1786–1791. [Google Scholar] [CrossRef]
- Martin, J.; Wang, Z.Q.; Zhang, X.H.; Wachtel, D.; Volaufova, J.; Matthews, D.E.; Cefalu, W.T. Chromium Picolinate Supplementation Attenuates Body Weight Gain and Increases Insulin Sensitivity in Subjects with Type 2 Diabetes. Diabetes Care 2006, 29, 1826–1832. [Google Scholar] [CrossRef][Green Version]
- Vincent, J.B. The Nutritional Biochemistry of Chromium (III), 1st ed.; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar] [CrossRef]
- Jacquamet, L.; Sun, Y.; Hatfield, J.; Gu, W.; Cramer, S.P.; Crowder, M.W.; Lorigan, G.A.; Vincent, J.B.; Latour, J.-M. Characterization of Chromodulin by X-ray Absorption and Electron Paramagnetic Resonance Spectroscopies and Magnetic Susceptibility Measurements. J. Am. Chem. Soc. 2003, 125, 774–780. [Google Scholar] [CrossRef]
- Kircheva, N.; Toshev, N.; Dudev, T. Holo-chromodulin: Competition between the native Cr3+ and other biogenic cations (Fe3+, Fe2+, Mg2+, and Zn2+) for the binding sites. Metallomics 2022, 14, mfac082. [Google Scholar] [CrossRef]
- Kooshki, F.; Tutunchi, H.; Vajdi, M.; Karimi, A.; Niazkar, H.R.; Shoorei, H.; Gargari, B.P. A Comprehensive insight into the effect of chromium supplementation on oxidative stress indices in diabetes mellitus: A systematic review. Clin. Exp. Pharmacol. Physiol. 2021, 48, 291–309. [Google Scholar] [CrossRef]
- Johnson, D.A. Some Thermodynamic Aspects of Inorganic Chemistry. In Inorganic Chemistry; Weller, M., Overton, T., Armstrong, F.A., Eds.; Oxford University Press: Oxford, UK, 2014. [Google Scholar]
- Malinauskaite, L.; Quick, M.; Reinhard, L.; Lyons, J.A.; Yano, H.; Javitch, J.; Nissen, P. A mechanism for intracellular release of Na+ by neurotransmitter/sodium symporters. Nat. Struct. Mol. Biol. 2014, 21, 1006–1012. [Google Scholar] [CrossRef]
- Neubig, R.R.; Siderovski, D. Regulators of G-Protein signalling as new central nervous system drug targets. Nat. Rev. Drug Discov. 2002, 1, 187–197. [Google Scholar] [CrossRef]
- Amara, S.G.; Sonders, M.S. Neurotransmitter transporters as molecular targets for addictive drugs. Drug Alcohol Depend. 1998, 51, 87–96. [Google Scholar] [CrossRef]
- Dudev, T.; Mazmanian, K.; Lim, C. Competition between Li+ and Na+ in sodium transporters and receptors: Which Na+-binding sites are “therapeutic” Li+ targets? Chem. Sci. 2018, 9, 4093–4103. [Google Scholar] [CrossRef]
- Mulrooney, S.B.; Hausinger, R. Nickel uptake and utilization by microorganisms. FEMS Microbiol. Rev. 2003, 27, 239–261. [Google Scholar] [CrossRef]
- Ragsdale, S.W. Nickel-Based Enzyme Systems. J. Biol. Chem. 2009, 284, 18571–18575. [Google Scholar] [CrossRef]
- Boer, J.L.; Mulrooney, S.B.; Hausinger, R.P. Nickel-dependent metalloenzymes. Arch. Biochem. Biophys. 2014, 544, 142–152. [Google Scholar] [CrossRef]
- Alfano, M.; Cavazza, C. Structure, function, and biosynthesis of nickel-dependent enzymes. Protein Sci. 2020, 29, 1071–1089. [Google Scholar] [CrossRef]
- Maroney, M.J.; Ciurli, S. Nonredox Nickel Enzymes. Chem. Rev. 2014, 114, 4206–4228. [Google Scholar] [CrossRef]
- Clugston, S.L.; Barnard, J.F.J.; Kinach, R.; Miedema, D.; Ruman, R.; Daub, E.; Honek, J.F. Overproduction and Characterization of a Dimeric Non-Zinc Glyoxalase I from Escherichia coli: Evidence for Optimal Activation by Nickel Ions. Biochemistry 1998, 37, 8754–8763. [Google Scholar] [CrossRef]
- Mazzei, L.; Cianci, M.; Vara, A.G.; Ciurli, S. The structure of urease inactivated by Ag(i): A new paradigm for enzyme inhibition by heavy metals. Dalton Trans. 2018, 47, 8240–8247. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, J.F.; Kistiakowsky, G.B.; Kridl, A.G. Inhibition of Urease by Silver Ions. J. Am. Chem. Soc. 1951, 73, 1232–1236. [Google Scholar] [CrossRef]
- Krajewska, B. Mono- (Ag, Hg) and di- (Cu, Hg) valent metal ions effects on the activity of jack bean urease. Probing the modes of metal binding to the enzyme. J. Enzym. Inhib. Med. Chem. 2008, 23, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Dobrev, S.; Kircheva, N.; Nikolova, V.; Angelova, S.; Dudev, T. Competition between Ag+ and Ni2+ in nickel enzymes: Implications for the Ag+ antibacterial activity. Comput. Biol. Chem. 2022, 101, 107785. [Google Scholar] [CrossRef] [PubMed]
- Dudev, T.; Lim, C. Competition between Li+ and Mg2+ in Metalloproteins. Implications for Lithium Therapy. J. Am. Chem. Soc. 2011, 133, 9506–9515. [Google Scholar] [CrossRef]
- Grauffel, C.; Weng, W.-H.; Dudev, T.; Lim, C. The Trinuclear Calcium Site in the C2 domain of PKCα/γ is Prone to Lithium Attack. ACS Omega 2021, 6, 20657–20666. [Google Scholar] [CrossRef]
- Martin, R.B. Aluminum: A Neurotoxic Product of Acid Rain. Acc. Chem. Res. 1994, 27, 204–210. [Google Scholar] [CrossRef]
- Macdonald, T.L.; Martin, R.B. Aluminum ion in biological systems. Trends Biochem. Sci. 1988, 13, 15–19. [Google Scholar] [CrossRef]
- Léonard, A.; Gerber, G.B. Mutagenicity, carcinogenicity and teratogenicity of aluminium. Mutat. Res. Genet. Toxicol. 1988, 196, 247–257. [Google Scholar] [CrossRef]
- Kavitha, A.V.; Jagadeesan, G. Role of Tribulus terrestris (Linn.) (Zygophyllacea) against mercuric chloride induced nephrotoxicity in mice, Mus musculus (Linn.). J. Environ. Biol. 2006, 27, 397–400. [Google Scholar]
- Martin, R. Fe3+ and Al3+ hydrolysis equilibria. Cooperativity in Al3+ hydrolysis reactions. J. Inorg. Biochem. 1991, 44, 141–147. [Google Scholar] [CrossRef]
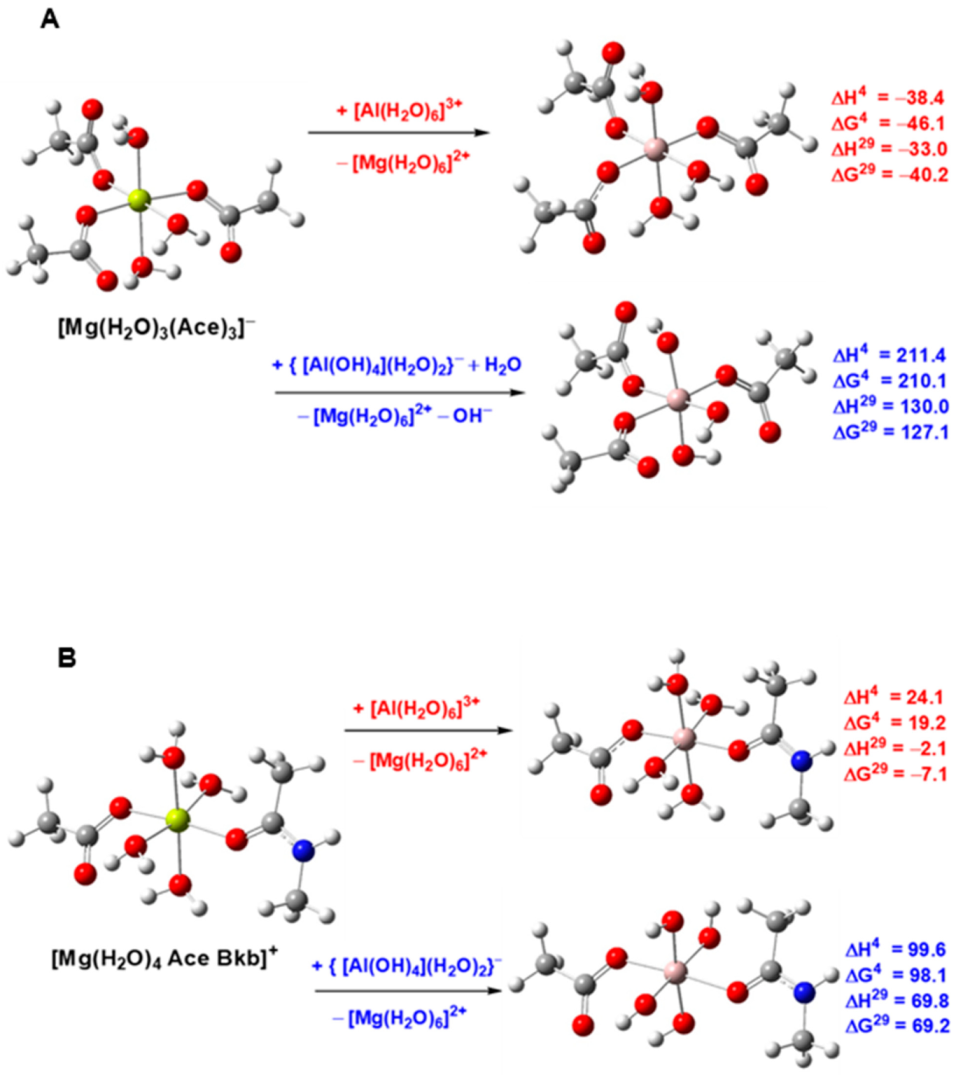
- Dudev, T.; Cheshmedzhieva, D.; Doudeva, L. Competition between abiogenic Al3+ and native Mg2+, Fe2+ and Zn2+ ions in protein binding sites: Implications for aluminum toxicity. J. Mol. Model. 2018, 24, 55. [Google Scholar] [CrossRef]
- Ohashi, K.; Fujiwara, S.; Mizuno, K. Roles of the cytoskeleton, cell adhesion and rho signalling in mechanosensing and mecha-notransduction. J. Biochem. 2017, 161, 245–254. [Google Scholar]
- Vining, K.H.; Mooney, D.J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 2017, 18, 728–742. [Google Scholar] [CrossRef]
- Dudev, T.; Frutos, L.M.; Castaño, O. How mechanical forces can modulate the metal affinity and selectivity of metal binding sites in proteins. Metallomics 2020, 12, 363–370. [Google Scholar] [CrossRef]


| Metal | Ligand | ||||
|---|---|---|---|---|---|
| CH3COO− a | CH3CONHCH3 b | OH− | CH3S− c | Imidazole d | |
| Ca2+ | −320.7 | −108.3 | −343.3 | −292.6 | −95.0 |
| Sr2+ | −297.9 | −93.9 | −321.3 | −273.2 | −81.8 |
| Fe2+ | −399.0 | −163.0 | −416.8 | −388.1 | −152.3 |
| Mg2+ | −375.1 | −140.4 | −383.6 | −357.3 | −134.7 |
| Mn2+ | −382.4 | −147.8 | −398.8 | −372.1 | −142.6 |
| Zn2+ | −414.0 | −168.9 | −417.5 | −423.5 | −174.1 |
| Cr3+ | −742.5 | −440.6 | −756.3 | −770.4 | −449.1 |
| Fe3+ | −740.0 | −436.0 | −736.0 | −777.8 | −453.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dudev, T. How Theoretical Evaluations Can Generate Guidelines for Designing/Engineering Metalloproteins with Desired Metal Affinity and Selectivity. Molecules 2023, 28, 249. https://doi.org/10.3390/molecules28010249
Dudev T. How Theoretical Evaluations Can Generate Guidelines for Designing/Engineering Metalloproteins with Desired Metal Affinity and Selectivity. Molecules. 2023; 28(1):249. https://doi.org/10.3390/molecules28010249
Chicago/Turabian StyleDudev, Todor. 2023. "How Theoretical Evaluations Can Generate Guidelines for Designing/Engineering Metalloproteins with Desired Metal Affinity and Selectivity" Molecules 28, no. 1: 249. https://doi.org/10.3390/molecules28010249
APA StyleDudev, T. (2023). How Theoretical Evaluations Can Generate Guidelines for Designing/Engineering Metalloproteins with Desired Metal Affinity and Selectivity. Molecules, 28(1), 249. https://doi.org/10.3390/molecules28010249





