Abstract
We designed and synthesized the 1,3,4-thiadiazole derivatives differing in the structure of the substituents in C2 and C5 positions. The cytotoxic activity of the obtained compounds was then determined in biological studies using MCF-7 and MDA-MB-231 breast cancer cells and normal cell line (fibroblasts). The results showed that in both breast cancer cell lines, the strongest anti-proliferative activity was exerted by 2-(2-trifluorometylophenylamino)-5-(3-methoxyphenyl)-1,3,4-thiadiazole. The IC50 values of this compound against MCF-7 and MDA-MB-231 breast cancer cells were 49.6 µM and 53.4 µM, respectively. Importantly, all new compounds had weaker cytotoxic activity on normal cell line than on breast cancer cell lines. In silico studies demonstrated a possible multitarget mode of action for the synthesized compounds. The most likely mechanism of action for the new compounds is connected with the activities of Caspase 3 and Caspase 8 and activation of BAX proteins.
1. Introduction
Breast cancer affects 14% of all women living in the world [1]. It is the most frequently diagnosed neoplasm in female patients. The basic available treatments for breast cancer are surgery, radiotherapy, and chemotherapy, individually tailored to the patient. Unfortunately, one of the main problems of the pharmacotherapy of cancers, including breast cancer, is the rapidly developing drug resistance. For this reason, it is necessary to search for new anticancer drugs [2].
Estrogens are listed by the WHO as one of the most important factors stimulating the development of breast cancer. These hormones can promote the development of breast cancer by stimulating proliferation and altering gene expression. Estrogens can also initiate the process of carcinogenesis through reactive metabolites. Many of the pharmacological treatments for breast cancer currently available, such as tamoxifen, mainly target the estrogen receptors (ER). As a consequence, these drugs are not effective in the treatment of non-estrogen-dependent cases of breast cancer. Research is ongoing to find new therapeutic and chemopreventive agents acting independently of estrogen receptors, effective against both estrogen-dependent and non-estrogen-dependent breast cancer cells [3].
Over the past 20 years, tremendous efforts have been made to unravel the molecular basis of cancer. Several proteins involved in the development of various types of cancer have been described, such as the suppressor protein p53 or the protein S100A4, a metastasis promoter [4]. This newly gained knowledge about the processes in the neoplastic cell is used to design and synthesize new cytotoxic and cytostatic substances. This resulted in the significant development of cancer pharmacotherapy towards targeted therapies. New drugs are designed to target tumor-specific proteins. Such therapeutics tend to have fewer severe side effects than old drugs because they are more selective for neoplastic changes [5]. One of them with documented antitumor activity are thiadiazoles. Four isomeric forms can be distinguished among the thiadiazole derivatives: 1,3,4-thiadiazole; 1,2,3-thiadiazole; 1,2,4-thiadiazole; and 1,2,5-thiadiazole. Published studies show that 1,3,4-thiadiazole derivatives are the most promising group in terms of potential therapeutic activity [6]. Compounds belonging to the group of 1,3,4-thiadiazole derivatives have shown in studies the potential of antibacterial, antifungal, antituberculosis, anti-inflammatory, analgesic, antipsychotic, anticonvulsant, antidepressant, and anti-leishmanial properties. Many studies have been published showing the anticancer activity of substances from this group [7].
It has been shown that some 1,3,4-thiadiazole derivatives can interfere with processes related to DNA replication. The bioactive properties of these compounds are explained by the fact that their molecular structure contains a heterocyclic ring, which is a bioisostere of a pyrimidine, which is the backbone of the structures of three nucleobases. Probably due to this similarity, cytostatic active 1,3,4-thiadiazole molecules interfere with DNA synthesis and, consequently, inhibit replication of both human tumor and bacterial cells. This allows them to inhibit the multiplication of both bacterial and cancer cells. Therefore, among the 1,3,4-thiadiazole derivatives, candidates for new antibiotic and anticancer drugs are sought [6].
Among the 1,3,4-thiadiazole derivatives tested for antitumor activity, there are thiadiazole systems fused with other rings and simple 2,5-disubstituted rings. Significantly more published research results indicate the therapeutic potential of molecules containing a simple unfused ring. Some of them have been shown to have promising anticancer activity, exceeding the reference drugs in tests. Many studies have demonstrated the activity of compounds from this group against breast cancer cell lines (Figure 1) [8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29]. Therefore, we assumed that the search for cytotoxic compounds in the group of simple 2,5-disubstituted 1,3,4-thiadiazole derivatives is a promising direction of research.
Figure 1.
1,3,4-thiadiazole derivatives recently identified as possessing cytotoxic activity against MCF-7 cancer cell lines [10,12,14,17,18,19,21,22,23,25,26,28].
Thiadiazole derivatives act through various molecular targets, for example, CA IX carbonic anhydrase, Src and Abl kinases, and topoisomerase II [30,31,32].
In order to develop new 1,3,4-thiadiazole derivatives, we created a library of molecular patterns and reports on the antitumor activity demonstrated by them in tests. Based on the analysis of the collected data, we designed and synthesized a group of 1,3,4-thiadiazole derivatives differing in the structure of the substituents in C2 and C5 positions. The cytotoxic activity of the obtained compounds was then determined in biological studies using MCF-7 and MDA-MB-231 breast cancer cells. The MCF-7 cell line, derived from a pleural effusion of malignant breast cancer, is a widely studied model for hormone-dependent human breast cancer. In contrast, the MDA-MB-231 cell line provides a model for human breast cancer, which exhibits an estrogen-independent state and does not express estrogen receptors.
In addition, in silico studies, we determined the interaction of the obtained compounds with proteins involved in neoplastic processes, such as topoisomerase IIb, Caspase 3, Caspase 8, Bcl-xl, Bcl2, and BAX.
2. Results and Discussion
2.1. Chemistry
The title compounds ST1–ST15 were obtained in a two-step synthesis (Scheme 1).
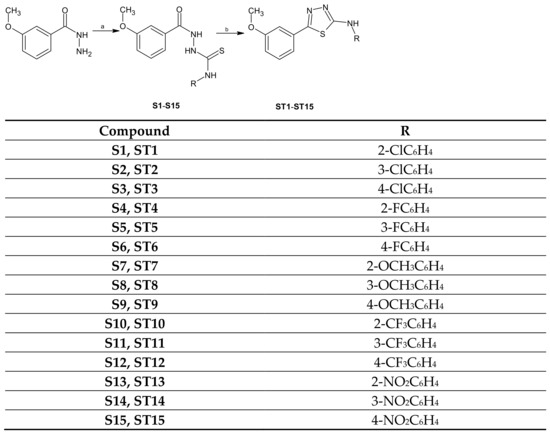
Scheme 1.
Synthesis of thiosemicarbazide and 1,3,4-thiadiazole derivatives. Reagents and conditions: (a) aryl isothiocyanates, 96% EtOH, reflux; (b) H2SO4 (conc), rt.
The reaction of 3-methoxybenzhydrazide with aryl isothiocyanates (2-, 3-, 4-chlorophenyl, 2-, 3-, 4-fluorophenyl, 2-, 3-, 4-methoxyphenyl, 2-, 3-, 4-trifluoromethylphenyl and 2-, 3-, 4-nitrophenyl) gave the corresponding 1,4-disubstituted thiosemicarbazides S1–S15, which in the reaction with concentrate sulfuric acid in room temperature lead to formation 1,3,4-thiadiazole derivatives ST1–ST15. The reaction yields ranged from 43 to 96% for thiosemicarbazide derivatives and 18–70% for 1,3,4-thiadiazoles. The structures of the new compounds were determined using IR, 1H NMR, 13C NMR, and elemental analyses.
The 1H NMR spectra showed the chemical shifts of the N1, N2, and N4 protons, which confirmed the formation of the thiosemicarbazide scaffold. The thiosemicarbazide protons were observed between 9.22 and 10.08, between 9.72 and 10.08, and between 10.49 and 10.73 ppm, respectively, as two or three singlets. The 1H NMR spectra confirmed the formation of thiadiazole derivatives. The N1, N2, and N4 protons of the thiosemicarbazide were not detected in the 1,3,4-thiadiazole compounds. Instead, N-H peaks of the amino group of cyclic compounds were observed between 8.26 and 11.32 ppm. Infrared spectra showed strong peaks in the range 1651–1688 cm−1 corresponding to the carbonyl group and 1316–1368 cm−1, which corresponds to the C=S group in thiosemicarbazide derivatives. In IR spectra of thiadiazole derivatives, absorption peaks in the range of 715–781 cm−1 from C-S were observed. In the 13C NMR spectra, the signals of a methyl group in the range of 53.13–57.40 ppm were observed. All other signals of carbons are adequate to the structure. The 1H NMR and 13C NMR spectra are presented in the Supplementary Materials file.
2.2. Bioactivity Studies
The cytotoxic activity of novel synthesized compounds (ST1–ST15) against two breast cancer cell lines was assessed using MTT assay and biosynthesis DNA. The cells were exposed to the tested compounds for 24 h. As a reference drug, etoposide was used. The obtained results revealed that all tested compounds exhibited cytotoxicity in concentration-dependent manner against MCF-7 and MDA-MB-231 (Figure 2).
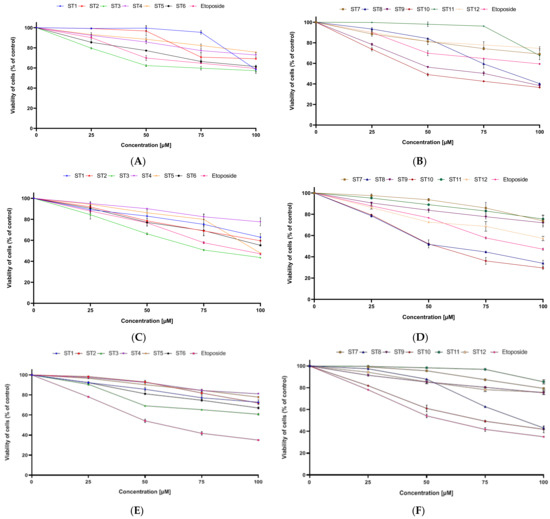
Figure 2.
The viability of MCF-7 (A,B) and MDA-MB-231 (C,D) cells and fibroblasts (E,F) treated for 24 h with various concentrations of tested compounds (ST1–ST12). Mean ± SD values from three independent experiments (n = 3) done in duplicate are presented.
The strongest anticancer activity against MCF-7 was exhibited by ST10, which had IC50 = 49.6 µM. A moderate inhibitory effect on the survival of MCF-7 cells was demonstrated by two compounds, ST8 and ST9, which had IC50 = 87.4 µM and 75.1 µM, correspondingly. It is worth noticing that those three compounds exerted higher anticancer activity against MCF-7 than etoposide. The IC50 of the reference drug and the other synthesized compounds was above 100 µM.
In the MDA-MB-231, ST10 also compound exerted the strongest anticancer activity. The concentration of ST10 needed to inhibit 50% viability of the cells was 53.4 µM. Similar anticancer activity had compound ST8 (IC50 = 56.4 µM). The moderate anticancer activity against this cell line was exhibited by ST3. The IC50 value was 78.6 µM. Other synthesized compounds had IC50 above 100 µM.
The results presented in this paper indicate that in both cell lines, the strongest cytotoxic properties were exhibited by compound ST10. However, high anticancer activity against MDA-MB-231 was also exerted by ST8. The biological activity of obtained thiadiazoles (ST1–ST15) depends on the position and kind of the substituent of N.
A significant problem with many anticancer compounds is their high toxicity to normal cells. Therefore, the effect of novel 1,3,4-thiadiazole derivatives on the viability of normal cell lines (fibroblasts) was investigated. It was shown that all new compounds had weaker cytotoxic activity on normal cell lines than on breast cancer cell lines.
Further research indicated that all tested compounds (ST1–ST15) lead to inhibition of DNA biosynthesis in MCF-7 and MDA-MB-231, and as the used concentration increased, the effect was enhanced (Figure 3).
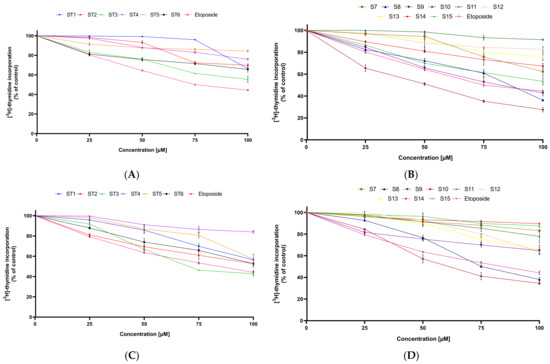
Figure 3.
The anti-proliferative effect of the tested compounds (ST1–ST15) against MCF-7 (A,B) and MDA-MB-231 (C,D). Mean ± SD values from three independent experiments (n = 3) done in duplicate are presented.
In the [3H]-thymidine incorporation assay, ST10 exhibited the strongest anti-proliferative activity against MCF-7 (IC50 = 51.5 µM), followed by ST9 (IC50 = 82 µM) and ST8 (IC50 = 86.5 µM). The rest of the synthesized compounds showed lower inhibitory activity against MCF-7 than the reference drug.
In MDA-MB-231, ST10 compound also exerted the strongest anti-proliferative activity. The IC50 value of ST10 against this cell line was 64.2 µM. Moreover, two compounds, ST3 and ST8, inhibited proliferation of this cell line higher than the reference drug (etoposide). The IC50 values were 73.8 µM and 75.2 µM, respectively. The other derivatives of 1,3,4-thiadiazole had weak anti-proliferative activity against the MDA-MB-231 breast cancer cell line. The IC50 value for each of them was above 100 µM.
These results showed that in both breast cancer cell lines, the strongest anti-proliferative activity was exerted by compound ST10. The moderate cytostatic activity against estrogen-dependent cell lines (MCF-7) had compounds ST8 and ST9, while against estrogen-independent cell line (MDA-MB-231), mild anti-proliferative activity had compound ST3 and compound ST8.
2.3. Docking
The docking simulations technique was performed using AutoDock Tools with 15 compounds. Each compound was docked into each of six different targets. The result of the docking study presented as binding energies and Estimated Inhibition Constant, Ki. From a total of 50 docking modes represented by LGA cluster analysis, the lowest energy docking mode with respective Ki prediction was selected from each docking simulation. For estimating inhibition activity to each target, we compared the binging energies of synthesized compounds and reference molecules from the downloaded spectrum (exception –BAX protein). All results are summarized in Table 1 and Table 2. Docking simulations demonstrated a possible multitarget mode of action for the synthesized compounds. Nevertheless, anticancer properties of ST1-ST15 probably are mainly connected with the activities of Caspase 3 and Caspase 8 and activation of BAX proteins. The most active compounds according to biological assays ST8 and ST15 also demonstrated good results during in silico simulations, which are closed to docking scores of the references ligands.

Table 1.
Docking simulations for activities of the ST1–ST15 to Topoisomerase IIb (PDB: 3qx3), Caspase 3 (PDB: 1GFW), and Caspase 8 (PDB: 3KJN).

Table 2.
Docking simulations for activities of the ST1–ST15 to Bcl-xl (PDB: 2YXJ), Bcl2 (PDB: 2W3L), and BAX (PDB: 1F16).
ST8 makes two hydrogen bonds with ARG260 (1.82 Å) and ARG413 (2.67 Å). Additionally, lipophilic amino acids ALA359, CYS360, and TRP410 are bound to the ligands by alkyl and Pi-alkyl interactions. Additionally, weak carbon–hydrogen bonds with SER316 and GLN358 increase the summary binding energy of ST8 with Caspase 8 (Figure 4).
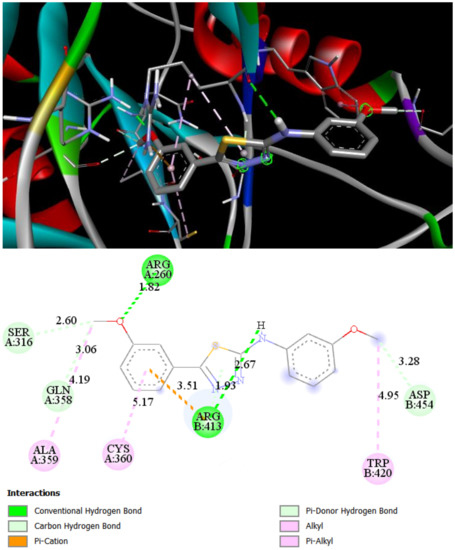
Figure 4.
2D and 3D interaction maps of ST8 with Caspase 8.
The best docking score in simulation with Caspase 8 was achieved by ST15 with a value of binding energy −8.28 kcal/mol and inhibition constant 0.852 μM. Compound ST15 bound to the active site by its oxygen of 4-NO2 with GLY318 as a hydrogen bond acceptor with a bond length 2.18 Å. 3-methoxy group forms the hydrogen bond to the TRP420 with a bond length of 2.14 Å. ARG413 forms with both phenyl rings of the molecules by Pi-cation and amide-Pi stacked interactions (Figure 5).
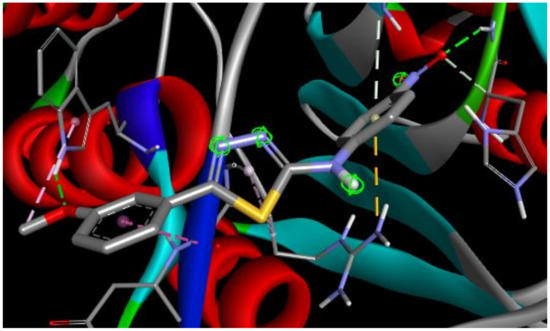
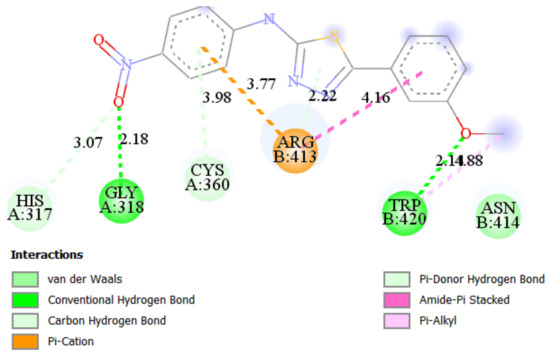
Figure 5.
2D and 3D interaction maps of ST15 with Caspase 8.
The most active according to the biological assays, ST10 shows the best binding affinities (−7.34 kcal/mol, Ki = 4.16 μM) for Caspase 8 and (−7.43 kcal/mol, Ki = 3.56 μM) for Bax-protein. In the ST10–Caspase8 complex, the hydrogen bonds are observed with ARG260 (1.93 Å) and TRP420 (2.89 Å) amino acid residues (Figure 6). The trifluoromethyl group forms a number of halogen interactions with electron-poor species of TYR12 and ARG413. Lipophilic aminoacids ALA359, CYS360, ARG413, and ARG413 connect to the ST10 by different types of hydrophobic interactions. In addition, carbon and Pi-donor hydrogen bonds contribute to the stabilization of the ST10–Caspase 8 complex.
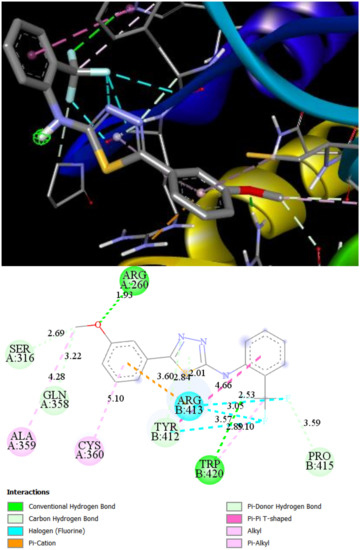
Figure 6.
2D and 3D interaction maps of ST10 with Caspase 8.
Docking simulations of ST10–Bax interaction proposed possible the molecule position within the channel, made by lipophilic amino acids from α2, α3, α5, and α9 helixes of Bax (Figure 7). Additionally, LEU120 forms the hydrogen bond with fluor with a length of 2.58 Å. Such a position of ST10 inside Bax would stabilize the whole protein and increase its apoptotic activity.
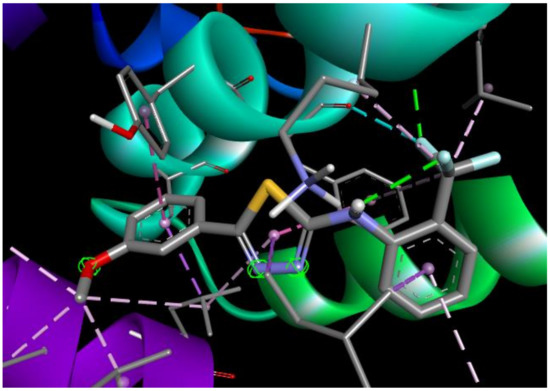
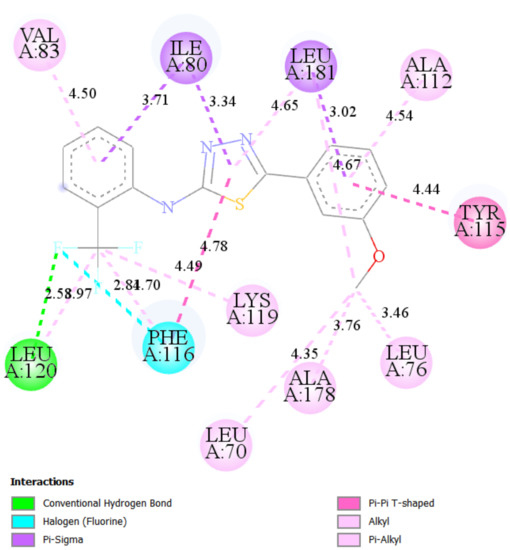
Figure 7.
2D and 3D interaction maps of ST10 with Bax.
From the in silico docking simulations, it is quite evident that 2-(phenylamino)-5-(3-methoxyphenyl)-1,3,4-thiadiazole scaffold is an interesting core for building new molecules with the affinity to Caspase 3 and Caspase 8 as potent anticancer agents.
2.4. ADMET
The drug-likeness calculation is a prediction that determines whether a particular pharmacological agent has properties consistent with its ability for using as an orally active drug. This prediction is based on an already established concept by Lipinski et al., called the Lipinski rule of five. Lipinski’s rules predict the molecular properties related to the pharmacokinetics of molecules. According to this rule, the compound that has MLogP <5, molecular weight <500, the number of hydrogen bond acceptors <10, and the number of hydrogen bond donors <5 possesses good oral bioavailability [33]. The SwissADME computed results demonstrated that ST1–ST15 satisfies Lipinski’s rule of five with zero violations. The predicted logP values revealed that they have optimal lipophilicity (ranging from 1.6 to 3.16) (Table 3). According to obtained data, all compounds are suited to Lipinski’s rule of five, possess high human intestinal absorption but do not get through the blood–brain barrier.

Table 3.
Prediction of the molecular descriptors (Lipinski rules of five) for compounds ST1–ST15 by Swissadme.
According to obtained data, all compounds are suited to Lipinski’s rule of five, which allows supposing ST1–ST15 as drug-like molecules.
Predictions of ADMET properties for ST1–ST15 also were made using the admetSAR portal (http://lmmd.ecust.edu.cn/admetsar2/ accessed on 20 November 2021), and the calculated ADMET parameters were highlighted in Table 4. The results indicated that all the synthesized compounds ST1–ST15 could be administrated orally. In addition, all the compounds demonstrate high CYP inhibitory promiscuity. Additionally, ST11–ST15 belongs to the III category of toxicity, which is characterized as slightly toxic. Category III includes compounds with LD50 values greater than 500 mg/kg but less than 5000 mg/kg, but ST8–ST9 and ST13–ST15 would possess carcinogenic activities according to in silico simulation results. Nevertheless, ST10 does not have this violation.

Table 4.
ADMET profile calculations for ST1–ST15.
3. Materials and Methods
3.1. Chemistry
All the substances were purchased from Sigma-Aldrich (Munich, Germany) and were used without further purification. The 1H and 13C NMR spectra were recorded on the BrukerAvance 300 (Bruker BioSpin GmbH, Rheinstetten, Germany) in DMSO-d6 with tetramethylsilane as the internal standard. IR spectra were recorded by Nicolet 6700 spectrometer (Thermo Scientific, Philadephia, PA, USA). The melting points were determined on the Stuart SMP50 melting point apparatus (Cole Parmer Ltd, Stone, UK) and are uncorrected. The purity of the compounds and the progress of the reaction were monitored by TLC (aluminum sheet 60 F254 plates (Merck Co., Kenilworth, NJ, USA). We used the solvent system CHCl3/EtOH (10:1, v/v). The elemental analyses were determined by a PerkinElmer 2400 series II CHNS/O analyzer (Waltham, MA, USA).
3.1.1. Synthesis of Thiosemicarbazide Derivatives
Synthesis of S1, S2, S3, S7, S8, S9, S11, and S13
First, 0.001 mol of 3-methoxybenzhydrazide and 5 mL of ethanol were placed in a round bottom flask. It was heated under reflux until a clear solution was obtained. An equimolar amount of the appropriate aryl isothiocyanate * was then added to the mixture. It was heated at the boiling point for 1 h. The solution was then cooled until the product precipitated completely. The resulting solid was filtered off and washed with diethyl ether and water.
* 2-chlorophenyl isothiocyanate for S1, 3-chlorophenyl isothiocyanate for S2, 4-chlorophenyl isothiocyanate for S3, 2-methoxyphenyl isothiocyanate for S7, 3-methoxyphenyl isothiocyanate for S8, 4-methoxyphenyl isothiocyanate for S9, 3-trifluoromethylphenyl isothiocyanate for S11, 2-nitrophenyl isothiocyanate for S13.
Synthesis of S4, S5, S6, S10, S12, S14, and S15
First, 0.001 mole of 3-methoxybenzhydrazide was dissolved in 5 mL of ethanol by heating under reflux. An equimolar amount of the appropriate aryl isothiocyanate * was then added to the mixture. The reaction mixture was heated until the product precipitated. Compounds S4, S14, and S15 immediately precipitated. Derivatives S5, S6, S10, and S12 were heated for 30 min until a solid precipitated. The resulting precipitate was filtered off and washed with diethyl ether and water.
* 2-fluorophenyl isothiocyanate for S4, 3-fluorophenyl isothiocyanate for S5, 4-fluorophenyl isothiocyanate for S6, 2-trifluoromethylphenyl isothiocyanate for S10, 4-trifluoromethylphenyl isothiocyanate for S12, 3-nitrophenyl isothiocyanate for S14, 4-nitrophenyl isothiocyanate for S15.
- 4-(2-Chlorophenyl)-1-(3-methoxyphenyl)thiosemicarbazide (S1)
- CAS 891376-78-6
Yield 77% (0.235 g), m.p. 168–170 °C. Spectral data were as follows: IR (cm–1) KBr: 3184 (NH), 2963 (CH aliph.), 1680 (C = O), 1583 (CH arom.), 1334 (C=S), 1267 (C-O-C). 1H NMR (DMSO-d6) δ (ppm): 3.82 (s, 3H, CH3), 7.16 (ddd, 1H, ArH, J = 8.2 Hz, J = 2.6 Hz, J = 1.0 Hz), 7.28 (t, 1H, ArH, J = 8.5 Hz), 7.35 (t, 1H, ArH, J = 5.8 Hz), 7.42 (t, 1H, ArH, J = 8.0 Hz), 7.45–7.56 (m, 4H, ArH), 9.66 (s, 1H, NH), 9.87 (s, 1H, NH), 10.61 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.83, 113.69, 118.15, 120.72, 127.51, 128.42, 129.74, 129.88, 131.30, 131.30, 131.76, 134.27, 137.32, 159.51, 166.43, 182,37. Elemental analysis for C15H14ClN3O2S. Calculated: C 53.65; H 4.20; N 12.51. Found: C 53.35; H 4.10; N 12.40.
- 4-(3-Chlorophenyl)-1-(3-methoxyphenyl)thiosemicarbazide (S2)
- CAS 903081-85-6
Yield 75% (0.252 g), m.p. 170–173 °C. Spectral data were as follows: IR (cm–1) KBr: 3181 (NH), 2969 (CH aliph.), 1664 (C = O), 1587 (CH arom.), 1336 (C=S), 1259 (C-O-C). 1H NMR (DMSO-d6) δ (ppm): 3.82 (s, 3H, CH3), 7.16 (dd, 1H, ArH, J = 8.3 Hz, J = 1.7 Hz), 7.32–7.34 (m, 1H, ArH), 7.36 (t, 2H, ArH, J = 8.1 Hz), 7.43 (t, 2H, ArH, J = 7.9 Hz), 7.47 (d, 1H, ArH, J = 8.0 Hz), 7.51–7.54 (m, 2H, ArH), 9.90 (s, 2H, 2NH), 10.57 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.83, 113.53, 118.21, 120.64, 124.75, 125.22, 125.75, 129.94, 134.23, 159.56, 166.22, 181.45. Elemental analysis for C15H14ClN3O2S. Calculated: C 53.65; H 4.20; N 12.51. Found: C 53.40; H 4.10; N 12.43.
- 4-(4-Chlorophenyl)-1-(3-methoxyphenyl)thiosemicarbazide (S3)
- CAS 891376-71-9
Yield 78% (0.262 g), m.p. 173–175 °C. Spectral data were as follows: IR (cm–1) KBr: 3151 (NH), 2963 (CH aliph.), 1680 (C = O), 1583 (CH arom.), 1334 (C=S), 1267 (C-O-C). 1H NMR (DMSO-d6) δ (ppm): 3.87 (s, 3H, CH3), 7.21 (dd, 1H, ArH, J = 8.2 Hz, J = 3.6 Hz), 7.44 (d, 2H, ArH, J = 8.8 Hz), 7.48 (t, 2H, ArH, J = 7.9 Hz), 7.53–7.60 (m, 3H, ArH), 9.89 (s, 2H, 2NH), 10.61 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.83, 113.58, 118.18, 120.62, 128.32, 129.91, 134.27, 138.73, 159.55, 166.20, 181.58. Elemental analysis for C15H14ClN3O2S. Calculated: C 53.65; H 4.20; N 12.51. Found: C 53.52; H 4.20; N 12.45.
- 4-(2-Fluorophenyl)-1-(3-methoxyphenyl)thiosemicarbazide (S4)
- CAS 891102-96-8
Yield 93% (0.297g), m.p. 182–185 °C. Spectral data were as follows: IR (cm–1) KBr: 3153 (NH), 2973 (CH aliph.), 1662 (C = O), 1588 (CH arom.), 1336 (C=S), 1241 (C-O-C). 1H NMR (DMSO-d6) δ (ppm): 3.81 (s, 3H, CH3), 7.15 (ddd, 1H, ArH, J = 8.2 Hz, J = 2.6 Hz), 7.19 (t, 1H, ArH, J = 8.0 Hz), 7.22–7.25 (m, 1H, ArH), 7.27–7.34 (m, 2H, ArH), 7.51 (t, 1H, ArH, J = 7.9 Hz), 7.66–7.71 (m, 2H, ArH), 9.58 (s, 1H, NH), 9.86 (s, 1H, NH),10.59 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.82, 113.65, 116.15 (d, J = 19.9 Hz), 118.16, 120.70, 124.35, 127.73, 128.59, 129.87, 131.13, 134.26, 157.05, 159.51, 166.31, 182.68. Elemental analysis for C15H14FN3O2S. Calculated: C 56.41; H 4.42; N 13.16. Found: C 56.35; H 4.32; N 13.10.
- 4-(3-Fluorophenyl)-1-(3-methoxyphenyl)thiosemicarbazide (S5)
- CAS 891103-04-1
Yield 65% (0.208 g), m.p. 180–182 °C. Spectral data were as follows: IR (cm–1) KBr: 3171 (NH), 2962 (CH aliph.), 1664 (C = O), 1571 (CH arom.), 1356 (C=S), 1261 (C-O-C). 1H NMR (DMSO-d6) δ (ppm): 3.83 (s, 3H, CH3), 6.98–7.01 (m, 1H, ArH), 7.16 (dd, 1H, ArH, J = 8.2 Hz, J = 2.6 Hz), 7.31–7.32 (m, 1H, ArH), 7.34–7.38 (m, 1H, ArH), 7.43 (t, 1H, ArH, J = 7.9 Hz), 7.52–7.55 (m, 2H, ArH), 9.88 (s, 2H, NH), 10.56 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.82, 113.31, 113.54, 118.20, 120.59, 129.93, 134.23, 141.43, 141.57, 159,54, 166.18, 181.09. Elemental analysis for C15H14FN3O2S. Calculated: C 56.41; H 4.42; N 13.16. Found: C 56.30; H 4.35; N 13.15.
- 4-(4-Fluorophenyl)-1-(3-methoxyphenyl)thiosemicarbazide (S6)
- CAS 905231-63-2
Yield 95% (0.303 g), m.p. 175–177 °C. Spectral data were as follows: IR (cm–1) KBr: 3173 (NH), 2829 (CH aliph.), 1667 (C = O), 1582 (CH arom.), 1316 (C=S), 1280 (C-O-C). 1H NMR (DMSO-d6) δ (ppm): 3.81 (s, 3H, CH3), 7.15–7.18 (m, 3H, ArH), 7.42 (t, 3H, ArH, J = 7.9 Hz), 7.52–7.55 (m, 2H, ArH), 9.77 (s, 2H, NH),10.54 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 57.40, 113.58, 115.15, 117.73, 120.63, 128.66, 129.89, 134.31, 136.05, 159.53, 160.75, 166.22, 181.88. Elemental analysis for C15H14FN3O2S. Calculated: C 56.41; H 4.42; N 13.16. Found: C 56.40; H 4.35; N 13.15.
- 1-(3-Methoxyphenyl)-4-(2-methoxyphenyl)thiosemicarbazide (S7)
- CAS 891370-81-3
Yield 89% (0.295 g), m.p. 135–137 °C. Spectral data were as follows: IR (cm–1) KBr: 3166 (NH), 2967 (CH aliph.), 1688 (C = O), 1593 (CH arom.), 1356 (C=S), 1279 (C-O-C). 1H NMR (DMSO-d6) δ (ppm): 3.83 (s, 3H, CH3), 3.88 (s, 3H, CH3), 6.98 (d, 1H, ArH; J = 8.4 Hz), 9.22 (s, 1H, NH), 7.04–7.08 (m, 1H, ArH), 7.28 (dd, 1H, ArH, J = 8.3 Hz, J = 2.0 Hz), 7.35–7.45 (m, 4H, ArH), 8.58 (d, 1H, NH, J = 2.0 Hz), 9.90 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.81, 56.20, 107.20, 113.37, 118.25, 119.74, 126.16, 128.26, 130.05, 133.29, 151.84, 158.87, 166.48. Elemental analysis for C16H17N3O3S. Calculated: C 57.99; H 5.17; N 12.68. Found: C 57.75; H 5.12; N 12.50.
- 1-(3-Methoxyphenyl)-4-(3-methoxyphenyl)thiosemicarbazide (S8)
- CAS 891369-83-8
Yield 87% (0.288 g), m.p. 172–175 °C. Spectral data were as follows: IR (cm–1) KBr: 3173 (NH), 2944 (CH aliph.), 1667 (C = O), 1575 (CH arom.), 1357 (C=S), 1251 (C-O-C). 1H NMR (DMSO-d6) δ (ppm): 3.73 (s, 3H, CH3), 3.81 (s, 3H, CH3), 6.73 (dd, 1H, ArH, J = 8.1 Hz, J = 2.5 Hz), 7.02–7.05 (m, 1H, ArH), 7.15 (ddd, 1H, ArH, J = 8.2 Hz, J = 2.7 Hz), 7.22 (t, 1H, ArH, J = 8.1 Hz), 7.41 (t, 1H, ArH, J = 7.9 Hz), 7.50–7.54 (m, 2H, ArH), 9.72 (s, 2H, 2NH), 10.51 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 54.92, 55.78, 107.20, 113.01, 116.27, 121.37, 127.79, 129.89, 133.29, 140.81, 158.87, 167.54. Elemental analysis for C16H17N3O3S. Calculated: C 57.99; H 5.17; N 12.68. Found: C 57.78; H 5.10; N 12.60.
- 1-(3-methoxyphenyl)-4-(4-methoxyphenyl)thiosemicarbazide (S9) [34]
All data are the same as in our previous work.
- 4-(2-Trifluoromethylphenyl)1-(3-methoxyphenyl)thiosemicarbazide (S10)
- CAS 891611-85-1
Yield 43% (0.159 g), m.p. 153–155 °C. Spectral data were as follows: IR (cm–1) KBr: 3152 (NH), 2962 (CH aliph.), 1667 (C = O), 1583 (CH arom.), 1368 (C=S), 1266 (C-O-C). 1H NMR (DMSO-d6) δ (ppm): 3.82 (s, 3H, CH3), 7.15 (dd, 1H, ArH, J = 8.2 Hz, J = 2.6 Hz), 7.42 (t, 1H, ArH, J = 7.9 Hz), 7.44–7.46 (m, 1H, ArH), 7.49 (t, 1H, ArH, J = 7.7 Hz), 7.52–7.55 (m, 2H, ArH), 7.68–7.72 (m, 2H, ArH), 9.58 (s, 1H, NH), 9.86 (s, 1H, NH), 10.57 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.80, 113.69, 118.11, 120.71, 126.58,127.77, 129.86, 133.00, 134.30, 137.78, 138.27, 159.50, 166.53, 183.19. Elemental analysis for C16H14F3N3O2S. Calculated: C 52.03; H 3.82; N 11.38. Found: C 51.89; H 3.66; N 11.29.
- 4-(3-Trifluoromethylphenyl)-1-(3-methoxyphenyl)thiosemicarbazide (S11)
- CAS 903815-86-1
Yield 95% (0.351 g), m.p. 178–180 °C. Spectral data were as follows: IR (cm–1) KBr: 3170 (NH), 2838 (CH aliph.), 1660 (C = O), 1579 (CH arom.), 1333 (C=S), 1287 (C-O-C). 1H NMR (DMSO-d6) δ (ppm): 3.83 (s, 3H, CH3), 7.17 (dd, 1H, ArH, J = 8.3 Hz, J = 2.6 Hz), 7.44 (t, 2H, ArH, J = 7.9 Hz), 7.59 (m, 3H, ArH), 7.85 (d, 2H, ArH, J = 7.8 Hz) 9.99 (s, 2H, 2NH), 10.61 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.83, 113.60, 118.23, 120.63, 121.85, 122.37, 123.65, 125.45, 129.50, 129.96, 134.15, 140.57, 159.58, 166.22, 181.54. Elemental analysis for C16H14F3N3O2S. Calculated: C 52.03; H 3.82; N 11.38. Found: C 51.93; H 3.75; N 11.32.
- 4-(4-Trifluoromethylphenyl)-1-(3-methoxyphenyl)thiosemicarbazide (S12)
Yield 65% (0.240 g), m.p. 195–197 °C. Spectral data were as follows: IR (cm–1) KBr: 3161 (NH), 2969 (CH aliph.), 1669 (C = O), 1579 (CH arom.), 1322 (C=S), 1289 (C-O-C). 1H NMR (DMSO-d6) δ (ppm): 3.83 (s, 3H, CH3), 7.17 (dd, 1H, ArH, J = 8.5 Hz, J = 2.9 Hz), 7.43 (t, 1H, ArH, J = 7.9 Hz), 7.52–7.55 (m, 2H, ArH), 7.70 (d, 2H, ArH, J = 7.9 Hz), 7.76 (bs, 2H, ArH), 9.98 (s, 2H, 2NH), 10.60 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.83, 113.59, 118.24, 120.63, 123.91, 125.60 (d, J = 34.8 Hz), 126.24, 129.95, 134.20, 143.56, 159.58, 166.22, 181.46. Elemental analysis for C16H14F3N3O2S. Calculated: C 52.03; H 3.82; N 11.38. Found: C 51.98; H 3.79; N 11.27.
- 1-(3-Methoxyphenyl)-4-(2-nitrophenyl)thiosemicarbazide (S13)
Yield 88% (0.305 g), m.p. 150–153 °C. Spectral data were as follows: IR (cm–1) KBr: 3177 (NH), 2836 (CH aliph.), 1661 (C = O), 1581 (CH arom.), 1337 (C=S), 1265 (C-O-C). 1H NMR (DMSO-d6) δ (ppm): 3.83 (s, 3H, CH3), 7.18 (d, 1H, ArH, J = 8.3 Hz), 7.44 (t, 2H, ArH, J = 7.9 Hz), 7.54–7.57 (m, 2H, ArH), 7.72–7.75 (m, 1H, ArH), 7.95 (d, 1H, ArH, J = 8.2 Hz), 8.04 (dd, 1H, ArH, J = 8.2 Hz, J = 1.5 Hz), 10.15 (s, 1H, NH), 10.19 (s, 1H, NH), 10.73 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.83, 113.60, 118.36, 120.66, 125.26, 126.87, 129.70, 130.02, 134.00, 144.20, 159.58, 166.55, 181.51. Elemental analysis for C15H14N4O4S. Calculated: C 42.02; H 4.07; N 16.18. Found: C 41.98; H 4.02; N 16.10.
- 1-(3-Methoxyphenyl)-4-(3-nitrophenyl)thiosemicarbazide (S14)
- CAS 891561-77-6
Yield 96% (0.333 g), m.p. 159–162 °C. Spectral data were as follows: IR (cm–1) KBr: 3159 (NH), 2845 (CH aliph.), 1667 (C = O), 1579 (CH arom.), 1335 (C=S), 1261 (C-O-C). 1H NMR (DMSO-d6) δ (ppm): 3.82 (s, 3H, CH3), 7.17 (dd, 1H, ArH, J = 8.2 Hz, J = 2.9 Hz), 7.43 (t, 1H, ArH, J = 7.9 Hz), 7.52–7.56 (m, 2H, ArH), 7.61 (t, 1H, ArH, J = 8.2 Hz), 8.01 (d, 2H, ArH, J = 8.2 Hz), 8.41–8.45 (m, 1H, ArH), 10.08 (s, 2H, 2NH), 10.63 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.85, 113.59, 118.29, 119.97, 120.64, 129.22, 132.27, 134.13, 141.01, 147.12, 159.60, 166.22, 181.44. Elemental analysis for C15H14N4O4S. Calculated: C 42.02; H 4.07; N 16.18. Found: C 41.95; H 4.00; N 16.18.
- 1-(3-Methoxyphenyl)-4-(4-nitrophenyl)thiosemicarbazide (S15)
- CAS 891561-93-6
Yield 92% (0.319 g), m.p. 165–167 °C. Spectral data were as follows: IR (cm–1) KBr: 3159 (NH), 2842 (CH aliph.), 1651 (C = O), 1563 (CH arom.), 1328 (C=S), 1258 (C-O-C). 1H NMR (DMSO-d6) δ (ppm): 3.82 (s, 3H, CH3), 7.16 (dd, 1H, ArH, J = 8.7 Hz, J = 3.2 Hz), 7.43 (t, 1H, ArH, J = 7.9 Hz), 7.50–7.55 (m, 2H, ArH), 7.90 (d, 2H, ArH, J = 8.4 Hz), 8.21 (d, 2H, ArH, J = 9.1 Hz), 9.98 (s, 2H, 2NH), 10.60 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 53.13, 113.59, 118.29, 119.74, 120.23, 120.64, 124.06, 125,37, 128.90, 130.00, 132.28, 134.11, 141.01, 158.48, 165.67, 180.15. Elemental analysis for C15H14N4O4S. Calculated: C 42.02; H 4.07; N 16.18. Found: C 41.95; H 4.00; N 16.15.
3.1.2. Synthesis of 1,3,4-thiadiazoles ST1–ST15
First, 0.2 g of the thiosemicarbazide derivatives obtained in stage I was weighed and placed in conical flasks. Then, 0.5 mL of concentrated sulfuric acid (VI) was added dropwise with thorough stirring. The reaction mixture was stirred until the precipitate dissolved, and 2 h were given for cooling at room temperature. Then, finely crushed ice was added to the reaction flasks and mixed intensively. A solid precipitated. After the complete dissolution of the ice, the solid product was filtered off. The product was dried thoroughly with filter paper. Then, crystallization from butanol was performed to obtain pure ST1–ST15 compounds.
- 2-(2-Chlorophenylamino)-5-(3-methoxyphenyl)-1,3,4-thiadiazole (ST1)
Yield 35% (0.078 g), m.p. 115–118 °C. Spectral data were as follows: IR (cm–1) KBr: 3151 (NH), 2883 (CH aliph.), 1665 (C=N), 1584 (CH arom.), 1261 (C-O-C), 715 (C-S). 1H NMR (DMSO-d6) δ (ppm): 3.84 (s, 3H, CH3), 7.08 (d, 1H, ArH, J = 4.0 Hz), 7.12–7.15 (m, 1H, ArH), 7.40–7.45 (m, 4H, ArH), 7.53 (d, 1H, ArH, J = 4.0 Hz), 8.30 (bs, 1H, ArH), 10.03 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.77, 111.85, 116.59, 119.88, 122.64, 123.86, 125.01, 128.41, 130.23, 130.23, 130.95, 131.99, 137.77, 160.10, 165.36. Elemental analysis for C15H12ClN3OS. Calculated: C 56.69; H 3.81; N 13.22. Found: C 56.55; H 3.75; N 13.20.
- 2-(3-Chlorophenylamino)-5-(3-methoxyphenyl)-1,3,4-thiadiazole (ST2)
Yield 53% (0.127 g), m.p. 192–194 °C. Spectral data were as follows: IR (cm–1) KBr: 3059 (NH), 2808 (CH aliph.), 1625 (C=N), 1564 (CH arom.), 1268 (C-O-C), 760 (C-S). 1H NMR (DMSO-d6) δ (ppm): 3.84 (s, 3H, CH3), 7.06–7.11 (m, 2H, ArH), 7.36–7.47 (m, 5H, ArH), 7.95 (t, 1H, ArH, J = 2.1 Hz), 10.78 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.81, 111.98, 116.44, 116.75, 117.40, 120.03, 122.08, 131.01, 131.22, 131.82, 134.01, 142.23, 158.61, 160.14, 164.18. Elemental analysis for C15H12ClN3OS. Calculated: C 56.69; H 3.81; N 13.22. Found: C 56.65; H 3.78; N 13.15.
- 2-(4-Chlorophenylamino)-5-(3-methoxyphenyl)-1,3,4-thiadiazole (ST3)
- CAS 1202973-36-1
Yield 70% (0.174 g), m.p. 205–207 °C. Spectral data were as follows: IR (cm–1) KBr: 3148 (NH), 2833 (CH aliph.), 1582 (CH arom.), 1283 (C-O-C), 757 (C-S). 1H NMR (DMSO-d6) δ (ppm): 3.84 (s, 3H, CH3), 7.07–7.09 (m, 1H, ArH), 7.39 (s, 3H, ArH), 7.42 (t, 2H, ArH, J = 3.9 Hz), 7.73–7.67 (m, 2H, ArH), 10.70 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.79, 111.95, 116.16, 119.52 120.91, 124.86, 129.42, 130.56, 131.89, 139.86, 158.71, 160.11, 164.29. Elemental analysis for C15H12ClN3OS. Calculated: C 56.69; H 3.81; N 13.22. Found: C 56.59; H 3.55; N 13.18.
- 2-(2-Fluorophenylamino)-5-(3-methoxyphenyl)-1,3,4-thiadiazole (ST4)
Yield 45% (0.126 g), m.p. 125–128 °C. Spectral data were as follows: IR (cm–1) KBr: 3205 (NH), 2836 (CH aliph.), 1619 (C=N), 1581 (CH arom.), 1289 (C-O-C), 749 (C-S). 1H NMR (DMSO-d6) δ (ppm): 3.84 (s, 3H, CH3), 7.08–7.10 (m, 2H, ArH), 7.24 (t, 1H, ArH, J = 7.8 Hz), 7.31 (t, 1H, ArH, J = 9.8 Hz), 7.41–7.45 (m, 3H, ArH), 8.38–8.40 (m, 1H, ArH), 10.32 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.80, 111.87, 115.74, 115.86, 116.65, 119.94, 121.19, 123.72, 123.77, 125.28, 128.83, 128.90, 130.98, 131.97, 158.98, 160.12, 164.75. Elemental analysis for C15H12FN3OS. Calculated: C 59.79; H 4.01; N 13.95. Found: C 59.56; H 3.87; N 13.65.
- 2-(3-Fluorophenylamino)-5-(3-methoxyphenyl)-1,3,4-thiadiazole (ST5)
Yield 66% (0.129 g), m.p. 197–199 °C. Spectral data were as follows: IR (cm–1) KBr: 3168 (NH), 2837 (CH aliph.), 1620 (C=N), 1599 (CH arom.), 1217 (C-O-C), 747 (C-S). 1H NMR (DMSO-d6) δ (ppm): 3.84 (s, 3H, CH3), 6.81–6.87 (m, 1H, ArH), 7.08 (dt, 1H, ArH, J = 6.8 Hz, J = 2.5 Hz), 7.30–7.35 (m, 1H, ArH), 7.37–7.46 (m, 4H, ArH), 7.71 (dt, 1H, ArH, J = 11.8 Hz, J = 2.3 Hz), 10.80 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 54.79, 104.87 (d, J = 27.0 Hz), 108.82 (d, J = 21.1 Hz), 111.31, 113.44, 116.31, 120.02, 131.19 (d, J = 9.8 Hz), 142.54 (d, J = 11.5 Hz), 158.51, 160.13, 162.23, 163.83, 164.23. Elemental analysis for C15H12FN3OS. Calculated: C 59.79; H 4.01; N 13.95. Found: C 59.68; H 3.99; N 13.78.
- 2-(4-Fluorophenylamino)-5-(3-methoxyphenyl)-1,3,4-thiadiazole (ST6)
Yield 46% (0.132 g), m.p. 172–174 °C. Spectral data were as follows: IR (cm–1) KBr: 3177 (NH), 2831 (CH aliph.), 1628 (C=N), 1579 (CH arom.), 1280 (C-O-C), 774 (C-S). 1H NMR (DMSO-d6) δ (ppm): 3.83 (s, 3H, CH3), 7.07 (dt, 1H, ArH, J = 7.5 Hz, J = 2.0 Hz), 7.21 (t, 2H, ArH, J = 8.9 Hz), 7.38–7.45 (m, 3H, ArH), 7.65–7.70 (m, 2H, ArH), 10.57 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.80, 111.88, 116.17 (d, J = 22.2 Hz), 116.58, 119.71, 119.76, 119.92, 130.96, 131.99, 137.49, 157.04, 157.84, 158.63, 160.12, 164.74. Elemental analysis for C15H12FN3OS. Calculated: C 59.79; H 4.01; N 13.95. Found: C 59.69; H 3.99; N 13.69.
- 5-(3-Methoxyphenyl)-2-(2-methoxyphenylamino)-1,3,4-thiadiazole (ST7)
- CAS 1203294-57-8
Yield 59% (0.165 g), m.p. 185–188 °C Spectral data were as follows: IR (cm–1) KBr: 3264 (NH), 2941 (CH aliph.), 1598 (C=N), 1560 (CH arom.), 1285 (C-O-C), 767 (C-S). 1H NMR (DMSO-d6) δ (ppm): 3.83 (s, 3H, CH3), 3.88 (s, 3H, CH3), 6.70 (d, 1H, ArH, J = 4.0 Hz), 7.06 (dd, 1H, ArH, J = 0.6 Hz, J = 4.0 Hz), 7.28 (dd, 1H, ArH, J = 0.6 Hz, J = 4.0 Hz), 7.35–7.45 (m, 2H, ArH), 7.43 (t, 1H, ArH, J = 4.0 Hz), 8.59 (d, 1H, ArH, J = 1.0 Hz)), 9.90 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.78, 56.41, 110.32, 111.80, 116.48, 117.59, 119.84, 121.08, 130.95, 132.13, 141.25, 149.15, 158.49, 160.09, 165.02. Elemental analysis for C16H15N3O2S. Calculated: C 61.32; H 4.82; N 13.41. Found: C 61.24; H 4.59; N 13.29.
- 5-(3-Methoxyphenyl)-2-(3-methoxyphenylamino)-1,3,4-thiadiazole (ST8)
- CAS 1203389-12-1
Yield 47% (0.128 g), m.p. 153–155 °C.Spectral data were as follows: IR (cm–1) KBr: 3261 (NH), 2939 (CH aliph.), 1600 (C=N), 1560 (CH arom.), 1288 (C-O-C), 773 (C-S). 1H NMR (DMSO-d6) δ (ppm): 3.76 (s, 3H, CH3), 3.84 (s, 3H, CH3), 7.00 (dd, 1H, ArH, J = 8.4 Hz, J = 1.9 Hz), 7.07 (dt, 1H, ArH, J = 7.3 Hz, J = 2.4 Hz), 7.41–7.43 (m, 4H, ArH,), 7.62 (d, 1H, ArH, J = 8.4 Hz), 10.63 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.80, 55.86, 101.79, 108.03, 111.75, 116.71, 119.99, 129.71, 130.15, 130.96, 131.98, 142.70, 157.30, 158.08, 160.12, 164.50. Elemental analysis for C16H15N3O2S. Calculated: C 61.32; H 4.82; N 13.41. Found: C 61.34; H 4.79; N 13.49.
- 5-(3-Methoxyphenyl)-2-(4-methoxyphenylamino)-1,3,4-thiadiazole (ST9)
Yield 32% (0.078 g), m.p. 145–148 °C. Spectral data were as follows: IR (cm–1) KBr: 3077 (NH), 2838 (CH aliph.), 1624 (C=N), 1583 (CH arom.), 1283 (C-O-C), 779 (C-S). 1H NMR (DMSO-d6) δ (ppm):) δ (ppm): 3.83 (s, 6H, CH3), 7.08 (dt, 1H, ArH, J = 7.2 Hz, J = 2.4 Hz), 7.40–7.43 (m, 5H, Ar Hz), 7.70 (d, 2H, ArH, J = 8.9 Hz), 10.70 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 52.89, 60.83, 109.46, 110.71, 114.19, 116.49, 119.41, 120.11, 131.35, 132.07, 134.83, 154.52, 157.03, 159.27, 165.46. Elemental analysis for C16H15N3O2S. Calculated: C 61.32; H 4.82; N 13.41. Found: C 61.28; H 4.79; N 13.20.
- 2-(2-Trifluorometylophenylamino)-5-(3-methoxyphenyl)-1,3,4-thiadiazole (ST10)
Yield 22% (0.033 g), m.p. 167–169 °C. Spectral data were as follows: IR (cm–1) KBr: 3130 (NH), 2834 (CH aliph.), 1618 (C=N), 1587 (CH arom.), 1270 (C-O-C), 746 (C-S). 1H NMR (DMSO-d6) δ (ppm): 3.81 (s, 3H, CH3), 7.03–7.07 (m, 1H, ArH), 7.32–7.43 (m, 4H, ArH), 7.69–7.77 (m, 2H, ArH), 7.89 (bs, 1H, ArH), 9.92 (s, 1H, NH).13C NMR (DMSO-d6) δ (ppm): 53.92, 112.45, 115.93, 119.60, 123.33, 125.65, 127.06, 130.93, 130.61, 133.33, 161.25. Elemental analysis for C16H12N3F3OS. Calculated: C 54.70; H 3.44; N 11.96. Found: C 54.56; H 3.37; N 11.68.
- 2-(3-Trifluorometylophenylamino)-5-(3-methoxyphenyl)-1,3,4-thiadiazole (ST11)
Yield 63% (0.210 g), m.p. 183–185 °C. Spectral data were as follows: IR (cm–1) KBr: 3051 (NH), 2826 (CH aliph.), 1625 (C=N), 1582 (CH arom.), 1272 (C-O-C), 769 (C-S). 1H NMR (DMSO-d6) δ (ppm): 3.84 (s, 3H, CH3), 7.07–7.11 (m, 1H, ArH), 7.37 (d, 1H, ArH, J = 7.7 Hz), 7.42–7.44 (m, 3H, ArH), 7.60 (t, 1H, ArH, J = 8.0 Hz), 7.77 (d, 1H, ArH, J = 7.8 Hz), 8.27 (bs, 1H, ArH), 10.93 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.10, 111.97, 113.94, 116.82, 118.66, 120.07, 121.53, 123.74, 125.54, 130.34 (d, J = 31.4 Hz), 130.88 (d, J = 38.1 Hz), 131.78, 141.54, 158.77, 160.14, 164.17. Elemental analysis for C16H12N3F3OS. Calculated: C 54.70; H 3.44; N 11.96. Found: C 54.68; H 3.47; N 11.98.
- 2-(4-Trifluorometylophenylamino)-5-(3-methoxyphenyl)-1,3,4-thiadiazole (ST12)
- CAS 1203413-88-0
Yield 34% (0.078 g), m.p. 145–147 °C. Spectral data were as follows:IR (cm–1) KBr: 3060 (NH), 2874 (CH aliph.), 1614 (C=N), 1581 (CH arom.), 1258 (C-O-C), 747 (C-S). 1H NMR (DMSO-d6) δ (ppm): 3.78 (s, 3H, CH3), 7.07–7.12 (m, 1H, ArH), 7.42–7.47 (m, 3H, ArH), 7.73 (d, 2H, ArH, J = 8.8 Hz), 7.87 (d, 2H, ArH, J = 8.5 Hz), 10.79 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.81, 112.07, 117.76, 120.06, 122.26 (d, J = 32.0 Hz), 124.11, 125.91, 126.95, 131.00, 131.77, 144.18, 159.04, 160.14, 163.98. Elemental analysis for C16H12N3F3OS. Calculated: C 54.70; H 3.44; N 11.96. Found: C 54.66; H 3.47; N 11.98.
- 5-(3-Methoxyphenyl)-2-(2-nitrophenylamino)-1,3,4-thiadiazole (ST13)
Yield 18% (0.052 g), m.p. 155–158 °C. Spectral data were as follows: IR (cm–1) KBr: 3270 (NH), 2864 (CH aliph.), 1602 (C=N), 1587 (CH arom.), 1271 (C-O-C), 751 (C-S). 1H NMR (DMSO-d6) δ (ppm): 3.83 (s, 3H, CH3), 7.09 (dd, 1H, ArH, J = 7.4 Hz, J = 2.5 Hz), 7.26 (t, 1H, ArH, J = 7.4 Hz), 7.42–7.47 (m, 3H, ArH), 7.75 (t, 1H, ArH, J = 7.6 Hz), 8.07 (d, 1H, ArH, J = 8.2 Hz), 8.26 (bs, 1H, ArH), 10.62 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.82, 111.99, 116.94, 120.05, 122.93, 123.73, 126.15, 131.06, 131.74, 135.08, 135.56, 138.58, 160.16. Elemental analysis for: C15H12N4O3S. Calculated: C 54.88; H 3.68; N 17.06. Found: C 54.67; H 3.49; N 17.00.
- 5-(3-Methoxyphenyl)-2-(3-nitrophenylamino)-1,3,4-thiadiazole (ST14) [35]
Yield 60% (0.189 g), m.p. 160–163 °C. Spectral data were as follows: IR (cm–1) KBr: 3265 (NH), 2889 (CH aliph.), 1624 (C=N), 1594 (CH arom.), 1272 (C-O-C), 781 (C-S). 1H NMR (DMSO-d6) δ (ppm): 3.84 (s, 3H, CH3), 7.08–7.12 (m, 1H, ArH), 7.44–7.45 (m, 3H, ArH), 7.65 (t, 1H, ArH, J = 8.2 Hz), 7.86–7.91 (m, 2H, ArH), 8.84 (t, 1H, ArH, J = 2.2 Hz), 11.15 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 56.37, 111.93, 112.05, 116.88, 120.11, 123.25, 130.92, 131.03, 131.71, 141.86, 148.86, 159.10, 160.15, 164.02. Elemental analysis for: C15H12N4O3S. Calculated: C 54.88; H 3.68; N 17.06. Found: C 54.77; H 3.55; N 17.02.
- 5-(3-Methoxyphenyl)-2-(4-nitrophenylamino)-1,3,4-thiadiazole (ST15)
- CAS 1203176-20-8
Yield 42% (0.127 g), m.p. 162–164 °C. Spectral data were as follows: IR (cm–1) KBr: 3217 (NH), 2838 (CH aliph.), 1622 (C=N), 1507 (CH arom.), 1219 (C-O-C), 747 (C-S). 1H NMR (DMSO-d6) δ (ppm): 3.89 (s, 3H, CH3), 7.10–7.12 (m, 1H, ArH), 7.44–7.46 (m, 3H, ArH), 7.88 (d, 2H, ArH, J = 9.4 Hz), 8.28 (d, 2H, ArH, J = 9.4 Hz), 11.32 (s, 1H, NH). 13C NMR (DMSO-d6) δ (ppm): 55.84, 112.20, 116.94, 117.53, 120.15, 126.02, 131.05, 131.59, 141.41, 146.63, 159.96, 160.15, 163.53. Elemental analysis for: C15H12N4O3S. Calculated: C 54.88; H 3.68; N 17.06. Found: C 54.87; H 3.48; N 17.02.
3.2. Cell Culture
Two breast cancer cell lines (MCF-7 and MDA-MB-231) and fibroblasts as normal cell lines, which were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA), were used to investigate the effects of the novel synthesized compounds. The cells were cultured in DMEM (Corning, Kennebunk, ME, USA), supplemented with FetalBovine Serum (Eurx, Gdansk, Poland) at concentration 10% and penicillin/streptomycin at 1%. Cells were cultured on 100 mm plates (Sarstedt, Newton, NC, USA) and then kept in an incubator that provided optimal growth conditions (5% CO2, temperature: 37 °C, humidity at 90–95% level). After reaching 80% confluence, cells were detached from the bottom of the plate using 0.05% trypsin supplemented with 0.2% EDTA (Gibco, San Diego, CA, USA). Then, using a hemocytometer, the number of cells was quantified and seeded at density 5 × 105 cells per well in six-well plates (Sarstedt, Newton, NC, USA) in 2 mL of DMEM. In this research, cells that obtained 80% of confluency were used.
3.3. Cell Viability Assay
The effect of novel synthesized derivatives (ST1-ST15) on the survival of breast cancer cells (MCF-7 and MDA-MB-231) and normal human cells(fibroblasts) was performed using the MTT assay according to the method described previously [36]. In short, both cancer cell lines were incubated 24 h with tested compounds and etoposide (reference drug) at the following concentrations: 25, 50, 75, and 100 µM. Following incubation, the medium was removed, and cells were washed three times with PBS. Then cells were incubated for 4 h with 1 mL PBS and 0.05 mL of MTT (5 mg/mL). The obtained product was dissolved in DMSO. The absorbance of the solutions was determined spectrophotometrically at λ = 570 nm using UV-VIS Helios Gamma (Unicam/ThermoFisher Scientific Inc., Cleveland, OH, USA). The compounds ST13–ST15 interfere with MTT and were tested only by [3H]-thymidine incorporation.
3.4. Proliferation [3H]-Thymidine
The anti-proliferative activity of novel synthesized compounds ST1–ST15 against breast cancer cell lines was analyzed by the method described in the literature [37]. The cells (MCF-7 and MDA-MB-231) were cultured on six-well plates (Sarstedt, Newton, NC, USA) and incubated with tested compounds and etoposide at various concentrations and 0.5 µCi [3H]-thymidine for 24 h at 37 °C. Then, the cells were rinsed twice with 1 mL of 0.05 M Tris/HCl buffer with 0.11 M NaCl, twice with 1 mL of 5% trichloroacetic acid (Stanlab, Lublin, Polska). After washing, the cells were solubilized with 1 mL of 0.1 NaOH containing 1% SDS (Sigma-Aldrich, St Louis, MO, USA). The solubilized cells were transferred to scintillation vials and filled with 2 mL of Ultima Gold XR. Reading occurred in a scintillation counter (1900 TR, TRI-CARB, Packard, Perkin Elmer, Inc., San Jose, CA, USA).
3.5. Docking
Aiming to investigate the docking scores of the bioactive conformations of the synthesized compounds and their specificity for different enzymes, which are potential anticancer targets, docking of compounds ST1–ST15 were performed at the active sites of the topoisomerase IIb (PDB code 3QX3), Caspase 3 (PDB code 1GFW), Caspase 8 (PDB code 3KJN), Bcl-xl (PDB code 2YXJ), Bcl2 (PDB: 2W3L), and BAX (PDB: 1F16). All structures were downloaded from the Protein Data Bank. With the use of AutoDock Tools version 4.2.6, all bound ligands and water molecules were removed, polar hydrogen atoms were added, nonpolar hydrogen atoms were merged, and rotatable bonds were defined. Kollman charges were added. The chemical structures of the compounds were drawn by Biovia Draw, and their 3D structures were optimized by Hyperchem 7.5 using MM+ and PM3 quantum techniques, respectively. ST1–ST15 were allowed with active rotatable bonds making them flexible, and the Gasteiger charges are assigned. The three-dimensional grid boxes were created, the spacing between grid points was 0.375 Å, and the grid maps representing the intact ligand in the actual docking target site were made by Auto Grid Tool. For more accurate results, we changed Lamarckian genetic algorithm (LGA) parameters from the default setting to enhanced, which includes 50 runs, 300 conformational possibilities, 50 populations, 2,500,000 energy evaluations, a maximum number of 106 energy evaluations, a mutation rate of 0.02, and a crossover rate of 0.80. Results differing by <2.0 Å in positional root-mean-square deviation (RMSD) were clustered together and represented by the result with the most favorable free energy of binding. To ensure that the ligand orientations and positions obtained from the docking studies were likely to represent valid and reasonable potential binding modes of the inhibitors, the docking methods and parameters used were validated by redocking experiments. Reference ligands were docked into the native proteins to determine the ability of the AutoDock program to reproduce the orientation and position of the ligand observed in the crystal structure Figure 8. Only for BAX protein, Zinc 14750348 was selected as reference ligands according to reported data [38].
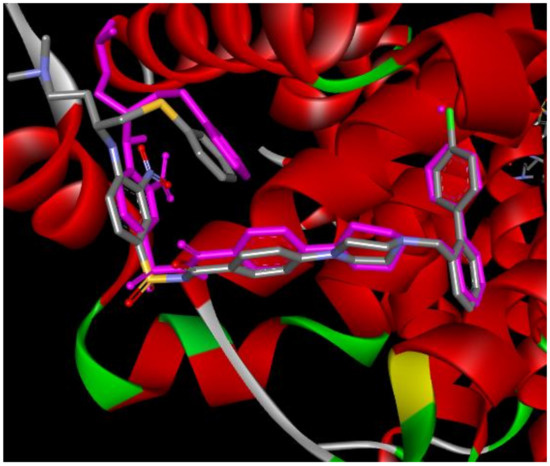
Figure 8.
Real and predicted (pink-colored) position of N3C inside Bcl-xl protein (PDB: 2YXJ, RMSD = 1.29).
3.6. In Silico Drug-Likeness Prediction
The drug-likeness of the synthesized compounds (ST1–ST15) were estimated by calculating molecular descriptors using freely accessible web server Swiss ADME (http://www.swissadme.ch/index.php accessed on 20 November 2021) [39]. ADMET properties were computed by the admetSAR portal (http://lmmd.ecust.edu.cn/admetsar2/ accesed on 20 November 2021).
4. Conclusions
New thiosemicarbazide and 1,3,4-thiadiazole derivatives were synthesized and evaluated for their in vitro anticancer activity. The results showed that in both breast cancer cell lines, the strongest anti-proliferative activity was exerted by compound ST10. The IC50 values of this compound against MCF-7 and MDA-MB-231 breast cancer cells were 49.6 µM and 53.4 µM, respectively. The moderate cytostatic activity against estrogen-dependent cell line (MCF-7) had compounds ST8 and ST9. While against estrogen-independent cell line (MDA-MB-231), mild anti-proliferative activity had compound ST3 and compound ST8. A significant problem with many anticancer compounds is no selectivity for cancer cells. New 1,3,4-thiadiazole derivatives had weaker cytotoxic activity on normal cell lines (fibroblasts) than on breast cancer cell lines. Docking simulations demonstrated a possible multitarget mode of action for the synthesized compounds. Nevertheless, anticancer properties of ST1–ST15 probably are mainly connected with the activities of Caspase 3 and Caspase 8 and activation of BAX proteins.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27061814/s1, 1H NMR, 13C NMR spectra.
Author Contributions
Conceptualization, M.W., A.B. and K.B.; methodology, M.W., A.B., A.S. and D.K.; investigation, S.J., D.K., A.G., A.S. and J.N.; data curation, M.W. and A.B.; writing—original draft preparation, M.W., S.J., D.K. and A.B.; writing—review and editing, K.B., A.B. and A.G.; supervision, M.W., K.B., A.B. and S.M.; visualization, D.K.; funding acquisition, M.W. and A.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Medical University of Lublin, Poland, grant number DS 15. This research was funded by the Medical University of Bialystok, Poland, grant number SUB/2/DN/21/002/2229.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The details of the data supporting the report results in this research were included in the paper and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are/ are not available from the authors upon request.
References
- Balasubramanian, R.; Rolph, R.; Morgan, C.; Hamed, H. Genetics of breast cancer: Management strategies and risk-reducing surgery. Br. J. Hosp. Med. 2019, 80, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Costa, B.; Amorim, I.; Gärtner, F.; Vale, N. Understanding breast cancer: From conventional therapies to repurposed drugs. Eur. J. Pharm. Sci. 2020, 151, 105401. [Google Scholar] [CrossRef] [PubMed]
- Licznerska, B.; Baer-Dubowska, W. Estrogenintracrinology: Therapy and chemoprevention of breast cancer. Postepy Hig. Med. Dosw. 2010, 64, 220–230. [Google Scholar]
- Liu, Z.; Delavan, B.; Roberts, R.; Tong, W. Lessons learned from two decades of anticancer drugs. Trends Pharmacol. Sci. 2017, 38, 852–872. [Google Scholar] [CrossRef] [Green Version]
- Friedman, R.; Boye, K.; Flatmark, K. Molecular modelling and simulations in cancer research. Biochim. Biophys. Acta 2013, 1836, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janowska, S.; Paneth, A.; Wujec, M. Cytotoxic Properties of 1, 3, 4-Thiadiazole Derivatives—A Review. Molecules 2020, 25, 4309. [Google Scholar] [CrossRef] [PubMed]
- Kalidhar, U.; Kau, A. 1, 3, 4-Thiadiazole derivatives and their biological activities: A Review. Res. J. Pharm. Biol. Chem. Sci. 2011, 2, 1091–1106. [Google Scholar]
- Morsy, S.M.; Badawi, A.M.; Cecchi, A.; Scozzafava, A.; Supuran, C.T. Carbonic anhydrase inhibitors. Biphenylsulfonamides with inhibitory action towards the transmembrane, tumor-associated isozymes IX possess cytotoxic activity against human colon, lung and breast cancer cell lines. J. Enzym. Inhib. Med. Chem. 2009, 24, 499–505. [Google Scholar] [CrossRef]
- El-Ashmaway, M.B.; El-Sherbeny, M.A.; El-Sayed, N.S. Synthesis, in vitro antitumor activity and DNA-binding affinity of novel thiadiazolopyrimidine and thiadiazoloquinazoline derivatives. Mans. J. Pharm. Sci. 2010, 26, 60–68. [Google Scholar]
- Yang, X.H.; Wen, Q.; Zhao, T.T.; Sun, J.; Li, X.; Xing, M.; Zhu, H.L. Synthesis, biological evaluation, and molecular docking studies of cinnamic acyl 1,3,4-thiadiazole amide derivatives as novel antitubulin agents. Bioorg. Med. Chem. 2012, 20, 1181–1187. [Google Scholar] [CrossRef]
- Hosseinzadeh, L.; Khorand, A.; Aliabadi, A. Discovery of 2-Phenyl-N-(5-(trifluoromethyl)-1,3,4-thiadiazol-2-yl) acetamide Derivatives as Apoptosis Inducers via the Caspase Pathway with Potential Anticancer Activity. Arch. Pharm. 2013, 346, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Dawood, K.M.; Eldebss, T.M.; El-Zahabi, H.S.; Yousef, M.H.; Metz, P. Synthesis of some new pyrazole-based 1,3-thiazoles and 1,3,4-thiadiazoles as anticancer agents. Eur. J. Med. Chem. 2013, 70, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Aliabadi, A.; Hasanvand, Z.; Kiani, A.; Mirabdali, S.S. Synthesis and In-vitro Cytotoxicity Assessment of N-(5-(Benzylthio)-1,3,4-thiadiazol-2-yl)-2-(4-(trifluoromethyl) phenyl) acetamide with Potential Anticancer Activity. Iran J. Pharm. Res. 2013, 12, 687. [Google Scholar] [PubMed]
- Gomha, S.M.; Salah, T.A.; Abdelhamid, A.O. Synthesis, characterization, and pharmacological evaluation of some novel thiadiazoles and thiazoles incorporating pyrazole moiety as anticancer agents. Monatsh. Chem. 2015, 146, 149–158. [Google Scholar] [CrossRef]
- Polkam, N.; Rayam, P.; Anireddy, J.S.; Yennam, S.; Anantaraju, H.S.; Dharmarajan, S.; Balasubramanian, S. Synthesis, in vitro anticancer and antimycobacterial evaluation of new 5-(2,5-dimethoxyphenyl)-1,3,4-thiadiazole-2-amino derivatives. Bioorg. Med. Chem. Lett. 2015, 25, 1398–1402. [Google Scholar] [CrossRef]
- Yadagiri, B.; Gurrala, S.; Bantu, R.; Nagarapu, L.; Polepalli, S.; Srujana, G.; Jain, N. Synthesis and evaluation ofbenzosuberone embedded with 1,3,4-oxadiazole, 1,3,4-thiadiazole and 1,2,4-triazole moieties as new potential anti proliferative agents. Bioorg. Med. Chem. Lett. 2015, 25, 2220–2224. [Google Scholar] [CrossRef]
- Li, S.; Jing, F.; Fu, X.; Zhao, J.; Wang, X.; Li, B.; Chen, B. Synthesis and antitumor activities of disulfide derivatives containing 1, 3, 4-thiadiazole moiety. Chin. J. Org. Chem. 2015, 35, 2624–2628. [Google Scholar] [CrossRef]
- Gomha, S.M.; Kheder, N.A.; Abdelhamid, A.O.; Mabkhot, Y.N. One pot single step synthesis and biological evaluation of some novel bis (1,3,4-thiadiazole) derivatives as potential cytotoxic agents. Molecules 2016, 21, 1532. [Google Scholar] [CrossRef] [Green Version]
- Abdelhamid, A.O.; Gomha, S.M.; Abdelriheem, N.A.; Kandeel, S.M. Synthesis of new 3-heteroarylindoles as potential anticancer agents. Molecules 2016, 21, 929. [Google Scholar] [CrossRef]
- Vudhgiri, S.; Koude, D.; Veeragoni, D.K.; Misra, S.; Prasad, R.B.N.; Jala, R.C.R. Synthesis and biological evaluation of 5-fatty-acylamido-1,3,4-thiadiazole-2-thioglycosides. Bioorg. Med. Chem. Lett. 2017, 27, 3370–3373. [Google Scholar] [CrossRef]
- Rezaei, Z.; Moghimi, S.; Javaheri, R.; Asadi, M.; Mahdavi, M.; Shabani, S.; Foroumadi, A. Synthesis and biological evaluation of 1,3,4-thiadiazole linked phthalimide derivatives as anticancer agents. Lett. Drugs Des. Discov. 2017, 14, 1138–1144. [Google Scholar] [CrossRef]
- Abdelhamid, A.O.; Gomha, S.M.; Abdelrehem, N.A.; Shalaby, A.M.; Kandeel, S.M. Synthesis and biological evaluation of some novel thiadiazole-benzofuran hybrids as potential antitumor agents. Synth. Comm. 2018, 48, 677–684. [Google Scholar] [CrossRef]
- Azaam, M.M.; Kenawy, E.R.; El-din, A.S.B.; Khamis, A.A.; El-Magd, M.A. Antioxidant and anticancer activities of α-aminophosphonates containing thiadiazole moiety. J. Saudi Chem. Soc. 2018, 22, 34–41. [Google Scholar] [CrossRef]
- Upadhyay, P.K.; Mishra, P. Synthesis, antimicrobial and anticancer activities of 5-(4-substituted phenyl)-1,3,4-thiadiazole-2-amines. Rasayan J. Chem. 2017, 10, 254–262. [Google Scholar]
- Nassar, I.F.; Att-Allah, S.R.; Hemdan, M.M. Utility of thiophene-2-carbonyl isothiocyanate as a precursor for the synthesis of 1,2,4-triazole, 1,3,4-oxadiazole and 1, 3, 4-thiadiazole derivatives with evaluation of their antitumor and antimicrobial activities. Phosphorus Sulfur Silicon Relat. Elem. 2018, 193, 630–636. [Google Scholar] [CrossRef]
- Rashdan, H.R.; Farag, M.M.; El-Gendey, M.S.; Mounier, M.M. Toward rational design of novel anti-cancer drugs based on targeting, solubility, and bioavailability exemplified by 1, 3, 4-thiadiazole derivatives synthesized under solvent-free conditions. Molecules 2019, 24, 2371. [Google Scholar] [CrossRef] [Green Version]
- Sekhar, D.C.; Rao, D.V.; Rao, A.T.; Kumar, U.L.; Jha, A. Design and synthesis of 1,3,4-thiadiazole derivatives as novel anticancer and antitubercular agents. Russ. J. Gen. Chem. 2019, 89, 770–779. [Google Scholar] [CrossRef]
- Mahapatra, D.K.; Dadure, K.M.; Haldar, A.G. Exploring the anti-breast cancer (against MCF-7 Cell Line) potentials of uracil substituted hippuric acid based 1, 3, 4-thiadiazole compound. Int. J. Pharm. Life Sci. 2019, 10, 6013–6015. [Google Scholar]
- Devi, E.R.; Sreenivasulu, R.; Rao, K.P.; Nadh, R.V.; Sireesha, M. Novel 1,3,4-Thiadiazole Linked Amide Derivatives of Pteridone: Synthesis and Study of Anticancer Activities. Lett. Org. Chem. 2020, 17, 54–60. [Google Scholar] [CrossRef]
- Almajan, G.L.; Innocenti, A.; Puccetti, L.; Manole, G.; Barbuceanu, S.; Saramet, I.; Supuran, C.T. Carbonic anhydrase inhibitors. Inhibition of the cytosolic and tumor-associated carbonic anhydrase isozymes I, II, and IX with a series of 1,3,4-thiadiazole-and 1,2,4-triazole-thiols. Bioorg. Med. Chem. Lett. 2005, 15, 2347–2352. [Google Scholar] [CrossRef]
- Esteves-Souza, A.; Rodrigues-Santos, C.E.; Del Cistia, C.D.N.; Silva, D.R.D.; Sant’Anna, C.M.R.; Echevarria, A. Solvent-free synthesis, DNA-topoisomerase II activity and molecular docking study of new asymmetrically N, N’-substituted ureas. Molecules 2012, 17, 12882–12894. [Google Scholar] [CrossRef]
- Radi, M.; Crespan, E.; Botta, G.; Falchi, F.; Maga, G.; Manetti, F.; Botta, M. Discovery and SAR of 1,3,4-thiadiazole derivatives as potent Abl tyrosine kinase inhibitors and cytodifferentiating agents. Bioorg. Med. Chem. Lett. 2008, 18, 1207–1211. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A. Lead-and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar]
- Popiołek, Ł.; Biernasiuk, A.; Paruch, K.; Malm, A.; Wujec, M. Synthesis and in vitro antimicrobial activity screening of new pipemidic acid derivatives. Arch. Pharm. Res. 2018, 41, 633–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Rideout, D.; Ramnarayan, K.; Tsai, C.-Y.; Yalamoori, V.V.; Wu, F.; Loweth, C.; Elabdellaoui, H.; Fung, L.; Brady, T.P. Preparation of Thiadiazoles and Other Organosulfur Inhibitors of Tyrosine Phosphatases. Patent, PCT International Application WO 2003032916 A2 20030424, 2003. [Google Scholar]
- Gornowicz, A.; Szymanowska, A.; Mojzych, M.; Bielawski, K.; Bielawska, A. The Effect of Novel 7-methyl-5-phenyl-pyrazolo[4,3-e]tetrazolo[4,5-b][1,2,4]triazine Sulfonamide Derivatives on Apoptosis and Autophagy in DLD-1 and HT-29 Colon Cancer Cells. Int. J. Mol. Sci. 2020, 21, 5221. [Google Scholar] [CrossRef]
- Hermanowicz, J.M.; Szymanowska, A.; Sieklucka, B.; Czarnomysy, R.; Pawlak, K.; Bielawska, A.; Pawlak, D. Exploration of novel heterofused 1,2,4-triazine derivative in colorectal cancer. J. Enzym. Inhib. Med. Chem. 2021, 36, 535–548. [Google Scholar] [CrossRef]
- Liu, Z.; Ding, Y.; Ye, N.; Wild, C.; Chen, H.; Zhou, J. Direct activation of Bax protein for cancer therapy. Med. Res. Rev. 2016, 36, 313–341. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).