High-Yield Characterization of Single Molecule Interactions with DeepTipTM Atomic Force Microscopy Probes
Abstract
1. Introduction
2. Results and Discussion
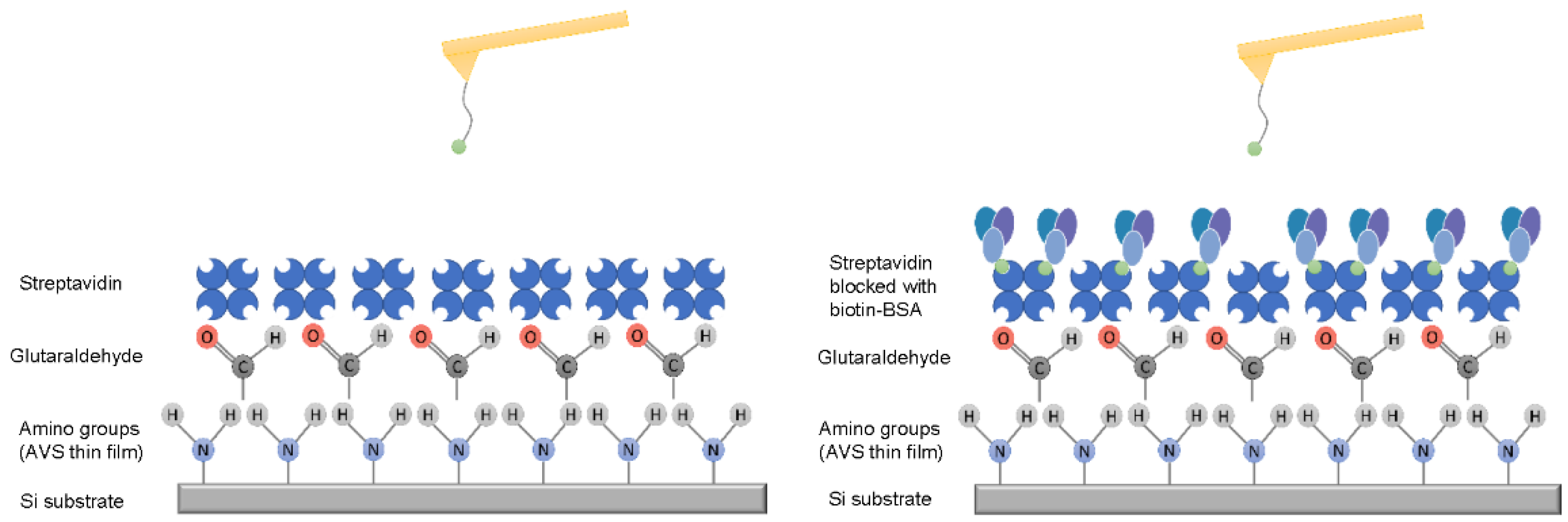
2.1. Amino Group Reactivity
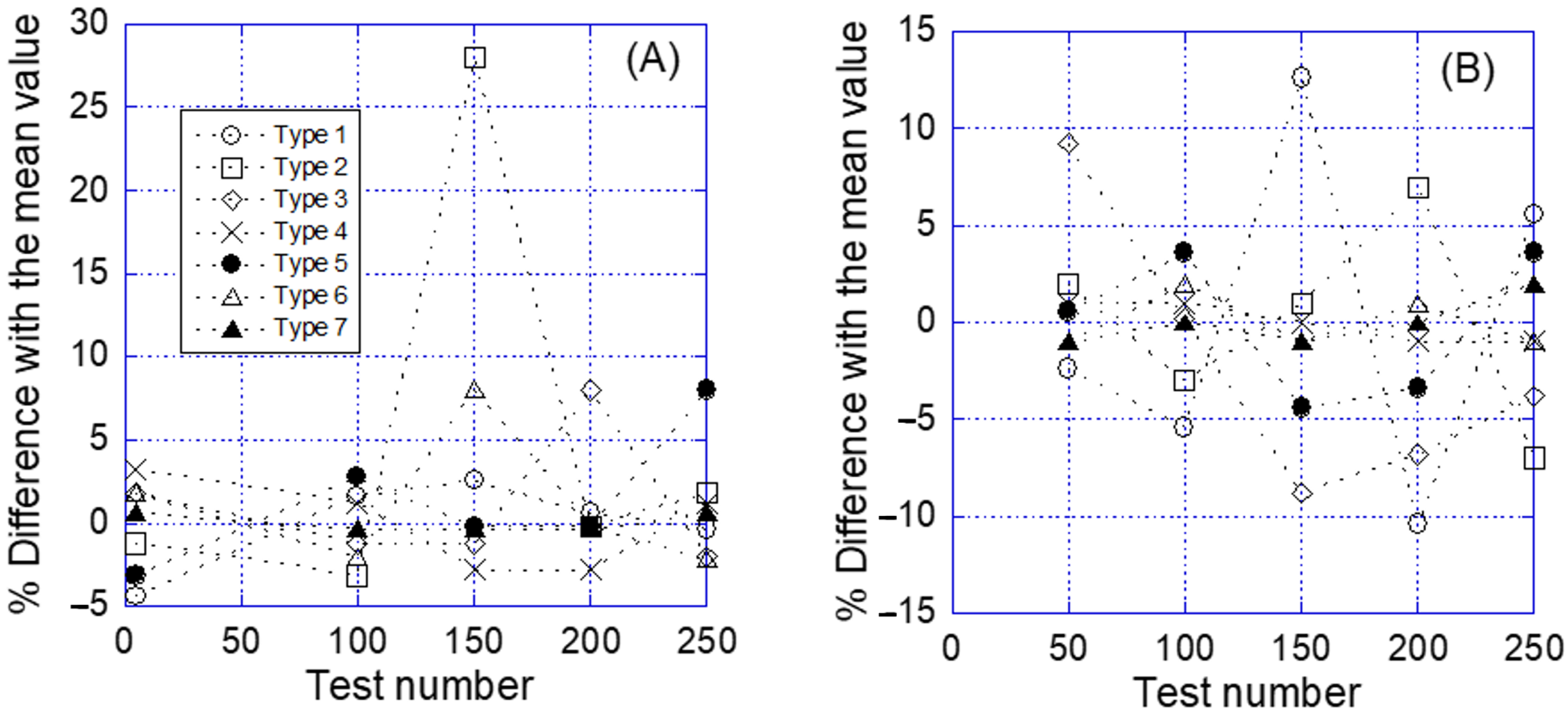
2.2. Resonance Frequency and Elastic Constant
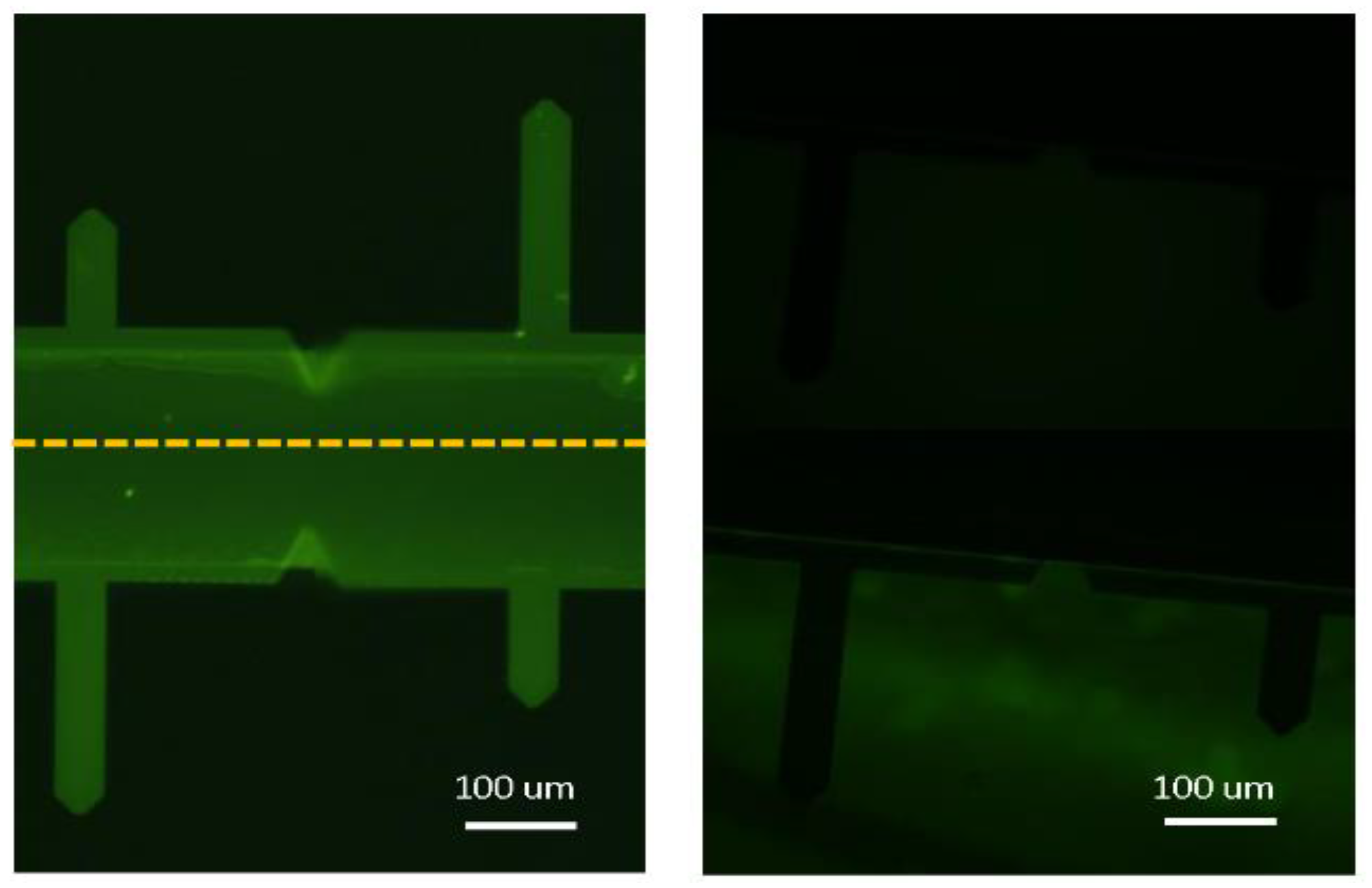
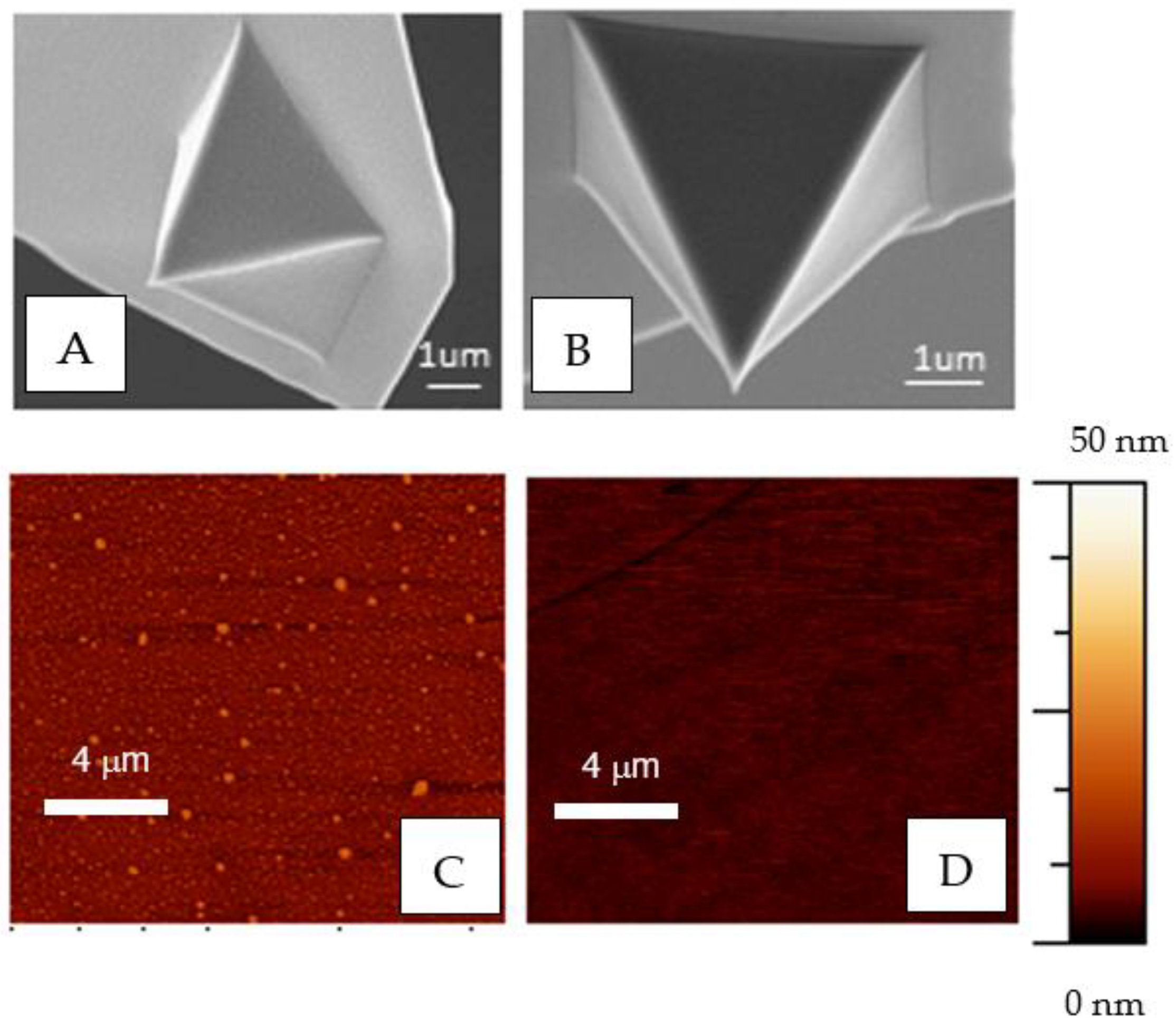
2.3. Surface Topography and Roughness
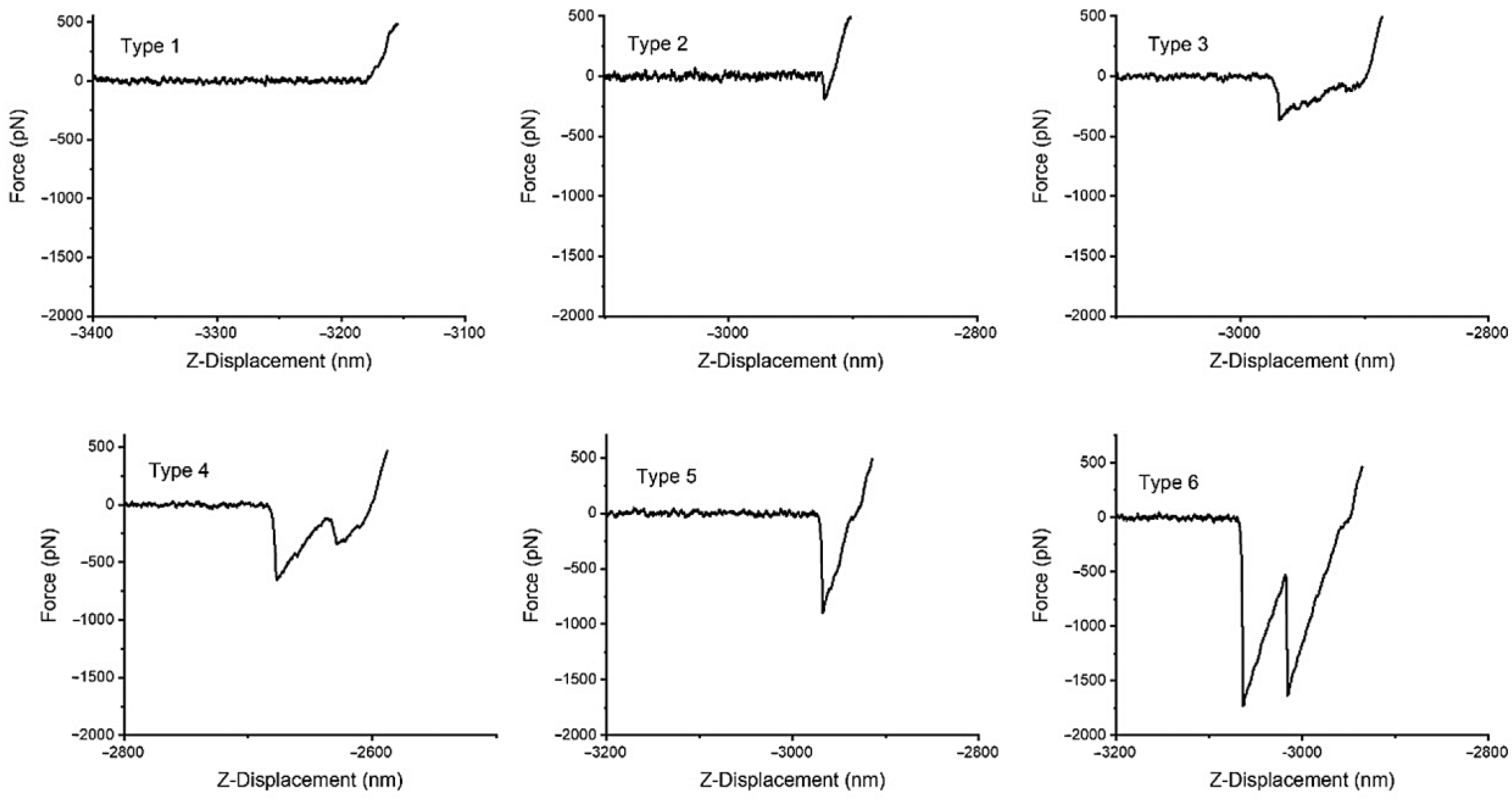
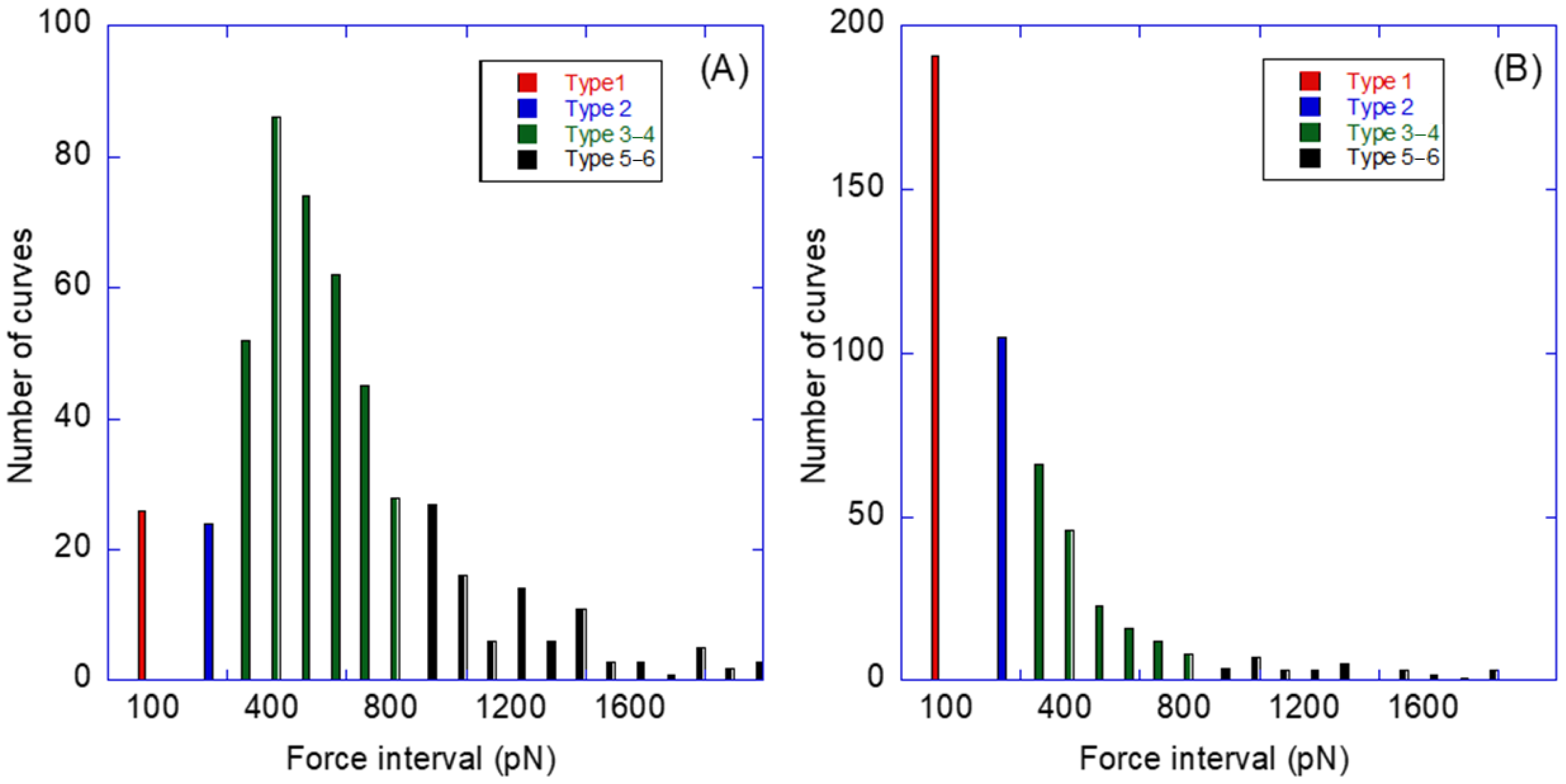
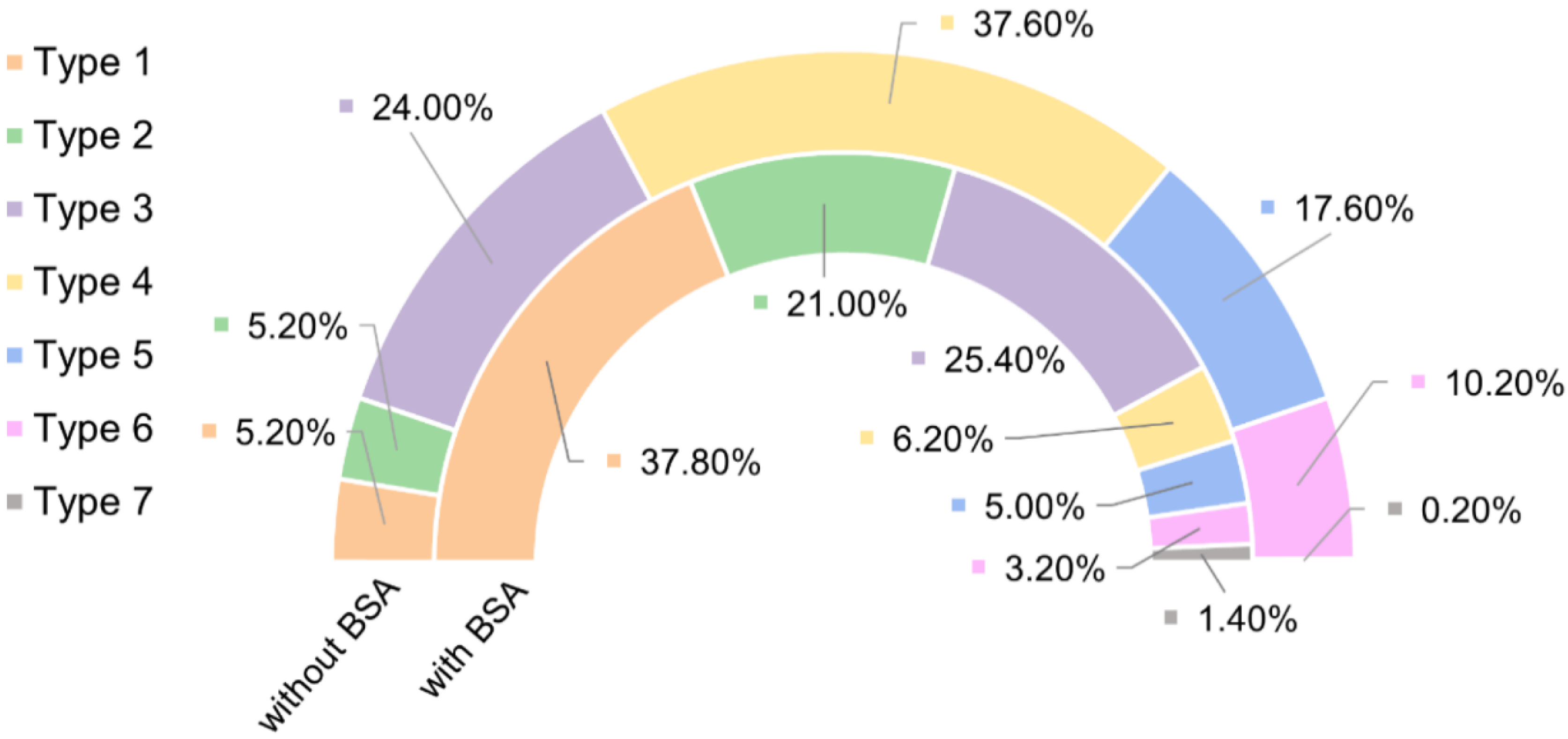
2.4. Biotin–Streptavidin Interaction Measured with the AFM
3. Materials and Methods
3.1. Sample Cleaning
3.2. Characterization of Amino Groups on the Surface of the Cantilever
3.3. Topography of the Surface
3.4. Characterization of the Resonance Frequency and Elastic Constant
3.5. Affinity Microscopy Measurements with a Biotin-Streptavidin Model System
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic Force Microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef] [PubMed]
- Hinterdorfer, P.; Baumgartner, W.; Gruber, H.J.; Schilcher, K.; Schindler, H. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. USA 1996, 93, 3477–3481. [Google Scholar] [CrossRef] [PubMed]
- Dumitru, A.C.; Conrard, L.; Giudice, C.L.; Henriet, P.; Veiga-Da-Cunha, M.; Derclaye, S.; Tyteca, D.; Alsteens, D. High-resolution mapping and recognition of lipid domains using AFM with toxin-derivatized probes. Chem. Commun. 2018, 54, 6903–6906. [Google Scholar] [CrossRef] [PubMed]
- Delguste, M.; Zeippen, C.; Machiels, B.; Mast, J.; Gillet, L.; Alsteens, D. Multivalent binding of herpesvirus to living cells is tightly regulated during infection. Sci. Adv. 2018, 4, eaat1273. [Google Scholar] [CrossRef]
- Yu, H.; Siewny, M.G.W.; Edwards, D.T.; Sanders, A.W.; Perkins, T.T. Hidden dynamics in the unfolding of individual bacteriorhodopsin proteins. Science 2017, 355, 945–950. [Google Scholar] [CrossRef]
- Pfreundschuh, M.; Alsteens, D.; Wieneke, R.; Zhang, C.; Coughlin, S.R.; Tampé, R.; Kobilka, B.K.; Müller, D.J. Identifying and quantifying two ligand-binding sites while imaging native human membrane receptors by AFM. Nat. Commun. 2015, 6, 8857. [Google Scholar] [CrossRef]
- Puntheeranurak, T.; Wildling, L.; Gruber, H.J.; Kinne, R.K.H.; Hinterdorfer, P. Ligands on the string: Single-molecule AFM studies on the interaction of antibodies and substrates with the Na+-glucose co-transporter SGLT1 in living cells. J. Cell Sci. 2006, 119, 2960–2967. [Google Scholar] [CrossRef]
- Dumitru, A.C.; Herruzo, E.T.; Rausell, E.; Ceña, V.; Garcia, R. Unbinding forces and energies between a siRNA molecule and a dendrimer measured by force spectroscopy. Nanoscale 2015, 7, 20267–20276. [Google Scholar] [CrossRef]
- Wang, D.; Stuart, J.D.; Jones, A.A.; Snow, C.D.; Kipper, M.J. Measuring interactions of DNA with nanoporous protein crystals by atomic force microscopy. Nanoscale 2021, 13, 10871–10881. [Google Scholar] [CrossRef]
- Bergkvist, M.; Cady, N.C. Chemical Functionalization and Bioconjugation Strategies for Atomic Force Microscope Cantilevers. In Bioconjugation Protocols: Strategies and Methods; Mark, S.S., Ed.; Humana Press: Totowa, NJ, USA, 2011; Volume 751, pp. 381–400. [Google Scholar] [CrossRef]
- Wildling, L.; Unterauer, B.; Zhu, R.; Rupprecht, A.; Haselgrübler, T.; Rankl, C.; Ebner, A.; Vater, D.; Pollheimer, P.; Pohl, E.E.; et al. Linking of Sensor Molecules with Amino Groups to Amino-Functionalized AFM Tips. Bioconjug. Chem. 2011, 22, 1239–1248. [Google Scholar] [CrossRef]
- Riener, C.K.; Stroh, C.M.; Ebner, A.; Klampfl, C.; Gall, A.A.; Romanin, C.; Lyubchenko, Y.L.; Hinterdorfer, P.; Gruber, H.J. Simple test system for single molecule recognition force microscopy. Anal. Chim. Acta 2003, 479, 59–75. [Google Scholar] [CrossRef]
- Kienberger, F.; Ebner, A.; Gruber, A.H.J.; Hinterdorfer, P. Molecular Recognition Imaging and Force Spectroscopy of Single Biomolecules. Acc. Chem. Res. 2006, 39, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Ditzler, L.R.; Sen, A.; Gannon, M.J.; Kohen, A.; Tivanski, A.V. Self-Assembled Enzymatic Monolayer Directly Bound to a Gold Surface: Activity and Molecular Recognition Force Spectroscopy Studies. J. Am. Chem. Soc. 2011, 133, 13284–13287. [Google Scholar] [CrossRef]
- Ebner, A.; Hinterdorfer, P.; Gruber, H.J. Comparison of different aminofunctionalization strategies for attachment of single antibodies to AFM cantilevers. Ultramicroscopy 2007, 107, 922–927. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Bash, R.; Yodh, J.G.; Hager, G.L.; Lohr, D.; Lindsay, S.M. Glutaraldehyde Modified Mica: A New Surface for Atomic Force Microscopy of Chromatin. Biophys. J. 2002, 83, 3619–3625. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Asakura, M.; Okahata, Y. Single-Molecule Force Spectroscopy for Studying Kinetics of Enzymatic Dextran Elongations. J. Am. Chem. Soc. 2011, 133, 5701–5703. [Google Scholar] [CrossRef]
- Barattin, R.; Voyer, N. Chemical modifications of AFM tips for the study of molecular recognition events. Chem. Commun. 2008, 1513–1532. [Google Scholar] [CrossRef]
- Dupres, V.; Menozzi, F.D.; Locht, C.; Clare, B.H.; Abbott, N.L.; Cuenot, S.; Bompard, C.; Raze, D.; Dufrêne, Y.F. Nanoscale mapping and functional analysis of individual adhesins on living bacteria. Nat. Methods 2005, 2, 515–520. [Google Scholar] [CrossRef]
- Schmitt, L.; Ludwig, M.; Gaub, H.E.; Tampé, R. A Metal-Chelating Microscopy Tip as a New Toolbox for Single-Molecule Experiments by Atomic Force Microscopy. Biophys. J. 2000, 78, 3275–3285. [Google Scholar] [CrossRef]
- Zhang, F.; Srinivasan, M.P. Self-Assembled Molecular Films of Aminosilanes and Their Immobilization Capacities. Langmuir 2004, 20, 2309–2314. [Google Scholar] [CrossRef]
- Casalini, S.; Dumitru, A.C.; Leonardi, F.; Bortolotti, C.A.; Herruzo, E.T.; Campana, A.; de Oliveira, R.F.; Cramer, T.; Garcia, R.; Biscarini, F. Multiscale Sensing of Antibody–Antigen Interactions by Organic Transistors and Single-Molecule Force Spectroscopy. ACS Nano 2015, 9, 5051–5062. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Huang, J.; Tan, J.; Gao, G.; Liu, S.; Wang, H.; Chen, Y. The study of single anticancer peptides interacting with HeLa cell membranes by single molecule force spectroscopy. Nanoscale 2012, 4, 1283–1286. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Ma, S.; Nie, L.; Shang, X.; Hao, X.; Tang, Z.; Wang, H. Size-dependent endocytosis of single gold nanoparticles. Chem. Commun. 2011, 47, 8091–8093. [Google Scholar] [CrossRef]
- Kao, F.-S.; Ger, W.; Pan, Y.-R.; Yu, H.-C.; Hsu, R.-Q.; Chen, H.-M. Chip-based protein-protein interaction studied by atomic force microscopy. Biotechnol. Bioeng. 2012, 109, 2460–2467. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, F.; Cai, J.-Y.; Yang, P.-H.; Liang, Z.-H. In-situ detection of resveratrol inhibition effect on epidermal growth factor receptor of living MCF-7 cells by Atomic Force Microscopy. Biosens. Bioelectron. 2014, 56, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Neundlinger, I.; Poturnayova, A.; Karpisova, I.; Rankl, C.; Hinterdorfer, P.; Snejdarkova, M.; Hianik, T.; Ebner, A. Characterization of Enhanced Monovalent and Bivalent Thrombin DNA Aptamer Binding Using Single Molecule Force Spectroscopy. Biophys. J. 2011, 101, 1781–1787. [Google Scholar] [CrossRef]
- Calvo, J.N.-M.; Elices, M.; Guinea, G.V.; Pérez-Rigueiro, J.; Arroyo-Hernández, M. Stability and activity of lactate dehydrogenase on biofunctional layers deposited by activated vapor silanization (AVS) and immersion silanization (IS). Appl. Surf. Sci. 2017, 416, 965–970. [Google Scholar] [CrossRef]
- Guo, S.; Ray, C.; Kirkpatrick, A.; Lad, N.; Akhremitchev, B.B. Effects of Multiple-Bond Ruptures on Kinetic Parameters Extracted from Force Spectroscopy Measurements: Revisiting Biotin-Streptavidin Interactions. Biophys. J. 2008, 95, 3964–3976. [Google Scholar] [CrossRef]
- Sedlak, S.M.; Schendel, L.C.; Gaub, H.E.; Bernardi, R.C. Streptavidin/biotin: Tethering geometry defines unbinding mechanics. Sci. Adv. 2020, 6, eaay5999. [Google Scholar] [CrossRef]
- Rico, F.; Moy, V.T. Energy landscape roughness of the streptavidin–biotin interaction. J. Mol. Recognit. 2007, 20, 495–501. [Google Scholar] [CrossRef]
- Rico, F.; Russek, A.; González, L.; Grubmüller, H.; Scheuring, S. Heterogeneous and rate-dependent streptavidin–biotin unbinding revealed by high-speed force spectroscopy and atomistic simulations. Proc. Natl. Acad. Sci. USA 2019, 116, 6594–6601. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, M.; Dettmann, W.; Gaub, H. Atomic force microscope imaging contrast based on molecular recognition. Biophys. J. 1997, 72, 445–448. [Google Scholar] [CrossRef] [PubMed]
- Moy, V.T.; Florin, E.-L.; Gaub, H.E. Intermolecular Forces and Energies Between Ligands and Receptors. Science 1994, 266, 257–259. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-K.; Wang, Y.-M.; Huang, L.-S.; Lin, S. Atomic force microscopy: Determination of unbinding force, off rate and energy barrier for protein–ligand interaction. Micron 2007, 38, 446–461. [Google Scholar] [CrossRef] [PubMed]
- Sedlak, S.; Bauer, M.; Kluger, C.; Schendel, L.C.; Milles, L.F.; Pippig, D.A.; Gaub, H.E. Monodisperse measurement of the biotin-streptavidin interaction strength in a well-defined pulling geometry. PLoS ONE 2017, 12, e0188722. [Google Scholar] [CrossRef]
- Grubmüller, H.; Heymann, B.; Tavan, P. Ligand Binding: Molecular Mechanics Calculation of the Streptavidin-Biotin Rupture Force. Science 1996, 271, 997–999. [Google Scholar] [CrossRef]
- Bioactive Surfaces S.L. Available online: www.bioactivesurfaces.com (accessed on 30 November 2022).
- Flater, E.E.; Zacharakis-Jutz, G.E.; Dumba, B.G.; White, I.A.; Clifford, C.A. Towards easy and reliable AFM tip shape determination using blind tip reconstruction. Ultramicroscopy 2014, 146, 130–143. [Google Scholar] [CrossRef]
- Wong, J.; Chilkoti, A.; Moy, V.T. Direct force measurements of the streptavidin–biotin interaction. Biomol. Eng. 1999, 16, 45–55. [Google Scholar] [CrossRef]
- Senapati, S.; Biswas, S.; Manna, S.; Ros, R.; Lindsay, S.; Zhang, P. A Y-Shaped Three-Arm Structure for Probing Bivalent Interactions between Protein Receptor–Ligand Using AFM and SPR. Langmuir 2018, 34, 6930–6940. [Google Scholar] [CrossRef]
- Yuan, C.; Chen, A.; Kolb, A.P.; Moy, V.T. Energy Landscape of Streptavidin−Biotin Complexes Measured by Atomic Force Microscopy. Biochemistry 2000, 39, 10219–10223. [Google Scholar] [CrossRef]
- Teulon, J.-M.; Delcuze, Y.; Odorico, M.; Chen, S.-w.W.; Parot, P.; Pellequer, J.-L. Single and multiple bonds in (strept)avidin-biotin interactions. J. Mol. Recognit. 2011, 24, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Piramowicz, M.D.; Czuba, P.; Targosz, M.; Burda, K.; Szymonski, M. Dynamic force measurements of avidin-biotin and strep-tavdin-biotin interactions using AFM. Acta Biochim. Pol. 2006, 53, 93–100. [Google Scholar] [CrossRef]
- Lecot, S.; Chevolot, Y.; Phaner-Goutorbe, M.; Yeromonahos, C. Impact of Silane Monolayers on the Adsorption of Streptavidin on Silica and Its Subsequent Interactions with Biotin: Molecular Dynamics and Steered Molecular Dynamics Simulations. J. Phys. Chem. B 2020, 124, 6786–6796. [Google Scholar] [CrossRef] [PubMed]
- Puntheeranurak, T.; Neundlinger, I.; Kinne, R.K.H.; Hinterdorfer, P. Single-molecule recognition force spectroscopy of transmembrane transporters on living cells. Nat. Protoc. 2011, 6, 1443–1452. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Liu, Z.; Liu, H.; Nash, M.A. Next Generation Methods for Single-Molecule Force Spectroscopy on Polyproteins and Receptor-Ligand Complexes. Front. Mol. Biosci. 2020, 7, 85. [Google Scholar] [CrossRef]
- Horcas, I.; Fernández, R.; Gómez-Rodriguez, J.M.; Colchero, J.; Gomez-Herrero, J.; Baro, A.M. WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007, 78, 13705. [Google Scholar] [CrossRef] [PubMed]
- Sader, J.E.; Chon, J.; Mulvaney, P. Calibration of rectangular atomic force microscope cantilevers. Rev. Sci. Instrum. 1999, 70, 3967–3969. [Google Scholar] [CrossRef]
- Arroyo-Hernández, M.; Daza, R.; Pérez-Rigueiro, J.; Elices, M.; Nieto-Márquez, J.; Guinea, G.V. Optimization of functionalization conditions for protein analysis by AFM. Appl. Surf. Sci. 2014, 317, 462–468. [Google Scholar] [CrossRef]
- Zimmermann, J.L.; Nicolaus, T.; Neuert, G.; Blank, K.G. Thiol-based, site-specific and covalent immobilization of biomolecules for single-molecule experiments. Nat. Protoc. 2010, 5, 975–985. [Google Scholar] [CrossRef]
| Dimensions | Resonance Frequency (Hz) | Elastic Constant (N/m) |
|---|---|---|
| 100 × 40 μm (DeepTip) | 45,200 ± 400 | 0.18 ± 0.01 |
| 200 × 40 μm (DeepTip) | 12,550 ± 50 | 0.026 ± 0.001 |
| 100 × 40 μm (Control) | 46,700 ± 100 | 0.22 ± 0.01 |
| 200 × 40 μm (Control) | 12,740 ± 30 | 0.032 ± 0.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corregidor, D.; Tabraue, R.; Colchero, L.; Daza, R.; Elices, M.; Guinea, G.V.; Pérez-Rigueiro, J. High-Yield Characterization of Single Molecule Interactions with DeepTipTM Atomic Force Microscopy Probes. Molecules 2023, 28, 226. https://doi.org/10.3390/molecules28010226
Corregidor D, Tabraue R, Colchero L, Daza R, Elices M, Guinea GV, Pérez-Rigueiro J. High-Yield Characterization of Single Molecule Interactions with DeepTipTM Atomic Force Microscopy Probes. Molecules. 2023; 28(1):226. https://doi.org/10.3390/molecules28010226
Chicago/Turabian StyleCorregidor, Daniel, Raquel Tabraue, Luis Colchero, Rafael Daza, Manuel Elices, Gustavo V. Guinea, and José Pérez-Rigueiro. 2023. "High-Yield Characterization of Single Molecule Interactions with DeepTipTM Atomic Force Microscopy Probes" Molecules 28, no. 1: 226. https://doi.org/10.3390/molecules28010226
APA StyleCorregidor, D., Tabraue, R., Colchero, L., Daza, R., Elices, M., Guinea, G. V., & Pérez-Rigueiro, J. (2023). High-Yield Characterization of Single Molecule Interactions with DeepTipTM Atomic Force Microscopy Probes. Molecules, 28(1), 226. https://doi.org/10.3390/molecules28010226





