Laurus nobilis L. Essential Oil-Loaded PLGA as a Nanoformulation Candidate for Cancer Treatment
Abstract
:1. Introduction
2. Results
2.1. LNEO Composition
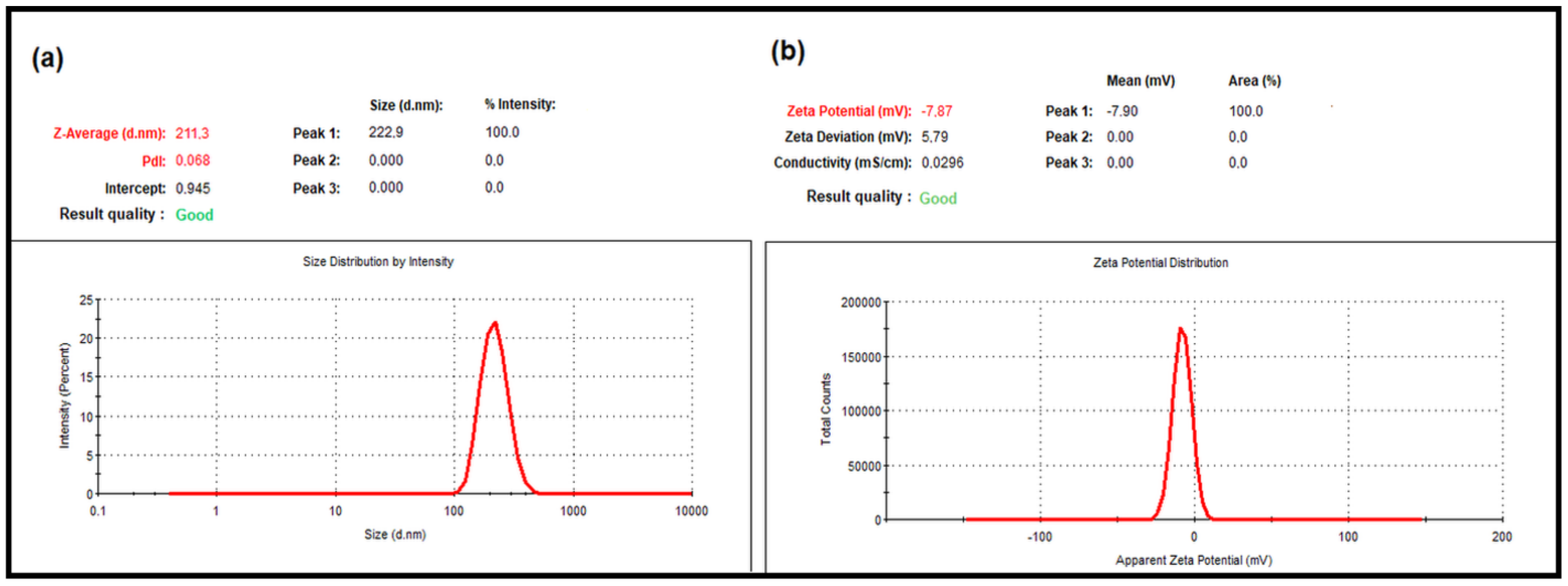
2.2. Average Particle Size, PdI and Zeta Potential Analysis Results
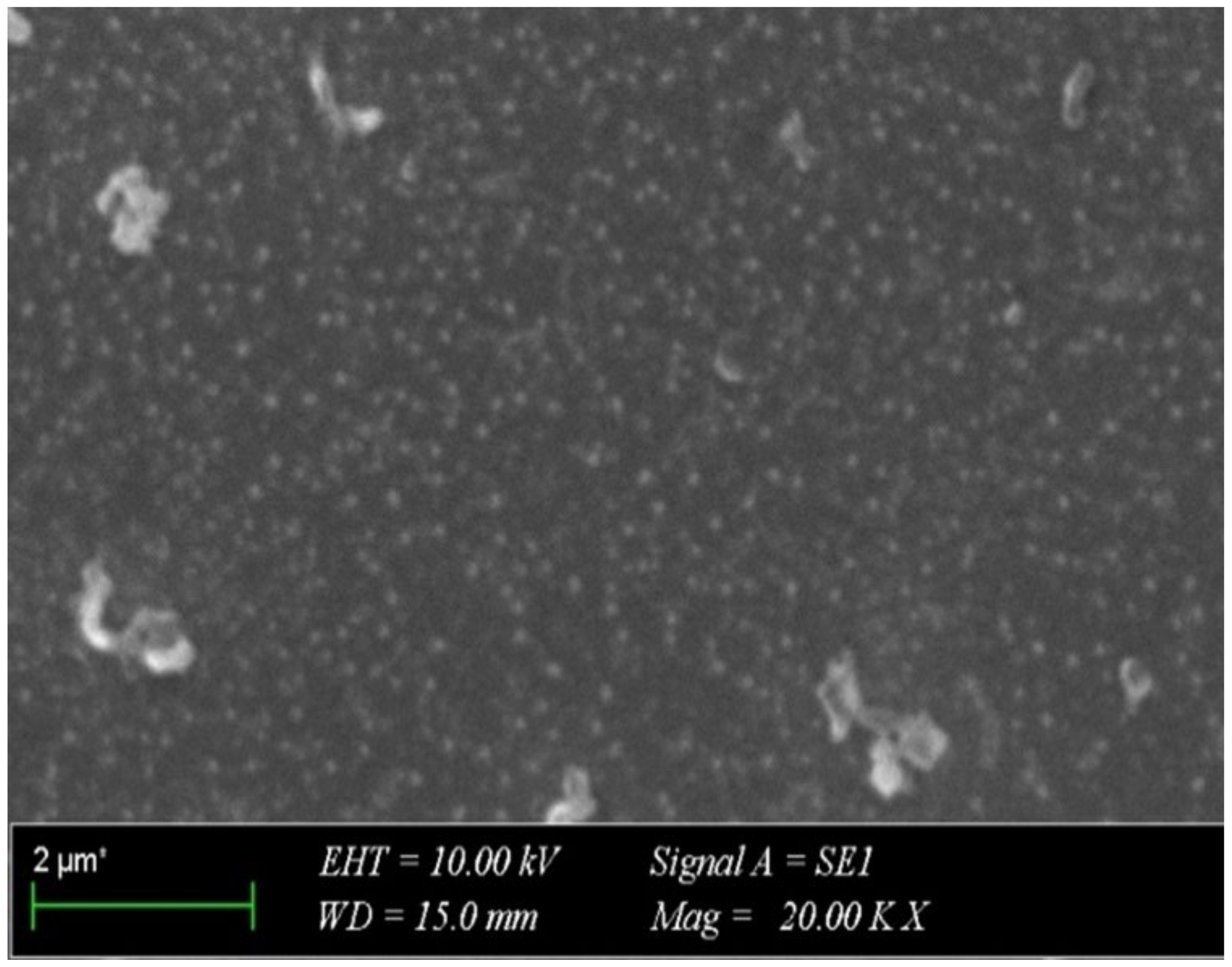
2.3. SEM Micrograph of the LNEO-NPs
2.4. Determination of the Encapsulation Efficiency and Loading Capacity
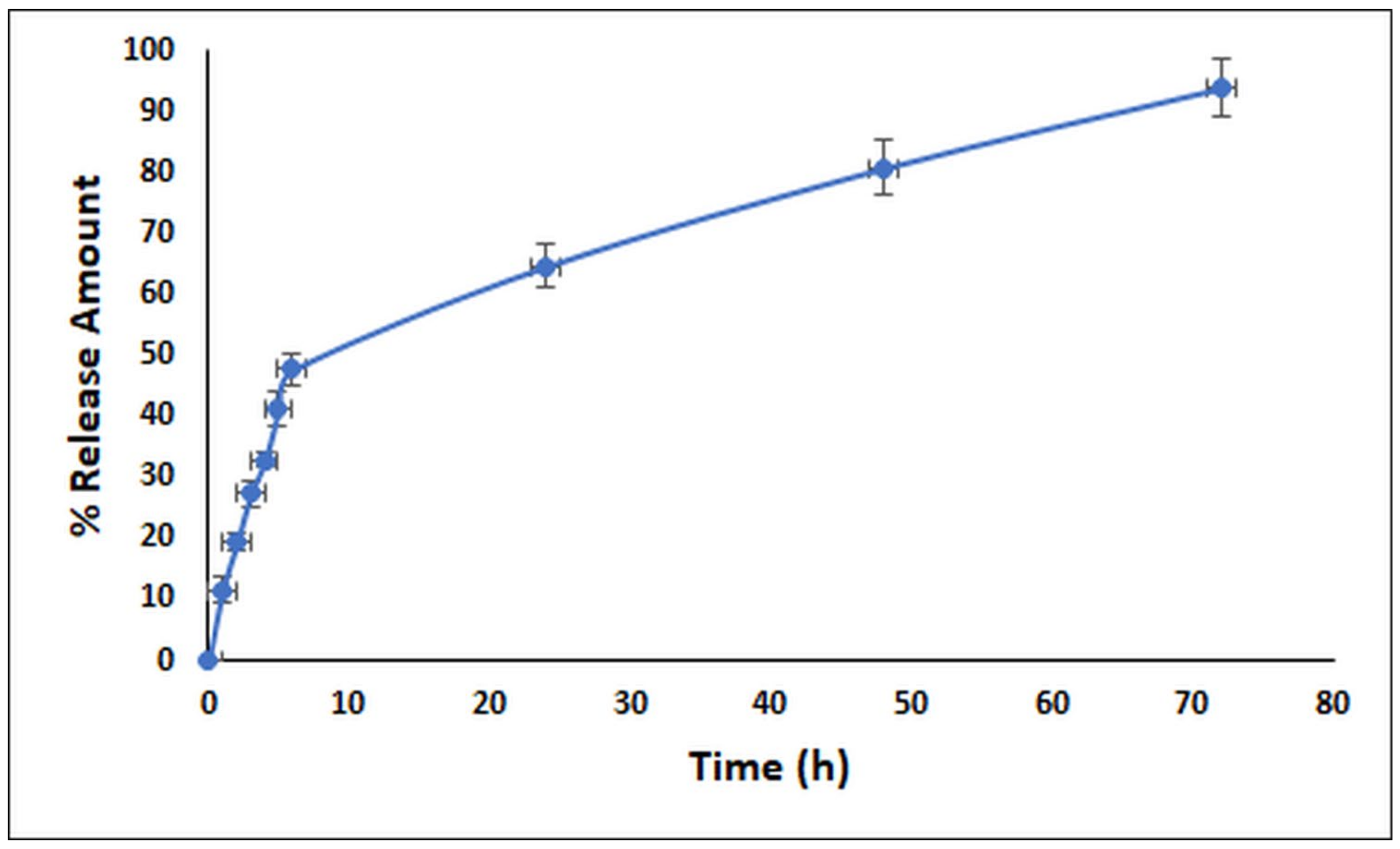
2.5. In Vitro Release Profile of the LNEO
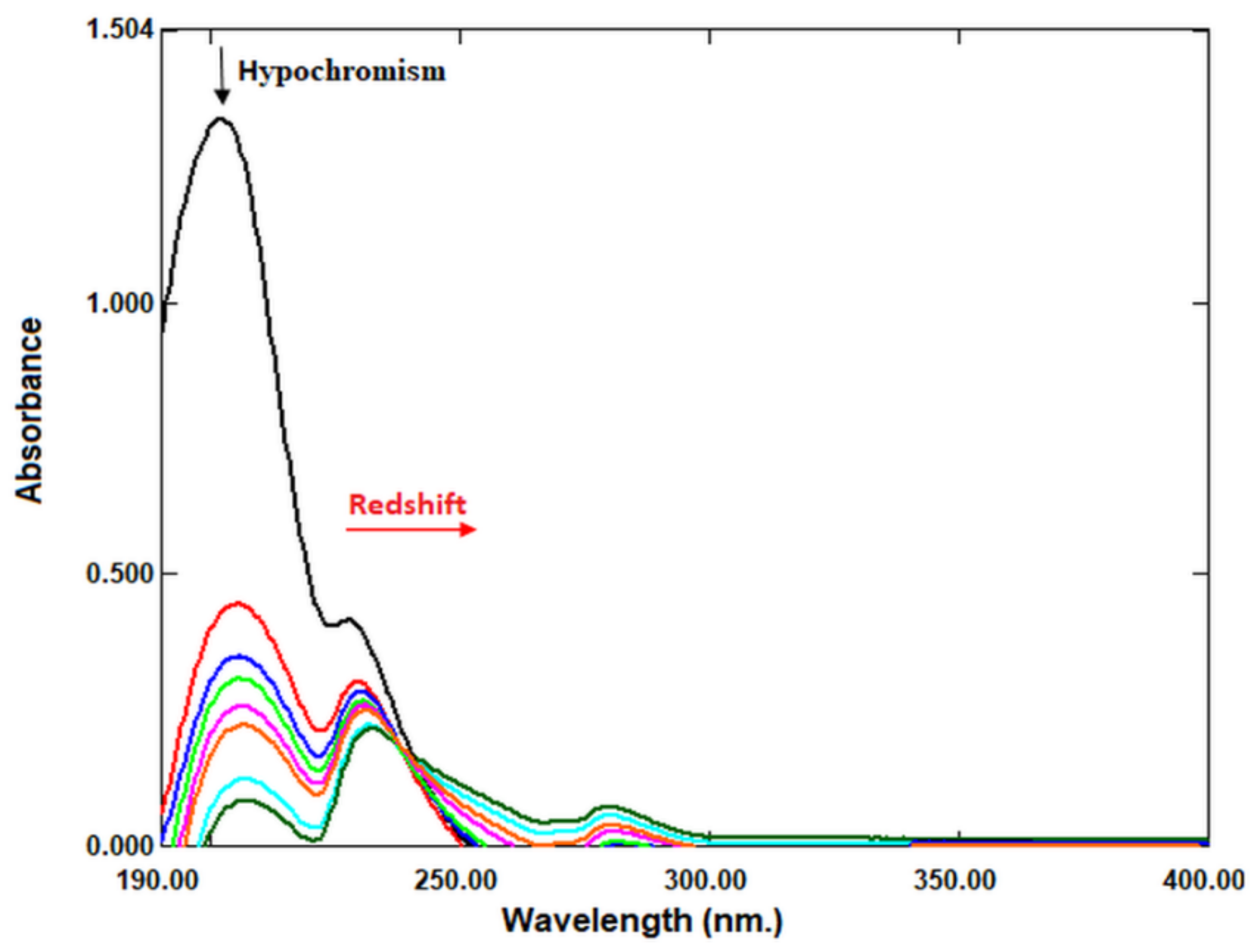
2.6. DNA Binding
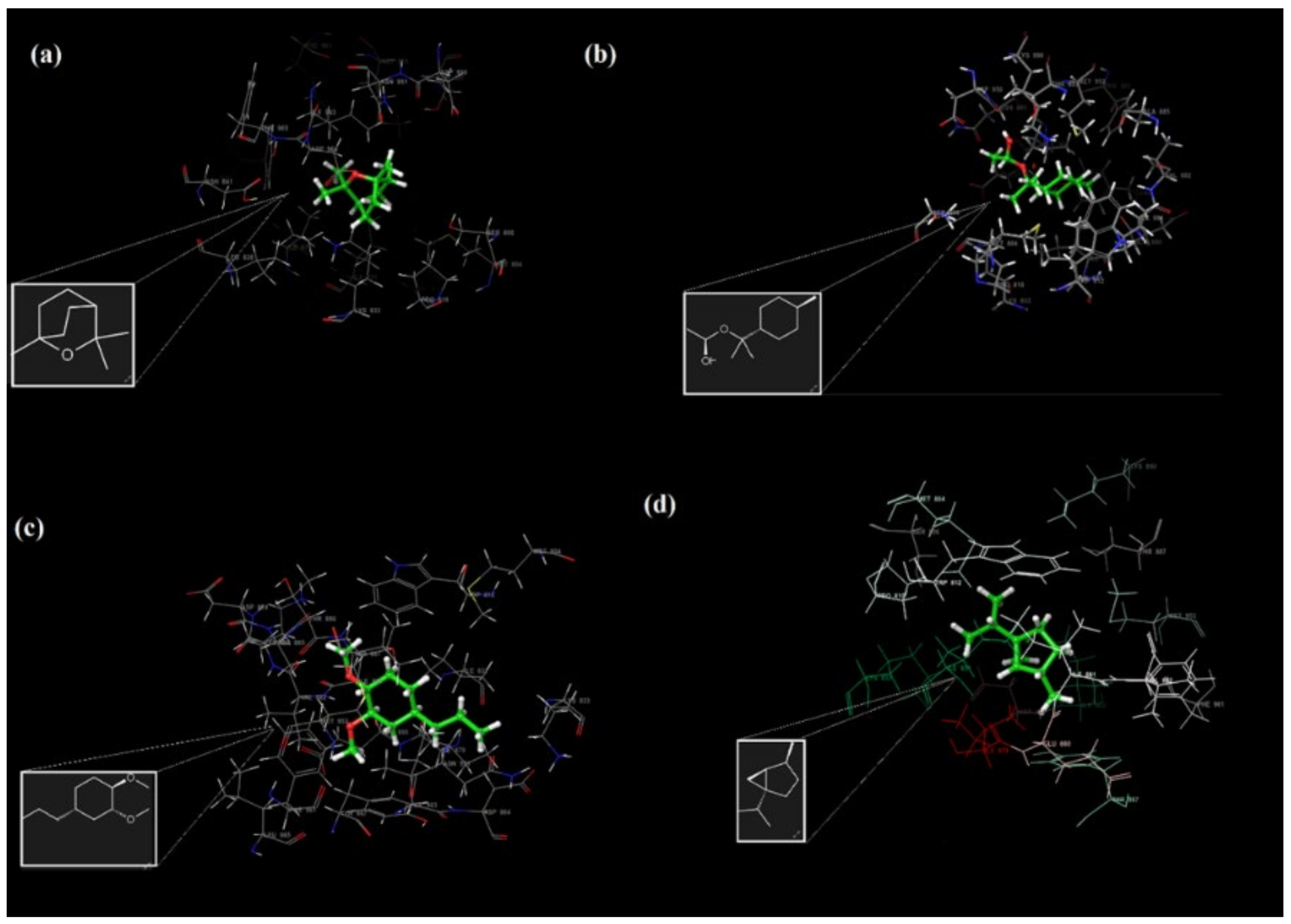
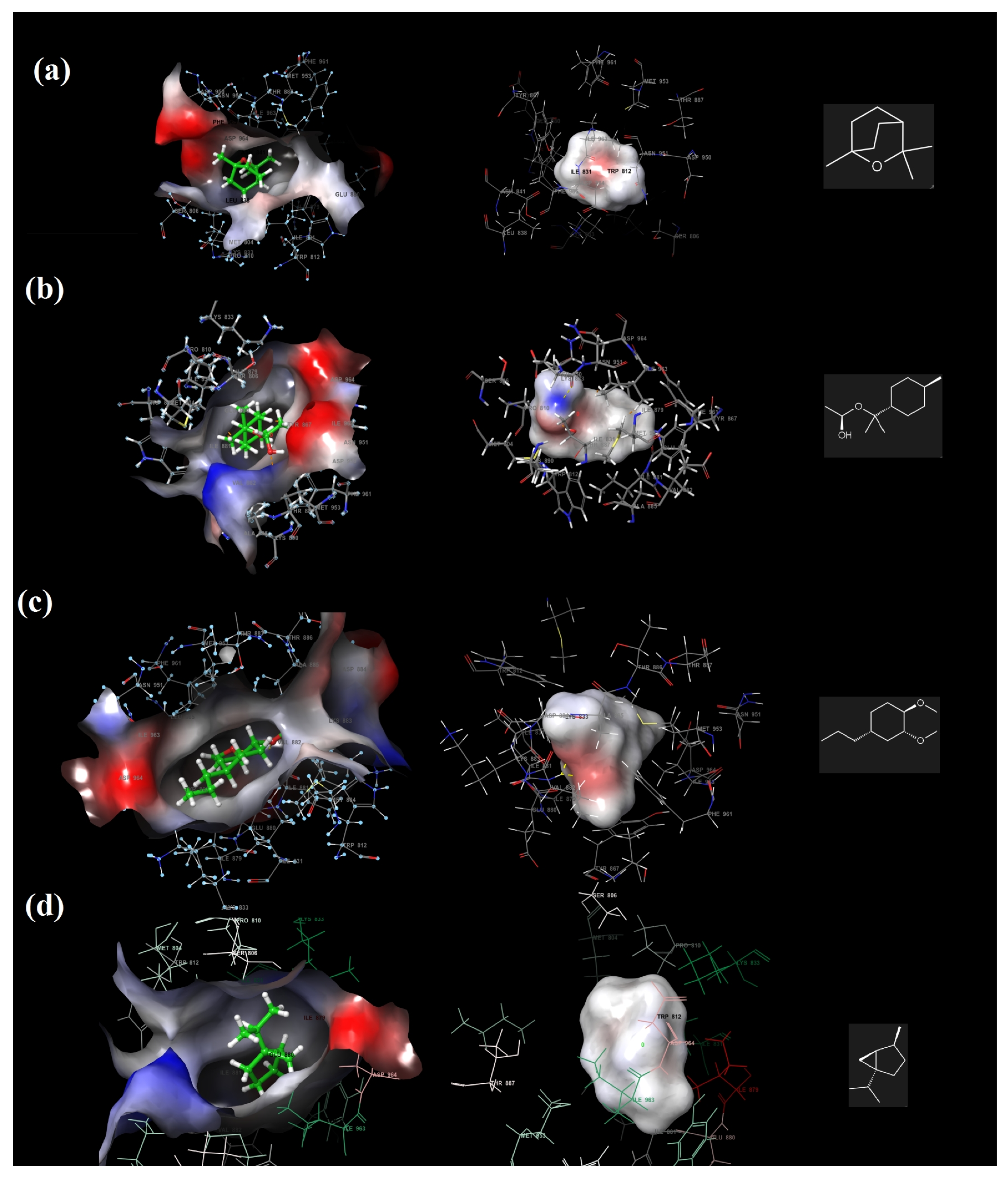
2.7. Molecular Docking and ADME Analysis Results
3. Discussion
4. Materials and Methods
4.1. Materials
Instrumentation and Chemicals
4.2. Methods
4.2.1. Laurus nobilis Essential Oil
4.2.2. Analysis of the Essential Oil
4.2.3. Preparation of the LNEO-NPs
4.2.4. Spectrophotometric Analysis of the LNEO
4.2.5. Average Particle Size, Polydispersity Index (PdI) and Zeta Potential Analyses of the LNEO-NPs
4.2.6. Scanning Electron Microscopy (SEM)
4.2.7. Determination of the Encapsulation Efficiency and Loading Capacity
4.2.8. In Vitro Release Profile of the LNEO-NPs
4.2.9. DNA Binding
4.2.10. Molecular Docking and ADME Analysis
5. Conclusions
6. Recommendations and Future Works
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LNEO | Laurus nobilis L. essential oil |
| PLGA | poly lactic-co-glycolic acid |
| NPs | nanoparticles |
| LNEO-NPs | Laurus nobilis L. essential oil loaded poly lactic-co-glycolic acid (PLGA) nanoparticles |
| GC-MS | gas chromatography-mass spectrometer |
| UV-Vis | ultraviolet–visible |
| DLS | dynamic light scattering |
| PdI | polydispersity index |
| PBS | phosphate-buffered saline |
| SD | standard deviation |
| SEM | scanning electron microscopy |
| CT-DNA | calf thymus DNA |
| PI3K/mTOR | phosphatidylinositol 3-kinase/mammalian rapamycin target |
| ADME | absorption, distribution, metabolism, excretion |
References
- Ferlay, J.; Soerjomataram, I.; Ervik, M.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.; Forman, D.; Bray, F. Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 Lyon; IARC Publications: Lyon, France, 2015; Available online: https://globocan.iarc.fr/Default.aspx (accessed on 2 September 2021).
- Plummer, M.; de Martel, C.; Vignat, J.; Ferlay, J.; Bray, F.; Franceschi, S. Global burden of cancers attributable to infections in 2012: A synthetic analysis. Lancet Glob. Health 2016, 4, e609–e616. [Google Scholar] [CrossRef] [Green Version]
- Stewart, B.W.; Bray, F.; Forman, D.; Ohgaki, H.; Straif, K.; Ullrich, A.; Wild, C.P. Cancer prevention as part of precision medicine: ‘plenty to be done’. Carcinogenesis 2016, 37, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Mehta, M.; Dhanjal, D.S.; Kaur, S.; Gupta, G.; Singh, H.; Thangavelu, L.; Rajeshkumar, S.; Tambuwala, M.; Bakshi, H.A. Emerging trends in the novel drug delivery approaches for the treatment of lung cancer. Chem. Biol. Interact. 2019, 309, 108720. [Google Scholar] [CrossRef] [PubMed]
- Rajivgandhi, G.; Saravanan, K.; Ramachandran, G.; Li, J.-L.; Yin, L.; Quero, F.; Alharbi, N.S.; Kadaikunnan, S.; Khaled, J.M.; Manoharan, N. Enhanced anti-cancer activity of chitosan loaded Morinda citrifolia essential oil against A549 human lung cancer cells. Int. J. Biol. Macromol. 2020, 164, 4010–4021. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, Y.; Gupta, V.K.; Jaitak, V. Anticancer activity of essential oils: A review. J. Sci. Food Agric. 2013, 93, 3643–3653. [Google Scholar] [CrossRef] [PubMed]
- Chahal, K.; Kaur, M.; Bhardwaj, U.; Singla, N.; Kaur, A.; Kaur, M.; Bhardwaj, U.; Singla, N.; Kaur, A. A review on chemistry and biological activities of Laurus nobilis L. essential oil. J. Pharmacogn. Phytochem. 2017, 6, 1153–1161. [Google Scholar]
- Demirbolat, I.; Karik, Ü.; Erçin, E.; Kartal, M. Gender Dependent Differences in Composition, Antioxidant and Antimicrobial Activities of Wild and Cultivated Laurus nobilis L. Leaf and Flower Essential Oils from Aegean Region of Turkey. J. Essent. Oil-Bear. Plants 2020, 23, 1084–1094. [Google Scholar] [CrossRef]
- Ozek, G.; Yaylacı, O.K.; Ozek, D.Y. Defne Yaprağı. TPJ 2018, 4, 31–35. [Google Scholar]
- Turek, C.; Stintzing, F.C. Stability of essential oils: A review. Compr. Rev. Food Sci. Food Saf. 2013, 12, 40–53. [Google Scholar] [CrossRef]
- Turasan, H.; Sahin, S.; Sumnu, G. Encapsulation of rosemary essential oil. LWT-Food Sci. Technol. 2015, 64, 112–119. [Google Scholar] [CrossRef]
- Asbahani, A.; Miladi, K.; Badri, W.; Sala, M.; Addi, E.A.; Casabianca, H.; Mousadik, A.; Hartmann, D.; Jilale, A.; Renaud, F.N.R.; et al. Essential oils: From extraction to encapsulation. Int. J. Pharm. 2015, 483, 220. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.C.; Oliveira, D.A.; Hill, L.E.; Zambiazi, R.C.; Borges, C.D.; Vizzotto, M.; Mertens-Talcott, S.; Talcott, S.; Gomes, C.L. Effect of nanoencapsulation using PLGA on antioxidant and antimicrobial activities of guabiroba fruit phenolic extract. Food Chem. 2018, 240, 396–404. [Google Scholar] [CrossRef]
- Wang, Y.; Qu, W.; Choi, S. FDA’s Regulatory Science Rogram for Generic PLA/PLGA-Based Drug Products. Am. Pharm. Rev. 2016, 20, 5–9. [Google Scholar]
- Wu, P.; Zhou, Q.; Zhu, H.; Zhuang, Y.; Bao, J. Enhanced antitumor efficacy in colon cancer using EGF functionalized PLGA nanoparticles loaded with 5-Fluorouracil and perfluorocarbon. BMC Cancer 2020, 20, 354. [Google Scholar] [CrossRef] [PubMed]
- Stevanovic, M.; Uskokovic, D. Poly (lactide-co-glycolide)-based micro and nanoparticles for the controlled drug delivery of vitamins. Curr. Nanosci. 2009, 5, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Tefas, L.R.; Tomuţă, I.; Achim, M.; Vlase, L. Development and optimization of quercetin-loaded PLGA nanoparticles by experimental design. Clujul Med. 2015, 88, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmkvist, A.D.; Friberg, A.; Nilsson, U.J.; Schouenborg, J. Hydrophobic ion pairing of a minocycline/Ca2+/AOT complex for preparation of drug-loaded PLGA nanoparticles with improved sustained release. Int. J. Pharm. 2016, 499, 351–357. [Google Scholar] [CrossRef] [Green Version]
- Almeida, K.B.; Ramos, A.S.; Nunes, J.B.; Silva, B.O.; Ferraz, E.R.; Fernandes, A.S.; Felzenszwalb, I.; Amaral, A.C.F.; Roullin, V.G.; Falcão, D.Q. PLGA nanoparticles optimized by Box-Behnken for efficient encapsulation of therapeutic Cymbopogon citratus essential oil. Colloids Surf. B 2019, 181, 935–942. [Google Scholar] [CrossRef]
- Badri, W.; El Asbahani, A.; Miladi, K.; Baraket, A.; Agusti, G.; Nazari, Q.A.; Errachid, A.; Fessi, H.; Elaissari, A. Poly (ε-caprolactone) nanoparticles loaded with indomethacin and Nigella sativa L. essential oil for the topical treatment of inflammation. J. Drug Deliv. Sci. Technol. 2018, 46, 234–242. [Google Scholar] [CrossRef]
- Meva, F.E.; Ntoumba, A.A.; Kedi, P.B.E.; Tchoumbi, E.; Schmitz, A.; Schmolke, L.; Klopotowski, M.; Moll, B.; Kökcam-Demir, Ü.; Mpondo, E.A.M. Silver and palladium nanoparticles produced using a plant extract as reducing agent, stabilized with an ionic liquid: Sizing by X-ray powder diffraction and dynamic light scattering. J. Mater. Res. Technol. 2019, 8, 1991–2000. [Google Scholar] [CrossRef]
- Budama-Kilinc, Y. Piperine nanoparticles for topical application: Preparation, characterization, in vitro and in silico evaluation. ChemistrySelect 2019, 4, 11693–11700. [Google Scholar] [CrossRef]
- Ponkarpagam, S.; Mahalakshmi, G.; Vennila, K.; Elango, K.P. Multi-spectroscopic, voltammetric and molecular docking studies on binding of anti-diabetic drug rosigiltazone with DNA. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 234, 118268. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.K.; Sahu, S.; Gupta, I.; Ghorai, T.K. Green synthesis of copper nanoparticles from an extract of Jatropha curcas leaves: Characterization, optical properties, CT-DNA binding and photocatalytic activity. RSC Adv. 2020, 10, 22027–22035. [Google Scholar] [CrossRef]
- Kecel-Gunduz, S.; Budama-Kilinc, Y.; Gok, B.; Bicak, B.; Akman, G.; Arvas, B.; Aydogan, F.; Yolacan, C. Computer-aided anticancer drug design: In vitro and in silico studies of new iminocoumarin derivative. J. Mol. Struct. 2021, 1239, 130539. [Google Scholar] [CrossRef]
- Le, P.T.; Cheng, H.; Ninkovic, S.; Plewe, M.; Huang, X.; Wang, H.; Bagrodia, S.; Sun, S.; Knighton, D.R.; Rogers, C.M.L. Design and synthesis of a novel pyrrolidinyl pyrido pyrimidinone derivative as a potent inhibitor of PI3Kα and mTOR. Bioorganic Med. Chem. Lett. 2012, 22, 5098–5103. [Google Scholar] [CrossRef] [PubMed]
- Kattan, S.W.; Nafie, M.S.; Elmgeed, G.A.; Alelwani, W.; Badar, M.; Tantawy, M.A. Molecular docking, anti-proliferative activity and induction of apoptosis in human liver cancer cells treated with androstane derivatives: Implication of PI3K/AKT/mTOR pathway. J. Steroid Biochem. Mol. Biol. 2020, 198, 105604. [Google Scholar] [CrossRef] [PubMed]
- Argyriou, A.A.; Bruna, J.; Marmiroli, P.; Cavaletti, G. Chemotherapy-induced peripheral neurotoxicity (CIPN): An update. Crit. Rev. Oncol. Hematol. 2012, 82, 51–77. [Google Scholar] [CrossRef] [PubMed]
- Monsuez, J.-J.; Charniot, J.-C.; Vignat, N.; Artigou, J.-Y. Cardiac side-effects of cancer chemotherapy. Int. J. Cardiol. 2010, 144, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Shipunova, V.O.; Sogomonyan, A.S.; Zelepukin, I.V.; Nikitin, M.P.; Deyev, S.M. PLGA Nanoparticles decorated with anti-HER2 affibody for targeted delivery and photoinduced cell death. Molecules 2021, 6, 3955. [Google Scholar] [CrossRef]
- Working, P.; Newman, M.; Huang, S.; Mayhew, E.; Vaage, J.; Lasic, D. Pharmacokinetics, biodistribution and therapeutic efficacy of doxorubicin encapsulated in Stealth® liposomes (Doxil®). J. Liposome Res. 1994, 4, 667–687. [Google Scholar] [CrossRef]
- Miele, E.; Spinelli, G.P.; Miele, E.; Tomao, F.; Tomao, S. Albumin-bound formulation of paclitaxel (Abraxane® ABI-007) in the treatment of breast cancer. Int. J. Nanomed. 2009, 4, 99. [Google Scholar] [CrossRef] [Green Version]
- Passero, F.C., Jr.; Grapsa, D.; Syrigos, K.N.; Saif, M.W. The safety and efficacy of Onivyde (irinotecan liposome injection) for the treatment of metastatic pancreatic cancer following gemcitabine-based therapy. Expert Rev. Anticancer Ther. 2016, 16, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Sani, N.S.; Onsori, H.; Akrami, S.; Rahmati, M. A Comparison of the Anti-Cancer Effects of Free and PLGA-PAA Encapsulated Hydroxytyrosol on the HT-29 Colorectal Cancer Cell Line. Anti-Cancer Agents Med. Chem. 2022, 22, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Elbatanony, R.S.; Parvathaneni, V.; Kulkarni, N.S.; Shukla, S.K.; Chauhan, G.; Kunda, N.K.; Gupta, V. Afatinib-loaded inhalable PLGA nanoparticles for localized therapy of non-small cell lung cancer (NSCLC)—development and in-vitro efficacy. Drug Deliv. Transl. Res. 2021, 11, 927–943. [Google Scholar] [CrossRef] [PubMed]
- Sharaf, N.S.; Shetta, A.; Elhalawani, J.E.; Mamdouh, W. Applying Box–Behnken Design for Formulation and Optimization of PLGA-Coffee Nanoparticles and Detecting Enhanced Antioxidant and Anticancer Activities. Polymers 2022, 14, 144. [Google Scholar] [CrossRef]
- Shabestarian, H.; Tabrizi, M.H.; Es-Haghi, A.; Khadem, F. The Brassica Napus Extract (BNE)-Loaded PLGA Nanoparticles as an Early Necroptosis and Late Apoptosis Inducer in Human MCF-7 Breast Cancer Cells. Nutr. Cancer 2021, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Shabestarian, H.; Homayouni Tabrizi, M.; Movahedi, M.; Neamati, A.; Sharifnia, F. Putative mechanism for cancer suppression by PLGA nanoparticles loaded with Peganum harmala smoke extract. J. Microencapsul. 2021, 38, 324–337. [Google Scholar] [CrossRef]
- Bao, S.; Zheng, H.; Ye, J.; Huang, H.; Zhou, B.; Yao, Q.; Lin, G.; Zhang, H.; Kou, L.; Chen, R. Dual targeting EGFR and STAT3 with Erlotinib and Alantolactone co-loaded PLGA nanoparticles for pancreatic cancer treatment. Front. Pharmacol. 2021, 12. [Google Scholar] [CrossRef]
- Moghaddam, F.A.; Ebrahimian, M.; Oroojalian, F.; Yazdian-Robati, R.; Kalalinia, F.; Tayebi, L.; Hashemi, M. Effect of thymoquinone-loaded lipid–polymer nanoparticles as an oral delivery system on anticancer efficiency of doxorubicin. J. Nanostructure Chem. 2021, 12, 33–44. [Google Scholar] [CrossRef]
- Santoyo, S.; Lloria, R.; Jaime, L.; Ibanez, E.; Senorans, F.; Reglero, G. Supercritical fluid extraction of antioxidant and antimicrobial compounds from Laurus nobilis L. Chemical and functional characterization. Eur. Food Res. Technol. 2006, 222, 565–571. [Google Scholar] [CrossRef]
- Ozcan, B.; Esen, M.; Sangun, M.K.; Coleri, A.; Caliskan, M. Effective antibacterial and antioxidant properties of methanolic extract of Laurus nobilis seed oil. J. Environ. Biol. 2010, 31, 637–641. [Google Scholar] [PubMed]
- Patrakar, R.; Mansuriya, M.; Patil, P. Phytochemical and pharmacological review on Laurus nobilis. Int. J. Pharm. Chem. Sci. 2012, 1, 595–602. [Google Scholar]
- Zargari, A. Medicinal plants. Tehran Univ. Med. J. 1990, 4, 574–578. [Google Scholar]
- Kosar, M.; Tunalier, Z.; Özek, T.; Kürkcüoglu, M.; Baser, K.H.C. A simple method to obtain essential oils from Salvia triloba L. and Laurus nobilis L. by using microwave-assisted hydrodistillation. Z. Nat. C 2005, 60, 501–504. [Google Scholar] [CrossRef]
- Sellami, I.H.; Wannes, W.A.; Bettaieb, I.; Berrima, S.; Chahed, T.; Marzouk, B.; Limam, F. Qualitative and quantitative changes in the essential oil of Laurus nobilis L. leaves as affected by different drying methods. Food Chem. 2011, 126, 691–697. [Google Scholar] [CrossRef]
- Maksimenko, O.; Malinovskaya, J.; Shipulo, E.; Osipova, N.; Razzhivina, V.; Arantseva, D.; Yarovaya, O.; Mostovaya, U.; Khalansky, A.; Fedoseeva, V. Doxorubicin-loaded PLGA nanoparticles for the chemotherapy of glioblastoma: Towards the pharmaceutical development. Int. J. Pharm. 2019, 524, 118733. [Google Scholar] [CrossRef]
- Sebaaly, C.; Jraij, A.; Fessi, H.; Charcosset, C.; Greige-Gerges, H. Preparation and characterization of clove essential oil-loaded liposomes. Food Chem. 2015, 178, 52–62. [Google Scholar] [CrossRef]
- Budama-Kilinc, Y.; Cakir-Koc, R.; Kecel-Gunduz, S.; Kokcu, Y.; Bicak, B.; Mutlu, H.; Ozel, A.E. Novel NAC-loaded poly (lactide-co-glycolide acid) nanoparticles for cataract treatment: Preparation, characterization, evaluation of structure, cytotoxicity, and molecular docking studies. PeerJ 2018, 6, e4270. [Google Scholar] [CrossRef] [Green Version]
- Shen, S.; Wu, Y.; Liu, Y.; Wu, D. High drug-loading nanomedicines: Progress, current status, and prospects. Int. J. Nanomed. 2017, 12, 4085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esfandyari-Manesh, M.; Ghaedi, Z.; Asemi, M.; Khanavi, M.; Manayi, A.; Jamalifar, H.; Atyabi, F.; Dinarvand, R. Study of antimicrobial activity of anethole and carvone loaded PLGA nanoparticles. J. Pharm. Res. 2013, 7, 290–295. [Google Scholar] [CrossRef]
- Schnoor, B.; Elhendawy, A.; Joseph, S.; Putman, M.; Chacón-Cerdas, R.; Flores-Mora, D.; Bravo-Moraga, F.; Gonzalez-Nilo, F.; Salvador-Morales, C. Engineering atrazine loaded poly (lactic-co-glycolic acid) nanoparticles to ameliorate environmental challenges. J. Agric. Food Chem. 2018, 66, 7889–7898. [Google Scholar] [CrossRef]
- Froiio, F.; Ginot, L.; Paolino, D.; Lebaz, N.; Bentaher, A.; Fessi, H.; Elaissari, A. Essential oils-loaded polymer particles: Preparation, characterization and antimicrobial property. Polymers 2019, 11, 1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraj, A.; Jaâfar, F.; Marti, M.; Coderch, L.; Ladhari, N. A comparative study of oregano (Origanum vulgare L.) essential oil-based polycaprolactone nanocapsules/microspheres: Preparation, physicochemical characterization, and storage stability. Ind. Crops Prod. 2019, 140, 111669. [Google Scholar] [CrossRef]
- Iannitelli, A.; Grande, R.; Stefano, A.D.; Giulio, M.D.; Sozio, P.; Bessa, L.J.; Laserra, S.; Paolini, C.; Protasi, F.; Cellini, L. Potential antibacterial activity of carvacrol-loaded poly (DL-lactide-co-glycolide)(PLGA) nanoparticles against microbial biofilm. Int. J. Mol. Sci. 2011, 12, 5039–5051. [Google Scholar] [CrossRef]
- Fonte, P.; Lino, P.R.; Seabra, V.; Almeida, A.J.; Reis, S.; Sarmento, B. Annealing as a tool for the optimization of lyophilization and ensuring of the stability of protein-loaded PLGA nanoparticles. Int. J. Pharm. 2016, 503, 163–173. [Google Scholar] [CrossRef]
- Zigoneanu, I.G.; Astete, C.E.; Sabliov, C.M. Nanoparticles with entrapped α-tocopherol: Synthesis, characterization, and controlled release. Nanotechnology 2008, 19, 105606. [Google Scholar] [CrossRef]
- Silva, L.M.; Hill, L.E.; Figueiredo, E.; Gomes, C.L. Delivery of phytochemicals of tropical fruit by-products using poly (DL-lactide-co-glycolide)(PLGA) nanoparticles: Synthesis, characterization, and antimicrobial activity. Food Chem. 2014, 165, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Chourasiya, V.; Bohrey, S.; Pandey, A. Formulation, optimization, characterization and in-vitro drug release kinetics of atenolol loaded PLGA nanoparticles using 33 factorial design for oral delivery. Mater. Discov. 2016, 5, 1–13. [Google Scholar] [CrossRef]
- Rancan, F.; Wiehe, A.; Nöbel, M.; Senge, M.O.; Al Omari, S.; Böhm, F.; John, M.; Röder, B. Influence of substitutions on asymmetric dihydroxychlorins with regard to intracellular uptake, subcellular localization and photosensitization of Jurkat cells. J. Photochem. Photobiol. B Biol. 2005, 78, 17–28. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Wang, L.; Wang, L.; Li, X.; Xu, Y. Spectroscopic studies on the interactions between novel bisnaphthalimide derivatives and calf thymus DNA. J. Photochem. Photobiol. B Biol. 2017, 166, 333–340. [Google Scholar] [CrossRef] [PubMed]
- <monospace>Zhang, G.; Guo, J.; Pan, J.; Chen, X.; Wang, J. Spectroscopic studies on the interaction of morin–Eu(III) complex with calf thymus DNA. J. Mol. Struct. 2009, 923, 114. [Google Scholar] [CrossRef]
- Long, E.C.; Barton, J.K. On demonstrating DNA intercalation. Acc. Chem. Res. 1990, 23, 271–273. [Google Scholar] [CrossRef]
- Frederick, C.A.; Williams, L.D.; Ughetto, G.; Van der Marel, G.A.; Van Boom, J.H.; Rich, A.; Wang, A.H. Structural comparison of anticancer drug-DNA complexes: Adriamycin and daunomycin. Biochemistry 1990, 29, 2538–2549. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.A. The manufacturing techniques of various drug loaded biodegradable poly (lactide-co-glycolide)(PLGA) devices. Biomaterials 2000, 21, 2475–2490. [Google Scholar] [CrossRef]
- Naghibzadeh, M.; Firoozi, S.; Nodoushan, F.S.; Adabi, M.; Khoradmehr, A.; Fesahat, F.; Esnaashari, S.S.; Khosravani, M.; Tavakol, S.; Pazoki-Toroudi, H. Application of electrospun gelatin nanofibers in tissue engineering. Biointerface Res. Appl. Chem. 2018, 1, 3048–3052. [Google Scholar]
- Javadzadeh, Y.; Ahadi, F.; Davaran, S.; Mohammadi, G.; Sabzevari, A.; Adibkia, K. Preparation and physicochemical characterization of naproxen–PLGA nanoparticles. Colloids Surf. B Biointerfaces 2010, 81, 498–502. [Google Scholar] [CrossRef]
- Fornaguera, C.; Feiner-Gracia, N.; Calderó, G.; García-Celma, M.; Solans, C. Galantamine-loaded PLGA nanoparticles, from nano-emulsion templating, as novel advanced drug delivery systems to treat neurodegenerative diseases. Nanoscale 2015, 7, 12076–12084. [Google Scholar] [CrossRef]
- Hosseini, S.F.; Zandi, M.; Rezaei, M.; Farahmandghavi, F. Two-step method for encapsulation of oregano essential oil in chitosan nanoparticles: Preparation, characterization and in vitro release study. Carbohydr. Polym. 2013, 95, 50–56. [Google Scholar] [CrossRef]
- Marmur, J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 1961, 16, 208–218. [Google Scholar] [CrossRef]
- Tariq, M.; Muhammad, N.; Ali, S.; Shirazi, J.H.; Tahir, M.N.; Khalid, N. Synthesis, spectroscopic, X-ray crystal structure, biological and DNA interaction studies of organotin (IV) complexes of 2-(4-ethoxybenzylidene) butanoic acid. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2014, 122, 356–364. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.; Trucks, G.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G. Gaussian 09, Revision d. 01, Gaussian. Inc. Wallingford CT 2009, 201. [Google Scholar]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 53, 1750–1759. [Google Scholar] [CrossRef]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Søndergaard, C.R.; Olsson, M.H.M.; Rostkowski, M.; Jensen, J.H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of p K a values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef]
- Schrödinger, L. Schrödinger, Release 2020-4: Maestro; Schrödinger LLC: New York, NY, USA, 2020. [Google Scholar]
- Ioakimidis, L.; Thoukydidis, L.; Mirza, A.; Naeem, S.; Reynisson, J. Benchmarking the reliability of QikProp. Correlation between experimental and predicted values. QSAR Comb. Sci. 2008, 27, 445–456. [Google Scholar] [CrossRef]
- Singh, D.B.; Gupta, M.K.; Kesharwani, R.K.; Misra, K. Comparative docking and ADMET study of some curcumin derivatives and herbal congeners targeting β-amyloid. Netw. Model. Anal. Health Inform. Bioinform. 2013, 2, 13–27. [Google Scholar] [CrossRef]






| No | Compound | R.T (Min) | R.IL | R.IC | Amount (%) |
|---|---|---|---|---|---|
| 1 | α-Pinene | 7.582 | 1035 | 1037 | 5.995 ± 0.0157 |
| 2 | Camphene | 8.864 | 1065 | 1068 | 0.928 ± 0.0086 |
| 3 | β-Myrcene | 10.406 | 1118 | 1120 | 1.127 ± 0.0043 |
| 4 | Sabinene | 10.898 | 1125 | 1129 | 9.767 ± 0.0129 |
| 5 | β-Pinene | 12.871 | 1167 | 1165 | 4.685 ± 0.0102 |
| 6 | Limonene | 15.454 | 1208 | 1206 | 1.395 ± 0.0021 |
| 7 | 1.8-Cineole | 16.525 | 1212 | 1218 | 62.370 ± 0.0162 |
| 8 | p-Cymene | 20.690 | 1280 | 1274 | 0.705 ± 0.0035 |
| 9 | Linalool | 40.983 | 1543 | 1549 | 4.386 ± 0.0078 |
| 10 | Terpinen-4-ol | 44.368 | 1601 | 1608 | 0.928 ± 0.0041 |
| 11 | α-Terpinyl Acetate | 49.992 | 1687 | 1690 | 4.485 ± 0.0017 |
| 12 | α-Terpineol | 50.061 | 1694 | 1693 | 0.137 ± 0018 |
| 13 | Methyleugenol | 66.680 | 2033 | 2036 | 0.201 ± 0.0021 |
| Total Identified | 94.411 | ||||
| Monoterpenes (1–6, and 8) * | 22.969 | ||||
| Oxygenated Monoterpenes (7, 9–12) * | 71.241 | ||||
| Phenylpropanoids (13) * | 0.201 | ||||
| Ligands | 1,8-Cineole | α-Terpinyl Acetate | Methyleugenol | Sabinene |
|---|---|---|---|---|
| Docking Score (Kcal/mol) | −4.89 | −4.68 | −5.97 | −5.26 |
| H-bonding Interaction (Angstrom) | - | Asp 950 (2.58) | Val 882(2.30) Val 882(2.43) | - |
| Salt Bridge Interaction | - | - | - | - |
| Cation-π Interaction | - | - | - | - |
| Hydrophobic Residues | Val 882, Ile 881, Ile 879, Trp 812, Pro 810, Met 804, Leu 838, Phe 832, Ile 831, Leu 838, Met 953, Phe 961, Ile 963, Phe 965, Tyr 867, Cys 869 | Val 882, Ile 881, Ile 879, Ala 885, Ala 889, Met 953, Ile 952, Tyr 867, Phe 961, Ile 963, Ile 831, Met 804, Ala 805, Trp 812, Pro 810 | Val 882, Ala 885, Met 804, Phe 965, Ile 963, Phe 961, Ile 879, Ile 881, Leu 865, Pro 866, Tyr 867, Leu 845, Ile 831, Leu 838, Pro 810, Trp 812, Met 953 | Val 882, Ile 881, Ile 879,Ala 885, Trp 812, Pro 810, Met 804, Phe 832, Ile 831, Phe 965, Ile 963, Phe 961, Leu 845, Met953, Tyr 867, Cys 869 |
| Polar Residues | His 962, Thr887, Asn 951, Ser 806 | Asn 946, Asn 951, Thr 887, Ser 806 | His 962, Ser 806, Asn 951, Thr 887, Thr 886 | Thr 887, Ser 806, His 962, Asn 951 |
| Charged (positive) Residues | Arg 947, Lys 890, Lys 833, Lys 808 | Lys 833, Lys 802, Lys 890 | Lys 883, Lys 890, Lys 833 | Lys 833, Lys 890, Lys 808 |
| Charged (negative) Residues | Asp 950, Asp 964, Ash 841, Asp 836, Glu 880 | Asp 950, Glu 880, Asp 964 | Asp 964, Glu 880, Ash8 41, Asp 884 | Asp 950, Asp 964, Glu 880 |
| Glycine | - | Gly 868 | Gly868 |
| Major Components of the LNEO | 1,8-Cineole | α-Terpinyl Acetate | MethyleuGenol | Sabinene | |
|---|---|---|---|---|---|
| Docking Score (kcal/mol) | −4.89 | −4.68 | −5.97 | −5.26 | |
| Principal Descriptors | (Range 95% of Drugs) | ||||
| Solute Molecular Weight | 154.252 | 200.320 | 186.294 | 138.252 | (130.0/725.0) |
| Solute Dipole Moment (D) | 1.624 | 2.112 | 2.633 | 0.160 | (1.0/12.5) |
| Solute Total SASA | 373.729 | 458.423 | 448.882 | 381.877 | (300.0/1000.0) |
| Solute Hydrophobic SASA | 373.729 | 412.688 | 448.882 | 381.877 | (0.0/750.0) |
| Solute Hydrophilic SASA | 0.000 | 45.735 | 0.000 | 0.000 | (7.0/330.0) |
| Solute Carbon Pi SASA | 0.000 | 0.000 | 0.000 | 0.000 | (0.0/450.0) |
| Solute Weakly Polar SASA | 0.000 | 0.000 | 0.000 | 0.000 | (0.0/175.0) |
| Solute Molecular Volume (Å3) | 618.965 | 796.485 | 752.599 | 626.954 | (500.0/2000.0) |
| Solute vdW Polar SA (PSA) | 7.264 | 28.091 | 17.239 | 0.000 | (7.0/200.0) |
| Solute No, of Rotatable Bonds | 0.000 | 4.000 | 4.000 | 1.000 | (0.0/15.0) |
| Solute as Donor—Hydrogen Bonds | 0.000 | 1.000 | 0.000 | 0.000 | (0.0/6.0) |
| Solute as Acceptor—Hydrogen Bonds | 0.750 | 1.000 | 3.400 | 0.000 | (2.0/20.0) |
| Solute Globularity (Sphere = 1) | 0.940 | 0.906 | 0.891 | 0.928 | (0.75/0.95) |
| Solute Ionization Potential (eV) | 10.363 | 10.705 | 10.465 | 10.610 | (7.9/10.5) |
| Solute Electron Affinity (eV) | −2.516 | −2.449 | −2.441 | −2.806 | (−0.9/1.7) |
| Predictions for Properties: | |||||
| QP Polarizability (Å3) | 18.617 | 23.153 | 21.397 | 18.295 | (13.0/70.0) |
| QP log P for hexadecane/gas | 4.348 | 6.379 | 5.285 | 4.311 | (4.0/18.0) |
| QP log P for octanol/gas | 5.449 | 9.483 | 7.702 | 4.817 | (8.0/35.0) |
| QP log P for water/gas | 1.357 | 4.129 | 3.266 | −1.113 | (4.0/45.0) |
| QP log P for octanol/water | 2.417 | 2.919 | 1.892 | 5.106 | (−2.0/6.5) |
| QP log S for aqueous solubility | −2.985 | −3.103 | −2.730 | −5.202 | (−6.5/0.5) |
| P log S—conformation independent | −3.615 | −2.256 | −2.730 | −5.202 | (−6.5/0.5) |
| QP log K HSA Serum Protein Binding | 0.214 | 0.152 | −0.277 | 0.396 | (−1.5/1.5) |
| QP log BB for brain/blood | 0.597 | −0.049 | −0.464 | 0.957 | (−3.0/1.2) |
| No, of Primary Metabolites | 1 | 0 | 0 | 0 | (1.0/8.0) |
| Predicted CNS Activity (-- to ++) | ++ | +/− | +/− | ++ | |
| HERG K+ Channel Blockage: log IC50 | −2.506M | −3.046 | −3.289 | −2.702M | (concern below −5) |
| Apparent Caco-2 Permeability (nm/sec) | 9906 | 3649 | 9906 | 9906 | (<25 poor, >500 great) |
| Apparent MDCK Permeability (nm/sec) | 5899 | 2004 | 5899 | 5899 | (<25 poor, >500 great) |
| QP log Kp for skin permeability | −0.923 | −1.978 | −1.331 | 1.079 | (Kp in cm/hr) |
| Jm, max transdermal transport rate | 4.476 | 1.661 | 16.199 | 10.415 | (micrograms/cm2-hr) |
| Lipinski Rule of 5 Violations | 0 | 0 | 0 | 1 | (maximum is 4) |
| Jorgensen Rule of 3 Violations | 0 | 0 | 0 | 0 | (maximum is 3) |
| % Human Oral Absorption in GI (±0%) | 100 | 100 | 100 | 100 | (<25% is poor) |
| Qual, Model for Human Oral Absorption | HIGH | HIGH | HIGH | HIGH | (>80% is high) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ercin, E.; Kecel-Gunduz, S.; Gok, B.; Aydin, T.; Budama-Kilinc, Y.; Kartal, M. Laurus nobilis L. Essential Oil-Loaded PLGA as a Nanoformulation Candidate for Cancer Treatment. Molecules 2022, 27, 1899. https://doi.org/10.3390/molecules27061899
Ercin E, Kecel-Gunduz S, Gok B, Aydin T, Budama-Kilinc Y, Kartal M. Laurus nobilis L. Essential Oil-Loaded PLGA as a Nanoformulation Candidate for Cancer Treatment. Molecules. 2022; 27(6):1899. https://doi.org/10.3390/molecules27061899
Chicago/Turabian StyleErcin, Esin, Serda Kecel-Gunduz, Bahar Gok, Tugba Aydin, Yasemin Budama-Kilinc, and Murat Kartal. 2022. "Laurus nobilis L. Essential Oil-Loaded PLGA as a Nanoformulation Candidate for Cancer Treatment" Molecules 27, no. 6: 1899. https://doi.org/10.3390/molecules27061899
APA StyleErcin, E., Kecel-Gunduz, S., Gok, B., Aydin, T., Budama-Kilinc, Y., & Kartal, M. (2022). Laurus nobilis L. Essential Oil-Loaded PLGA as a Nanoformulation Candidate for Cancer Treatment. Molecules, 27(6), 1899. https://doi.org/10.3390/molecules27061899






