Bovine Serum Albumin-Based Nanoparticles: Preparation, Characterization, and Antioxidant Activity Enhancement of Three Main Curcuminoids from Curcuma longa
Abstract
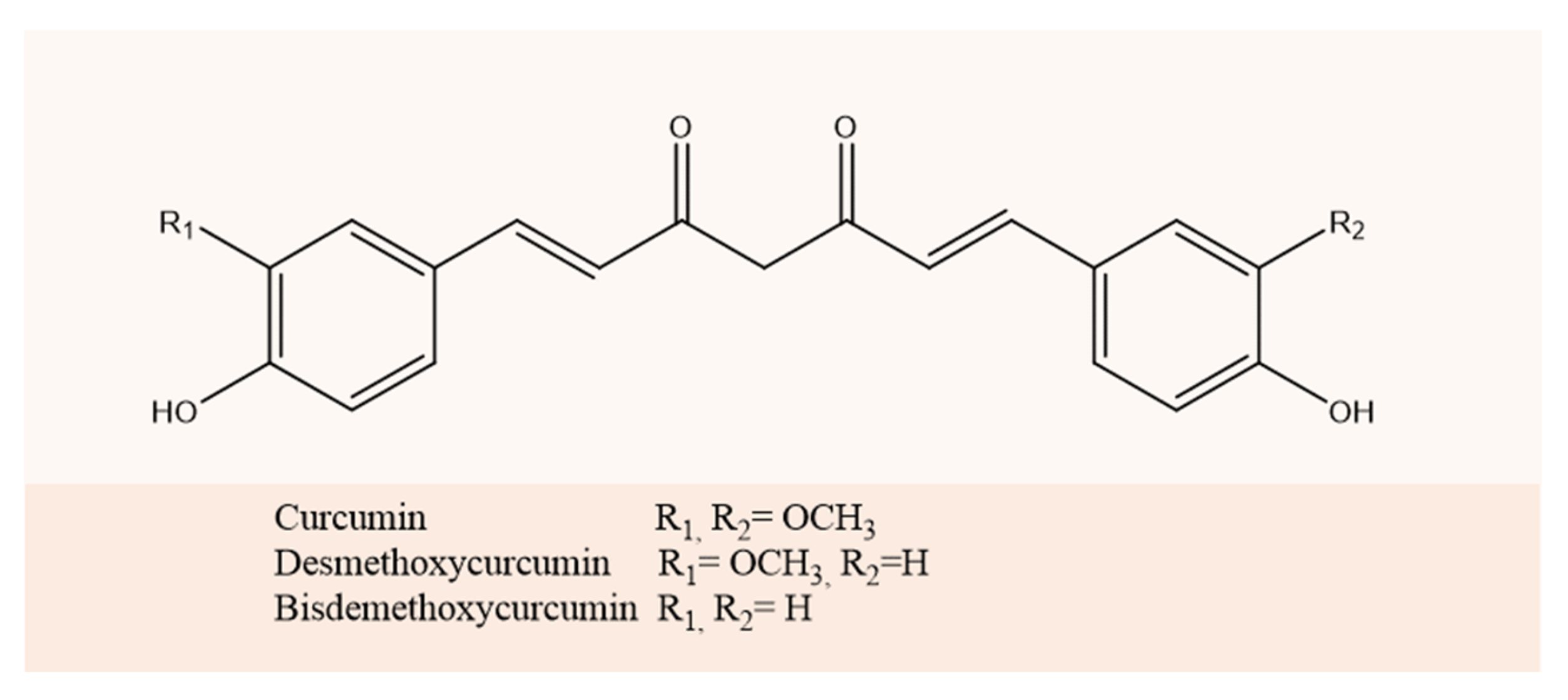
:1. Introduction
2. Results
2.1. Characterization of CUR, DMC, and BDM Nanoparticles
2.1.1. Encapsulation
2.1.2. Particle Size, Particle Size Distribution, Morphology, and Encapsulation Efficiency
2.1.3. Differential Scanning Calorimetry (DSC)
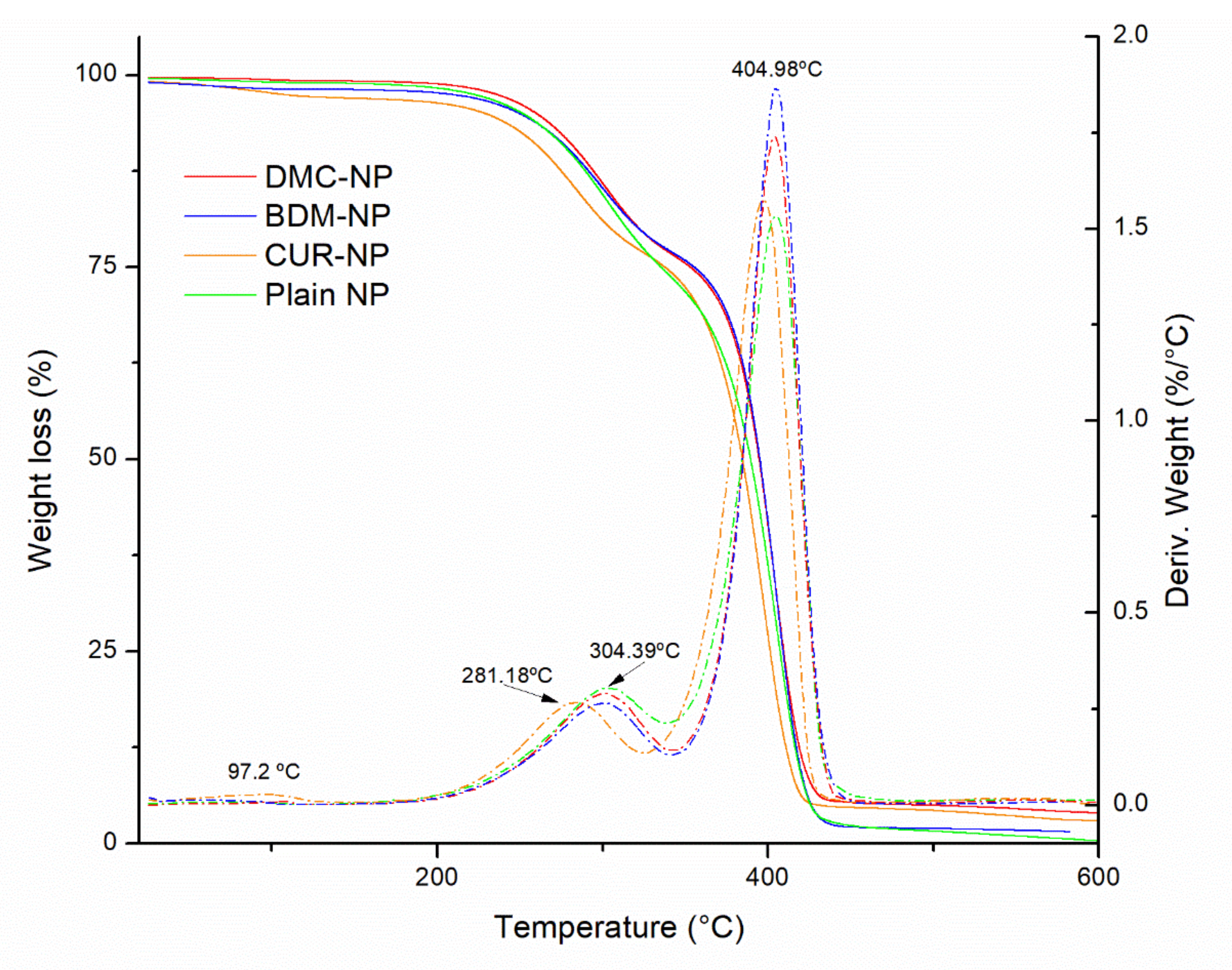
2.1.4. Thermogravimetric Analysis (TGA)
2.2. In Vitro Evaluation of CUR, DMC, and BDM Hybrid Nanoparticles
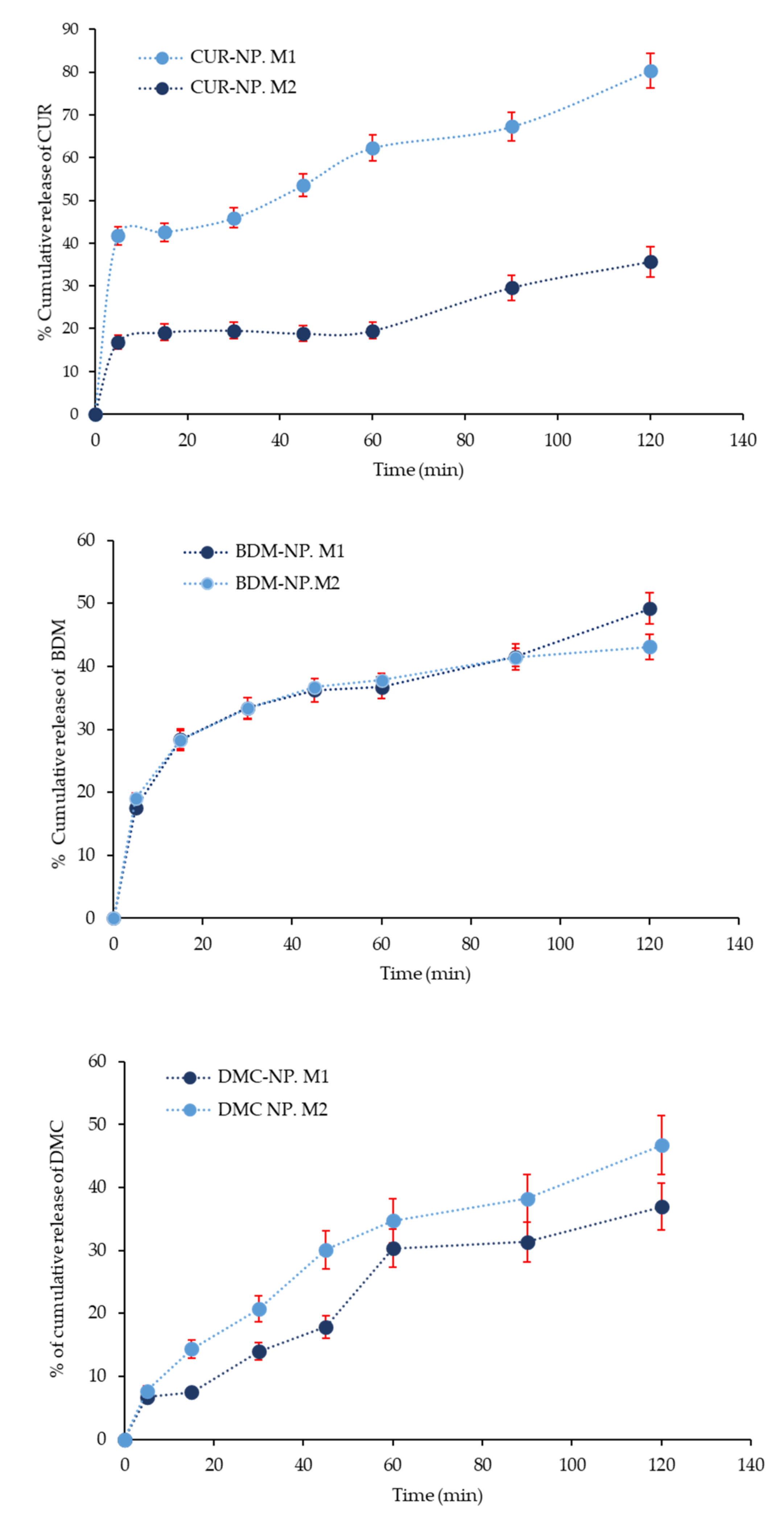
2.2.1. In Vitro Curcuminoids Release Studies
2.2.2. Kinetic Parameters of Drug Release
2.2.3. Antioxidant Activity Evaluation of Free and Curcuminoids Hybrid Nanoparticles
3. Materials and Methods
3.1. Preparation of CUR, DMC, and BDM Hybrid Nanoparticles
3.2. Physicochemical Characterization of CUR, DMC, and BDM Hybrid Nanoparticles
3.2.1. Fourier Transform Infrared Spectroscopy (FT-IR)
3.2.2. High Resolution Transmission Electron Microscopy (HR-TEM)
3.2.3. Dynamic Light Scattering (DLS)
3.2.4. Encapsulation Efficiency (EE)
3.2.5. Thermal Analyses
3.3. In Vitro studies of Curcuminoids Hybrid NP
3.3.1. Release profile of CUR, DMC, and BDM from Hybrid NP
3.3.2. Release Kinetic Models
- (i)
- The zero-order drug delivery model is expressed by the following equation [49].
- (ii)
- The first-order drug delivery model is expressed by the following equation [55]:where [A]t represents the % of drug release, k is the first-order release model constant, and [A]0 is the intersection with the axis.
- (iii)
- The Second-order drug delivery model is expressed by the following equation.
- (iv)
- The Ritger–Peppas model is given by the following [56]:where k is a release rate depending on the structural and geometrical characteristics of the release system, and n is the diffusional exponent defining the release mechanism.
- (v)
- The Higuchi equation is expressed by the following equation [57].
- (vi)
- The Weibull model equation is given by the following [58]:where a is the scale parameter of the equation and determines the process time scale. t is the location parameter, which shows the lag time before the start of the release process (often zero). b is the shape parameter, which has three cases:Case 1: b = 1, an exponential curve;Case 2: b > 1, the release curve is s shaped or sigmoid with an upward curvature followed by a turning point;Case 3: b < 1, a parabolic curve with a high slope at initial step and then a consistent exponential decay curve.
3.3.3. DPPH Radical-Scavenging Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Jiang, Y.; Lu, H.; Dag, A.; Hart-Smith, G.; Stenzel, M.H. Albumin–polymer conjugate nanoparticles and their interactions with prostate cancer cells in 2D and 3D culture: Comparison between PMMA and PCL. J. Mater. Chem. B 2016, 4, 2017–2027. [Google Scholar] [CrossRef] [PubMed]
- Deljoo, S.; Rabiee, N.; Rabiee, M. Curcumin-hybrid Nanoparticles in Drug Delivery System. Asian J. Nanosci. Mater. 2018, 2, 66–91. [Google Scholar] [CrossRef]
- Pan, K.; Chen, H.; Baek, S.J.; Zhong, Q. Self-assembled curcumin-soluble soybean polysaccharide nanoparticles: Physicochemical properties and in vitro anti-proliferation activity against cancer cells. Food Chem. 2018, 246, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, L.N.M.; Alcantara, A.C.S.; Franz-Montan, M.; Couto, V.M.; Nista, S.V.G.; de Paula, E. Nanostructured organic-organic bio-hybrid delivery systems. Biomed. Appl. Nanopart. 2019, 341–374. [Google Scholar] [CrossRef]
- Hong, S.; Choi, D.W.; Kim, H.N.; Park, C.G.; Lee, W.; Park, H.H. Protein-Based Nanoparticles as Drug Delivery Systems. Pharmaceutics 2020, 12, 604. [Google Scholar] [CrossRef]
- Elblbesy, M.A. Hemocompatibility of Albumin Nanoparticles as a Drug Delivery System—An In Vitro Study. J. Biomater. Nanobiotechnol. 2016, 7, 64–71. [Google Scholar] [CrossRef] [Green Version]
- Sadeghi, R.; Moosavi-Movahedi, A.A.; Emam-jomeh, Z.; Kalbasi, A.; Razavi, S.H.; Karimi, M.; Kokini, J. The effect of different desolvating agents on BSA nanoparticle properties and encapsulation of curcumin. J. Nanopart. Res. 2014, 16, 2565. [Google Scholar] [CrossRef]
- Gupta, R.; Jain, V.; Nagar, J.C.; Ansari, A.; Sharma, K.; Sarkar, A.; Khan, M.S. Bioavailability Enhancement Techniques for Poorly Soluble Drugs: A Review. Asian J. Pharm. Res. Dev. 2020, 8, 75–78. [Google Scholar] [CrossRef]
- Abdelaziz, H.M.; Freag, M.S.; Elzoghby, A.O. Solid Lipid Nanoparticle-Based Drug Delivery for Lung Cancer. In Nanotechnology-Based Targeted Drug Delivery Systems for Lung Cancer; Elsevier: Amsterdam, The Netherlands, 2019; pp. 95–121. [Google Scholar]
- Bourassa, P.; Kanakis, C.D.; Tarantilis, P.; Pollissiou, M.G.; Tajmir-Riahi, H.A. Resveratrol, Genistein, and Curcumin Bind Bovine Serum Albumin. J. Phys. Chem. B 2010, 114, 3348–3354. [Google Scholar] [CrossRef]
- Clark, A.H.; Judge, F.J.; Richards, J.B.; Stubbs, J.M.; Suggett, A. Electron microscopy of network structures in thermally-induced globular protein gels. Int. J. Pept. Protein Res. 2009, 17, 380–392. [Google Scholar] [CrossRef]
- Ko, S.; Gunasekaran, S. Preparation of sub-100-nm β-lactoglobulin (BLG) nanoparticles. J. Microencapsul. 2006, 23, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Wang, S.; Zhao, B.; Han, C.; Wang, M.; Zhou, W. Effect of PLGA as a polymeric emulsifier on preparation of hydrophilic protein-loaded solid lipid nanoparticles. Colloids Surf. B Biointerfaces 2008, 67, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Puglia, C.; Santonocito, D.; Bonaccorso, A.; Musumeci, T.; Ruozi, B.; Pignatello, R.; Carbone, C.; Parenti, C.; Chiechio, S. Lipid Nanoparticle Inclusion Prevents Capsaicin-Induced TRPV1 Defunctionalization. Pharmaceutics 2020, 12, 339. [Google Scholar] [CrossRef] [PubMed]
- García-Pinel, B.; Porras-Alcalá, C.; Ortega-Rodríguez, A.; Sarabia, F.; Prados, J.; Melguizo, C.; López-Romero, J.M. Lipid-Based Nanoparticles: Application and Recent Advances in Cancer Treatment. Nanomaterials 2019, 9, 638. [Google Scholar] [CrossRef] [Green Version]
- Kotha, R.R.; Luthria, D.L. Curcumin: Biological, Pharmaceutical, Nutraceutical, and Analytical Aspects. Molecules 2019, 24, 2930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Archivio, A.A.; Maggi, M.A. Investigation by response surface methodology of the combined effect of pH and composition of water-methanol mixtures on the stability of curcuminoids. Food Chem. 2017, 219, 414–418. [Google Scholar] [CrossRef]
- Lateh, L.; Kaewnopparat, N.; Yuenyongsawad, S.; Panichayupakaranant, P. Enhancing the water-solubility of curcuminoids-rich extract using a ternary inclusion complex system: Preparation, characterization, and anti-cancer activity. Food Chem. 2022, 368, 130827. [Google Scholar] [CrossRef]
- Kocher, A.; Schiborr, C.; Behnam, D.; Frank, J. The oral bioavailability of curcuminoids in healthy humans is markedly enhanced by micellar solubilisation but not further improved by simultaneous ingestion of sesamin, ferulic acid, naringenin and xanthohumol. J. Funct. Foods 2015, 14, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Nayak, A.P.; Tiyaboonchai, W.; Patankar, S.; Madhusudhan, B.; Souto, E.B. Curcuminoids-loaded lipid nanoparticles: Novel approach towards malaria treatment. Colloids Surf. B Biointerfaces 2010, 81, 263–273. [Google Scholar] [CrossRef]
- Duvoix, A.; Blasius, R.; Delhalle, S.; Schnekenburger, M.; Morceau, F.; Henry, E.; Dicato, M.; Diederich, M. Chemopreventive and therapeutic effects of curcumin. Cancer Lett. 2005, 223, 181–190. [Google Scholar] [CrossRef]
- Suresh, K.; Nangia, A. Curcumin: Pharmaceutical solids as a platform to improve solubility and bioavailability. CrystEngComm 2018, 20, 3277–3296. [Google Scholar] [CrossRef]
- Sadeghi-Ghadi, Z.; Behjou, N.; Ebrahimnejad, P.; Mahkam, M.; Goli, H.R.; Lam, M.; Nokhodchi, A. Improving Antibacterial Efficiency of Curcumin in Magnetic Polymeric Nanocomposites. J. Pharm. Innov. 2022, 1–16. [Google Scholar] [CrossRef]
- Fu, J.; Sun, C.; Tan, Z.; Zhang, G.; Chen, G.; Song, L. Nanocomplexes of curcumin and glycated bovine serum albumin: The formation mechanism and effect of glycation on their physicochemical properties. Food Chem. 2022, 368, 130651. [Google Scholar] [CrossRef]
- Gupta, U.; Singh, V.; Kumar, V.; Khajuria, Y. Spectroscopic Studies of Cholesterol: Fourier Transform Infra-Red and Vibrational Frequency Analysis. Mater. Focus 2014, 3, 211–217. [Google Scholar] [CrossRef]
- Zweers, M.L.T.; Grijpma, D.W.; Engbers, G.H.M.; Feijen, J. The preparation of monodisperse biodegradable polyester nanoparticles with a controlled size. J. Biomed. Mater. Res. 2003, 66B, 559–566. [Google Scholar] [CrossRef]
- Jinno, J.I.; Kamada, N.; Miyake, M.; Yamada, K.; Mukai, T.; Odomi, M.; Toguchi, H.; Liversidge, G.G.; Higaki, K.; Kimura, T. Effect of particle size reduction on dissolution and oral absorption of a poorly water-soluble drug, cilostazol, in beagle dogs. J. Control. Release 2006, 111, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.P.; Vaz, A.F.M.; Correia, M.T.S.; Cerqueira, M.A.; Vicente, A.A.; Carneiro-da-Cunha, M.G. Quercetin-Loaded Lecithin/Chitosan Nanoparticles for Functional Food Applications. Food Bioprocess Technol. 2014, 7, 1149–1159. [Google Scholar] [CrossRef] [Green Version]
- Valencia, M.S.; da Silva Júnior, M.F.; Xavier-Júnior, F.H.; de Oliveira Veras, B.; de Albuquerque, P.B.; de Oliveira Borba, E.F.; da Silva, T.G.; Xavier, V.L.; de Souza, M.P.; das Graças Carneiro-da-Cunha, M. Characterization of curcumin-loaded lecithin-chitosan bioactive nanoparticles. Carbohydr. Polym. Technol. Appl. 2021, 2, 100119. [Google Scholar] [CrossRef]
- Yen, F.-L.; Wu, T.-H.; Lin, L.-T.; Cham, T.-M.; Lin, C.-C. Nanoparticles formulation of Cuscuta chinensis prevents acetaminophen-induced hepatotoxicity in rats. Food Chem. Toxicol. 2008, 46, 1771–1777. [Google Scholar] [CrossRef]
- Hajj Ali, H.; Michaux, F.; Bouelet Ntsama, I.S.; Durand, P.; Jasniewski, J.; Linder, M. Shea butter solid nanoparticles for curcumin encapsulation: Influence of nanoparticles size on drug loading. Eur. J. Lipid Sci. Technol. 2016, 118, 1168–1178. [Google Scholar] [CrossRef]
- Chang, H.-I.; Su, Y.-H.; Lin, Y.-J.; Chen, P.-J.; Shi, C.-S.; Chen, C.-N.; Yeh, C.-C. Evaluation of the protective effects of curcuminoid (curcumin and bisdemethoxycurcumin)-loaded liposomes against bone turnover in a cell-based model of osteoarthritis. Drug Des. Devel. Ther. 2015, 9, 2285. [Google Scholar] [CrossRef] [Green Version]
- Zamarioli, C.M.; Martins, R.M.; Carvalho, E.C.; Freitas, L.A.P. Nanoparticles containing curcuminoids (Curcuma longa): Development of topical delivery formulation. Brazilian J. Pharmacogn. 2015, 25, 53–60. [Google Scholar] [CrossRef] [Green Version]
- Mazzarino, L.; Travelet, C.; Ortega-Murillo, S.; Otsuka, I.; Pignot-Paintrand, I.; Lemos-Senna, E.; Borsali, R. Elaboration of chitosan-coated nanoparticles loaded with curcumin for mucoadhesive applications. J. Colloid Interface Sci. 2012, 370, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Salehiabar, M.; Nosrati, H.; Javani, E.; Aliakbarzadeh, F.; Kheiri Manjili, H.; Davaran, S.; Danafar, H. Production of biological nanoparticles from bovine serum albumin as controlled release carrier for curcumin delivery. Int. J. Biol. Macromol. 2018, 115, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Amano, C.; Minematsu, H.; Fujita, K.; Iwashita, S.; Adachi, M.; Igarashi, K.; Hinuma, S. Nanoparticles Containing Curcumin Useful for Suppressing Macrophages In Vivo in Mice. PLoS ONE 2015, 10, e0137207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Péret-Almeida, L.; Cherubino, A.P.F.; Alves, R.J.; Dufossé, L.; Glória, M.B.A. Separation and determination of the physico-chemical characteristics of curcumin, demethoxycurcumin and bisdemethoxycurcumin. Food Res. Int. 2005, 38, 1039–1044. [Google Scholar] [CrossRef]
- Garti, N.; Karpuj, L.; Sarig, S. Correlation between crystal habit and the composition of solvated and nonsolvated cholesterol crystals. J. Lipid Res. 1981, 22, 785–791. [Google Scholar] [CrossRef]
- Kumbhar, D.D.; Pokharkar, V.B. Physicochemical investigations on an engineered lipid–polymer hybrid nanoparticle containing a model hydrophilic active, zidovudine. Colloids Surf. A Physicochem. Eng. Asp. 2013, 436, 714–725. [Google Scholar] [CrossRef]
- Das, S.; Chaudhury, A. Recent Advances in Lipid Nanoparticle Formulations with Solid Matrix for Oral Drug Delivery. AAPS PharmSciTech 2011, 12, 62–76. [Google Scholar] [CrossRef] [Green Version]
- Heffernan, C.; Ukrainczyk, M.; Gamidi, R.K.; Hodnett, B.K.; Rasmuson, Å.C. Extraction and Purification of Curcuminoids from Crude Curcumin by a Combination of Crystallization and Chromatography. Org. Process Res. Dev. 2017, 21, 821–826. [Google Scholar] [CrossRef] [Green Version]
- Manuel Bravo-Arredondo, J.; Moreno, A.; Mendoza, M.E. Crystal growth of cholesterol in hydrogels and its characterization. J. Cryst. Growth 2014, 401, 242–247. [Google Scholar] [CrossRef]
- Xing, R.; Guo, J.; Miao, C.; Liu, S.; Pan, H. Fabrication of protein-coated CdS nanocrystals via microwave-assisted hydrothermal method. J. Exp. Nanosci. 2014, 9, 582–587. [Google Scholar] [CrossRef]
- Gao, J.; Ming, J.; He, B.; Fan, Y.; Gu, Z.; Zhang, X. Preparation and characterization of novel polymeric micelles for 9-nitro-20(S)-camptothecin delivery. Eur. J. Pharm. Sci. 2008, 34, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.Y.; Tam, K.C.; Gan, L.H. Release kinetics of hydrophobic and hydrophilic model drugs from pluronic F127/poly(lactic acid) nanoparticles. J. Control. Release 2005, 103, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Vandenhaute, M.; Snoeck, D.; Vanderleyden, E.; De Belie, N.; Van Vlierberghe, S.; Dubruel, P. Stability of Pluronic® F127 bismethacrylate hydrogels: Reality or utopia? Polym. Degrad. Stab. 2017, 146, 201–211. [Google Scholar] [CrossRef]
- Craig, D.Q. The mechanisms of drug release from solid dispersions in water-soluble polymers. Int. J. Pharm. 2002, 231, 131–144. [Google Scholar] [CrossRef]
- Kour, P.; Afzal, S.; Gani, A.; Zargar, M.I.; Nabi Tak, U.; Rashid, S.; Dar, A.A. Effect of nanoemulsion-loaded hybrid biopolymeric hydrogel beads on the release kinetics, antioxidant potential and antibacterial activity of encapsulated curcumin. Food Chem. 2022, 376, 131925. [Google Scholar] [CrossRef]
- Malekjani, N.; Jafari, S.M. Release modeling of nanoencapsulated food ingredients by empirical and semiempirical models. In Release and Bioavailability of Nanoencapsulated Food Ingredients; Elsevier: Amsterdam, The Netherlands, 2020; pp. 211–246. [Google Scholar]
- Romero, K.W.; Quirós, M.I.; Huertas, F.V.; Vega-Baudrit, J.R.; Navarro-Hoyos, M.; Araya-Sibaja, A.M. Design of Hybrid Polymeric-Lipid Nanoparticles Using Curcumin as a Model: Preparation, Characterization, and In Vitro Evaluation of Demethoxycurcumin and Bisdemethoxycurcumin-Loaded Nanoparticles. Polymer 2021, 13, 4207. [Google Scholar] [CrossRef]
- Jayaprakasha, G.K.; Jaganmohan Rao, L.; Sakariah, K.K. Antioxidant activities of curcumin, demethoxycurcumin and bisdemethoxycurcumin. Food Chem. 2006, 98, 720–724. [Google Scholar] [CrossRef]
- Guo, X.; Li, W.; Wang, H.; Fan, Y.-Y.; Wang, H.; Gao, X.; Niu, B.; Gong, X. Preparation, characterization, release and antioxidant activity of curcumin-loaded amorphous calcium phosphate nanoparticles. J. Non. Cryst. Solids 2018, 500, 317–325. [Google Scholar] [CrossRef]
- Huang, X.; Huang, X.; Gong, Y.; Xiao, H.; McClements, D.J.; Hu, K. Enhancement of curcumin water dispersibility and antioxidant activity using core-shell protein-polysaccharide nanoparticles. Food Res. Int. 2016, 87, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos, P.D.F.; Francisco, C.R.L.; Coqueiro, A.; Leimann, F.V.; Pinela, J.; Calhelha, R.C.; Porto Ineu, R.; Ferreira, I.C.F.R.; Bona, E.; Gonçalves, O.H. The nanoencapsulation of curcuminoids extracted from: Curcuma longa L. and an evaluation of their cytotoxic, enzymatic, antioxidant and anti-inflammatory activities. Food Funct. 2019, 10, 573–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afzal, S.; Lone, M.S.; Maswal, M.; Dar, A.A. Modulation of surface tension and rheological behavior of methyl cellulose–Amino acid based surfactant mixture by hydrophobic drug rifampicin: An insight into drug stabilization and pH-responsive release. J. Mol. Liq. 2020, 319, 114353. [Google Scholar] [CrossRef]
- Ritger, P.L.; Peppas, N.A. A simple equation for description of solute release I. Fickian and non-fickian release from non-swellable devices in the form of slabs, spheres, cylinders or discs. J. Control. Release 1987, 5, 23–36. [Google Scholar] [CrossRef]
- Higuchi, T. Rate of Release of Medicaments from Ointment Bases Containing Drugs in Suspension. J. Pharm. Sci. 1961, 50, 874–875. [Google Scholar] [CrossRef] [PubMed]
- Weibull, W. A Statistical Distribution Function of Wide Applicability. J. Appl. Mech. 1951, 18, 293–297. [Google Scholar] [CrossRef]
- Navarro-Hoyos, M.; Arnáez-Serrano, E.; Quesada-Mora, S.; Azofeifa-Cordero, G.; Wilhelm-Romero, K.; Quirós-Fallas, M.I.; Alvarado-Corella, D.; Vargas-Huertas, F.; Sánchez-Kopper, A. Polyphenolic QTOF-ESI MS Characterization and the Antioxidant and Cytotoxic Activities of Prunus domestica Commercial Cultivars from Costa Rica. Molecules 2021, 26, 6493. [Google Scholar] [CrossRef]




| Formulation | Size Average (nm) 1 | PDI 1 | EE (%) 1 | LC (mg/g) | Yield (%) |
|---|---|---|---|---|---|
| CUR-NP | 15.83 ± 0.18 | 0.250 ± 0.02 | 98.76 ± 0.68 a | 19.03 ± 0.45 | 93.35 ± 1.57 |
| DMC-NP | 17.29 ± 3.34 | 0.393 ± 0.05 | 98.99 ± 0.98 a | 28.34 ± 0.97 | 62.85 ± 1.52 |
| BDM-NP | 15.14 ± 0.14 | 0.156 ± 0.004 | 96.85 ± 0.26 b | 20.35 ± 0.94 | 85.68 ± 2.78 |
| System | Zero Order | First Order | Second Order | Ritger-Peppas | Higuchi | Weibull | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| k | [A]0 | R2 | k | ln[A]0 | R2 | k | 1/[A]0 | R2 | k | n | R2 | k | R2 | b | a | R2 | |
| CUR-NP M1 | 20.617 | 38.022 | 0.9862 | 0.356 | 3.689 | 0.9825 | 0.112 | 3.059 | 0.9778 | 32.756 | 0.290 | 0.9270 | 52.902 | 0.9595 | 0.176 | 1.109 | 0.9350 |
| CUR-NP M2 | 9.569 | 14.709 | 0.9092 | 0.382 | 2.771 | 0.9274 | 0.001 | −0.048 | 0.9133 | 28.147 | 0.245 | 0.8462 | 15.549 | 0.7924 | 0.072 | 3.830 | 0.6109 |
| BDM-NP M1 | 10.781 | 24.886 | 0.7935 | 0.416 | 3.143 | 0.7541 | −0.014 | 0.043 | 0.6129 | 18.293 | 0.560 | 0.9853 | 88.423 | 0.9542 | 0.148 | 2.059 | 0.9629 |
| BDM-NP M2 | 13.585 | 22.909 | 0.8763 | 0.342 | 3.205 | 0.6946 | −0.011 | 0.041 | 0.5924 | 17.378 | 0.613 | 0.9966 | 20.810 | 0.9477 | 0.138 | 2.045 | 0.9824 |
| DMC-NP M1 | 2.750 | 21.266 | 0.9573 | 0.112 | 3.059 | 0.9778 | 0.005 | −0.047 | 0.9610 | 4.4428 | 0.985 | 0.9135 | 4.1546 | 0.9320 | 0.027 | 3.634 | 0.8259 |
| DMC-NP M2 | 3.192 | 20.799 | 0.9373 | 0.139 | 3.037 | 0.9262 | 0.006 | −0.048 | 0.9080 | 4.3549 | 1.080 | 0.9032 | 4.9457 | 0.9700 | 0.034 | 3.621 | 0.9391 |
| IC50 (µg/mL) 1,2,3 | |||
|---|---|---|---|
| EtOH 4 | Water 4 | BSA | |
| CUR | 9.60 a,* ± 0.12 | 2444.80 a,# ± 9.68 | 9.28 a,* ± 0.29 |
| DMC | 12.46 b,* ± 0.02 | 2143.07 b,# ± 0.61 | 11.70 b,* ± 0.13 |
| BDM | 17.94 c,* ± 0.06 | 1398.68 c,# ± 5.07 | 15.19 c,* ± 0.57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araya-Sibaja, A.M.; Wilhelm-Romero, K.; Quirós-Fallas, M.I.; Vargas Huertas, L.F.; Vega-Baudrit, J.R.; Navarro-Hoyos, M. Bovine Serum Albumin-Based Nanoparticles: Preparation, Characterization, and Antioxidant Activity Enhancement of Three Main Curcuminoids from Curcuma longa. Molecules 2022, 27, 2758. https://doi.org/10.3390/molecules27092758
Araya-Sibaja AM, Wilhelm-Romero K, Quirós-Fallas MI, Vargas Huertas LF, Vega-Baudrit JR, Navarro-Hoyos M. Bovine Serum Albumin-Based Nanoparticles: Preparation, Characterization, and Antioxidant Activity Enhancement of Three Main Curcuminoids from Curcuma longa. Molecules. 2022; 27(9):2758. https://doi.org/10.3390/molecules27092758
Chicago/Turabian StyleAraya-Sibaja, Andrea Mariela, Krissia Wilhelm-Romero, María Isabel Quirós-Fallas, Luis Felipe Vargas Huertas, José Roberto Vega-Baudrit, and Mirtha Navarro-Hoyos. 2022. "Bovine Serum Albumin-Based Nanoparticles: Preparation, Characterization, and Antioxidant Activity Enhancement of Three Main Curcuminoids from Curcuma longa" Molecules 27, no. 9: 2758. https://doi.org/10.3390/molecules27092758
APA StyleAraya-Sibaja, A. M., Wilhelm-Romero, K., Quirós-Fallas, M. I., Vargas Huertas, L. F., Vega-Baudrit, J. R., & Navarro-Hoyos, M. (2022). Bovine Serum Albumin-Based Nanoparticles: Preparation, Characterization, and Antioxidant Activity Enhancement of Three Main Curcuminoids from Curcuma longa. Molecules, 27(9), 2758. https://doi.org/10.3390/molecules27092758







