Self-Supplying Guide RNA-Mediated CRISPR/Cas12a Fluorescence System for Sensitive Detection of T4 PNKP
Abstract
1. Introduction
2. Results and Discussion
2.1. The Working Principle of the Proposed Method
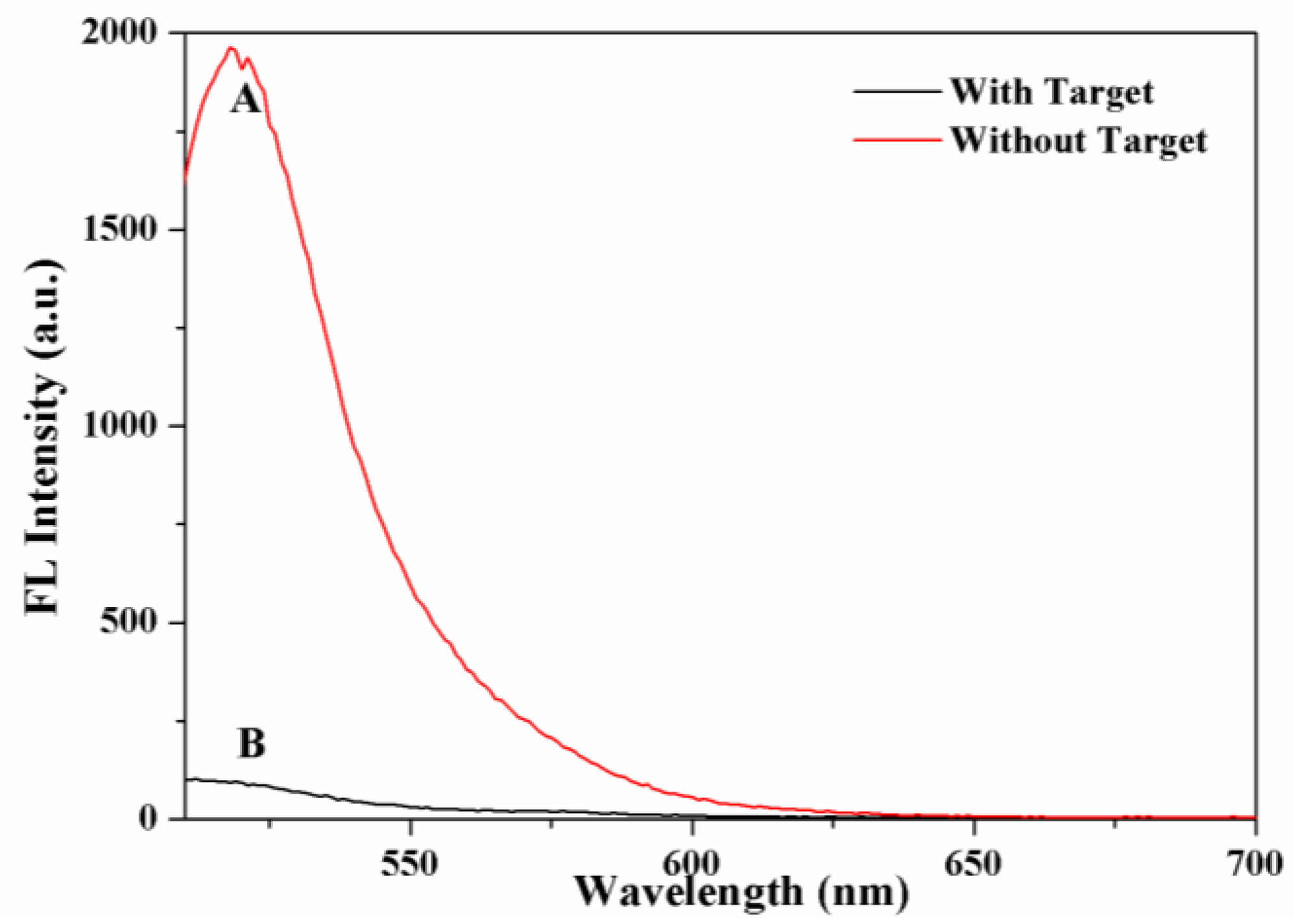
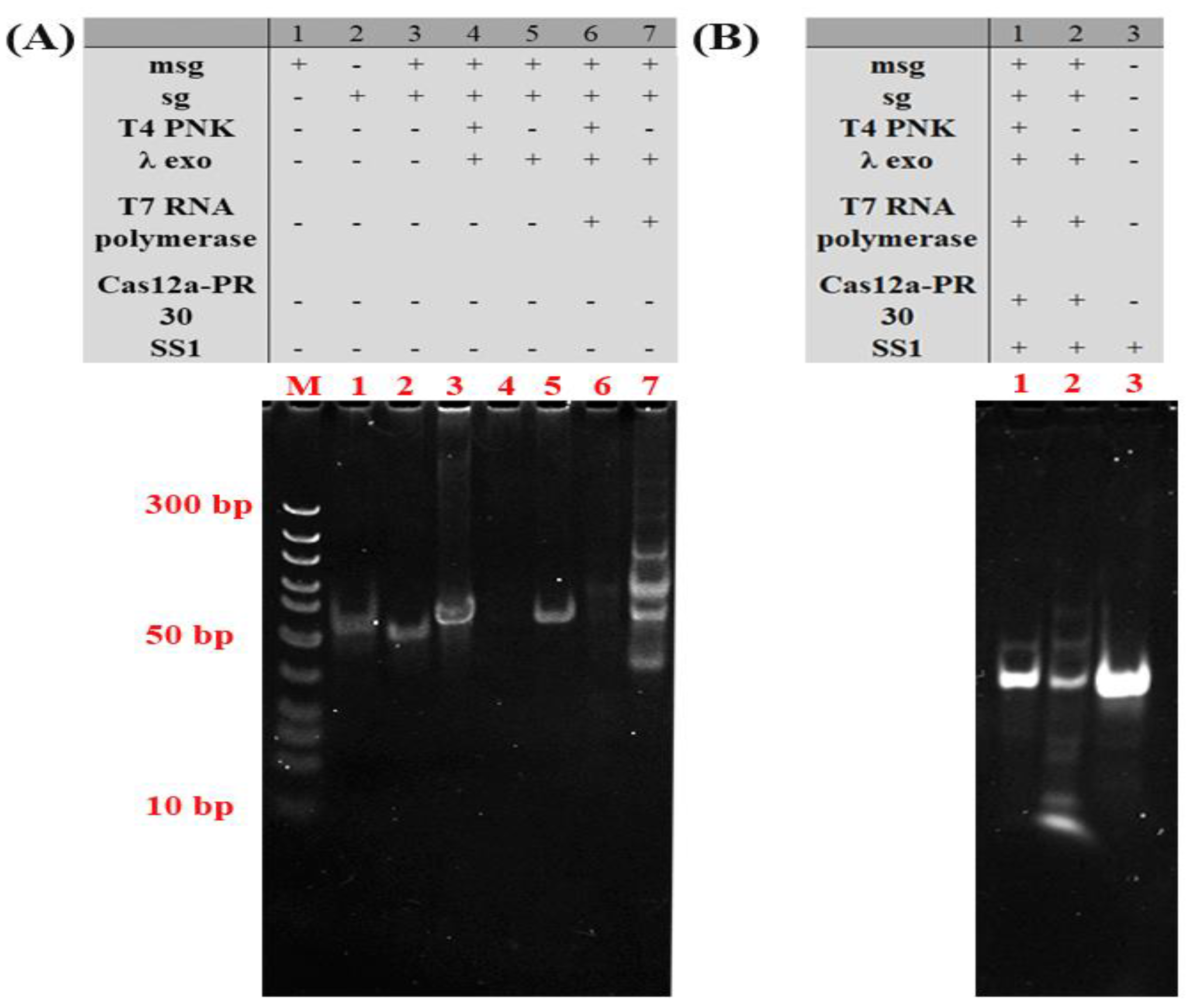
2.2. Feasibility Verification of the Sensing System
2.3. Optimization of Assay Conditions
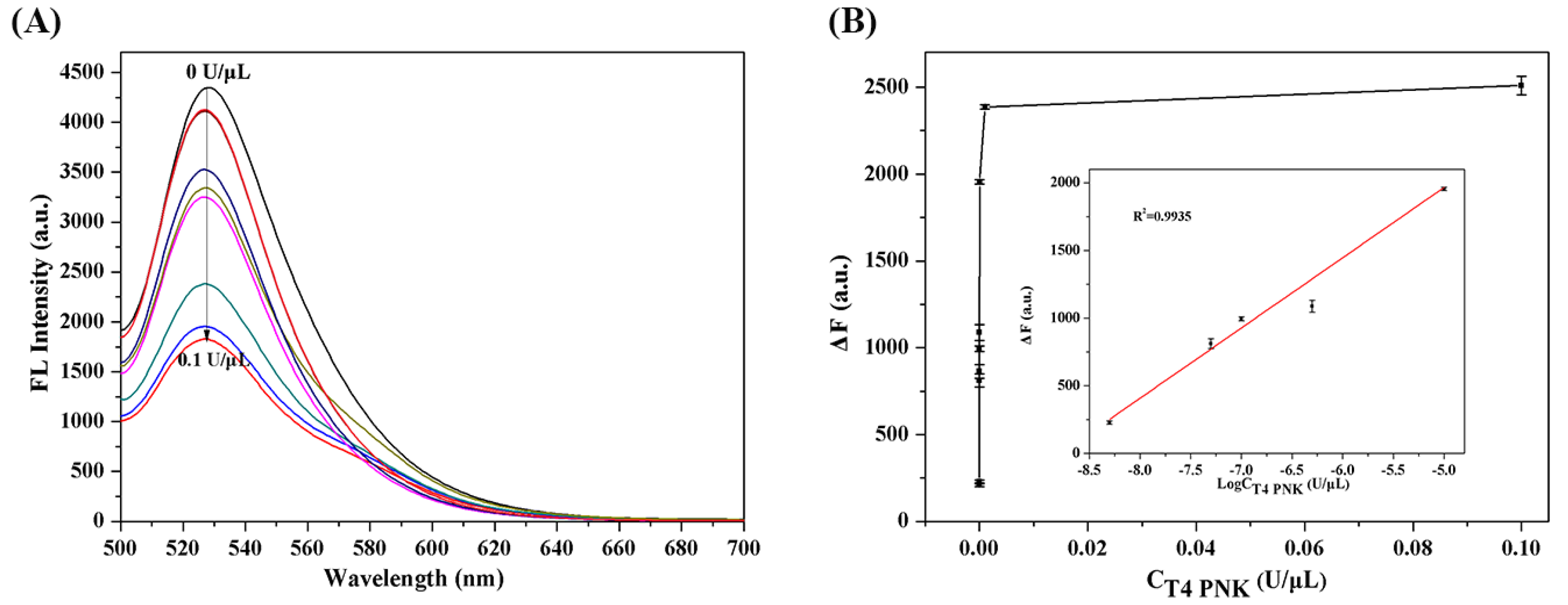
2.4. Analytical Performance of the T4 PNKP Detection
2.5. Analysis of Complex Biological Sample
3. Materials and Methods
3.1. Reagents and Instrumentation
3.2. T4 PNKP Activity Assay
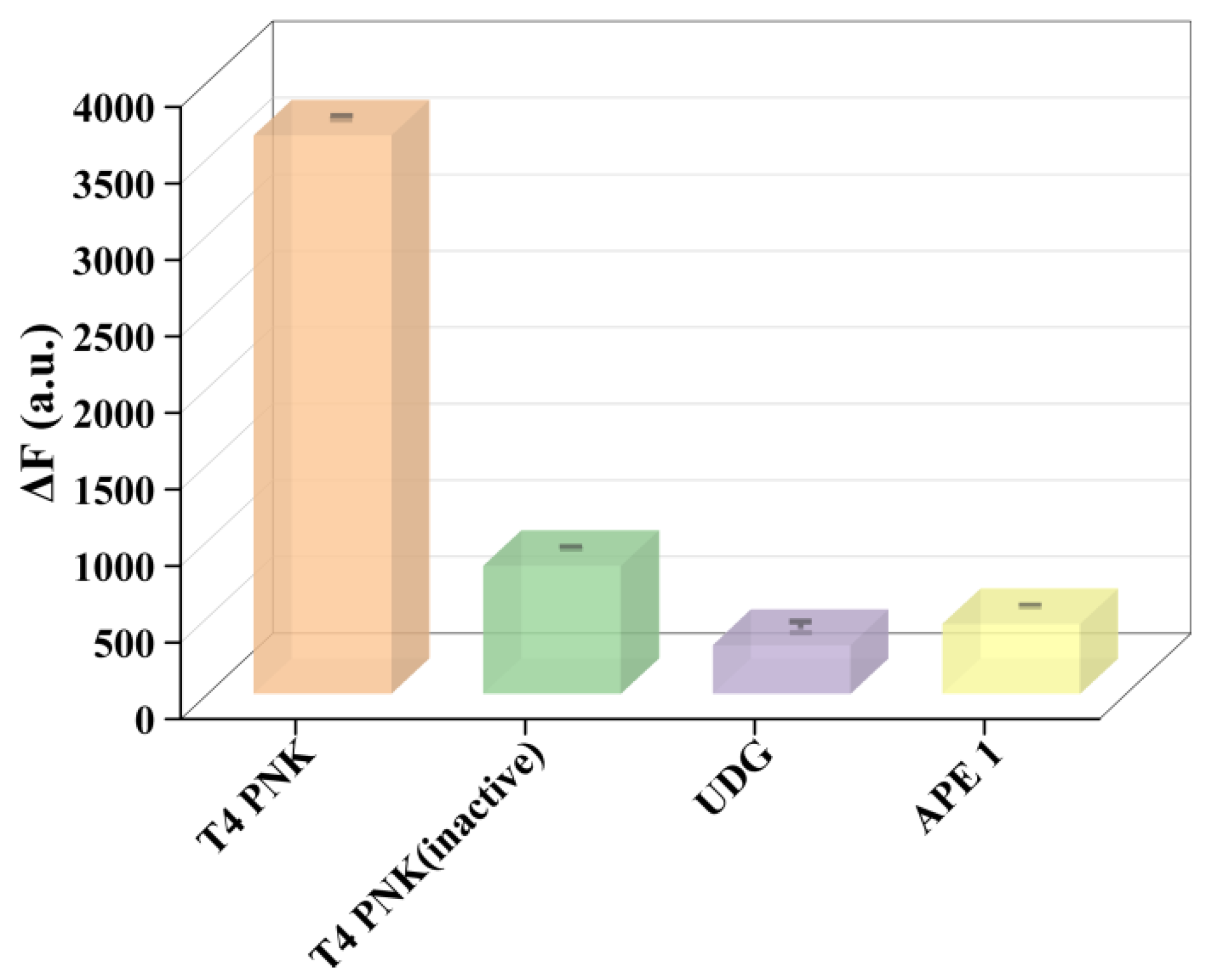
3.3. Selectivity Assay
3.4. Polyacrylamide Gel Electrophoresis Analysis
3.5. T4 PNKP Activity Detection in Diluted Cell Extracts
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mao, J.; Chen, X.; Xu, H.; Xu, X. DNAzyme-driven DNAWalker Biosensor for Amplified Electrochemical Detection of T4 polyNucleotide Kinase Activity and Inhibition. J. Electroanal. Chem. 2020, 874, 114470. [Google Scholar] [CrossRef]
- Luo, R.; Zhou, H.; Dang, W.; Long, Y.; Tong, C.; Xie, Q.; Daniyal, M.; Liu, B.; Wang, W. A DNAzyme-rGO Coupled Fluorescence Assay for T4 PNKP Activity in Vitro and Intracellular Imaging. Sens. Actuat B-Chem. 2020, 310, 127884. [Google Scholar] [CrossRef]
- Tahbaz, N.; Subedi, S.; Weinfeld, M. Role of Polynucleotide Kinase/phosphatase in Mitochondrial DNA Repair. Nucleic Acids Res. 2011, 40, 3484–3495. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.K. DNA Damage, Mutagenesis and Cancer. Int. J. Mol. Sci. 2018, 19, 970. [Google Scholar] [CrossRef]
- Lin, L.; Shi, D.; Li, Q.; Wang, G.F.; Zhang, X.J. Detection of T4 Polynucleotide Kinase Based on a MnO2 Nanosheet-3,3′,5,5′-Tetramethylbenzidine (TMB) Colorimetric System. Anal. Methods 2016, 8, 4119–4126. [Google Scholar] [CrossRef]
- Lin, M.; Wan, H.; Zhang, J.; Wang, Q.; Hu, X.; Xia, F. Electrochemical DNA Sensors Based on MoS2-AuNPs for Polynucleotide Kinase Activity and Inhibition Assay. ACS Appl. Mater. Interfaces 2020, 12, 45814–45821. [Google Scholar] [CrossRef]
- Song, Z.; Li, Y.; Teng, H.; Ding, C.; Xu, G.; Luo, X. Designed Zwitterionic Peptide Combined with Sacrificial Fe-MOF for Low Fouling and Highly Sensitive Electrochemical Detection of T4 Polynucleotide Kinase. Sens. Actuators B Chem. 2020, 305, 127329. [Google Scholar] [CrossRef]
- He, Y.; Cheng, L.; Yang, Y.; Chen, P.; Qiu, B.; Guo, L.; Wang, Y.; Lin, Z.; Hong, G. Label-free Homogeneous Electrochemical Biosensor for HPV DNA Based on Entropy-Driven Target Recycling and Hyperbranched Rolling Circle Amplification. Sens. Actuators B Chem. 2020, 320, 128407. [Google Scholar] [CrossRef]
- Zhang, L.; Fan, W.; Jia, D.; Feng, Q.; Ren, W.; Liu, C. Microchamber-Free Digital Flow Cytometric Analysis of T4 Polynucleotide Kinase Phosphatase Based on Single-Enzyme-to-Single-Bead Space-Confined Reaction. Anal. Chem. 2021, 93, 14828–14836. [Google Scholar] [CrossRef]
- Zhuang, J.Y.; Lai, W.Q.; Xu, M.D.; Zhou, Q.; Tang, D.P. Plasmonic AuNP/g-C3N4 Nanohybrid-based Photoelectrochemical Sensing Platform for Ultrasensitive Monitoring of Polynucleotide Kinase Activity Accompanying DNAzyme-Catalyzed Precipitation Amplification. ACS Appl. Mater. Interfaces 2015, 7, 8330–8338. [Google Scholar] [CrossRef]
- Li, P.P.; Cao, Y.; Mao, C.J.; Jin, B.K.; Zhu, J.J. TiO2/g-C3N4/CdS Nanocomposite-based Photoelectrochemical Biosensor for Ultrasensitive Evaluation of T4 Polynucleotide Kinase Activity. Anal. Chem. 2019, 91, 1563–1570. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Luo, R.; Xie, Q.; You, P.D.; Yu, H.H.; Qin, Y.; Tong, C.Y.; Danial, M.; Liu, B.; Wang, W. A New Fluorescence Method for Monitoring PNKP Activity in Vitro, Natural Compounds Screening and Intracellular Imaging. Sens. Actuators B Chem. 2021, 329, 129203. [Google Scholar] [CrossRef]
- Zhang, T.; Shen, Y.; Ge, J.; Wang, W.; Qu, L.; Li, Z. A highly Sensitive Fluorescence Method for the Detection of T4 polynucleotide Kinase Phosphatase Based on Polydopamine Nanotubes. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 267, 120594. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Cheng, X.; Xu, G.; Wei, F.; Bao, J.; Mei, J.; Ren, D.; Hu, Q.; Cen, Y. A Nanoplatform based on Metal–Organic Frameworks and Coupled Exonuclease Reaction for the Fluorimetric Determination of T4 Polynucleotide Kinase Activity and Inhibition. Mikrochim. Acta 2020, 187, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jie, G.; Li, C.; Zhao, Y.; Kuang, Q.; Niu, S. Fluorescent Mn: ZnCdS@ZnS and CdTe Quantum Dots Probes on SiO2 Microspheres for Versatile Detection of Carcinoembryonic Antigen and Monitoring T4 Polynucleotide Kinase Activity. ACS Appl. Nano Mater. 2019, 2, 4637–4645. [Google Scholar] [CrossRef]
- Wang, Y.S.; Chen, G.; Jia, H.; Cui, X.Y.; Xu, L.Y.; Cao, Z.; She, Y.X.; Jin, F.; Zhang, Y.D.; Wang, J.; et al. Hacimuftuoglu A., Jia H.Y., Lamu S.Q., Hammock B.D. A Competitive Immunoassay for Detecting Triazophos based on Fluorescent Catalytic Hairpin Self-Assembly. Microchim. Acta 2022, 189, 114. [Google Scholar] [CrossRef]
- Tao, J.; Liu, Z.; Zhu, Z.; Zhang, Y.; Wang, H.; Pang, P.; Yang, C.; Yang, W. Electrochemical Detection of T4 polynucleotide Kinase Activity Based on Magnetic Fe3O4@TiO2 Nanoparticles Triggered by a Rolling Circle Amplification Strategy. Talanta 2022, 241, 123272. [Google Scholar] [CrossRef]
- Jiang, H.; Xu, Y.; Dai, L.; Liu, X.; Kong, D. Ultrasensitive, Label-free Detection of T4 ligase and T4 polynucleotide KinaseBased on Target-triggered Hyper-branched Rolling Circle Amplification. Sens. Actuators B Chem. 2018, 260, 70–77. [Google Scholar] [CrossRef]
- Xia, X.; Ma, B.; Zhang, T.; Lu, Y.; Khan, M.R.; Hu, Y.; Lei, C.; Deng, S.; He, Q.; He, G.; et al. G-Quadruplex-Probing CRISPR-Cas12 Assay for Label-Free Analysis of Foodborne Pathogens and Their Colonization In Vivo. ACS Sens. 2021, 6, 3295–3302. [Google Scholar] [CrossRef]
- Peng, S.; Tan, Z.; Chen, S.; Lei, C.; Nie, Z. Integrating CRISPR-Cas12a with a DNA Circuit as a Generic Sensing Platform for Amplified Setection of MicroRNA. Chem. Sci. 2020, 11, 7362–7368. [Google Scholar] [CrossRef]
- Lv, Z.; Wang, Q.; Yang, M. Multivalent Duplexed-Aptamer Networks Regulated a CRISPR-Cas12a System for Circulating Tumor Cell Detection. Anal. Chem. 2021, 93, 12921–12929. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ye, Q.; Chen, M.; Zhou, B.; Zhang, J.; Pang, R.; Xue, L.; Wang, J.; Zeng, H.; Wu, S.; et al. An Ultrasensitive CRISPR/Cas12a based Electrochemical Biosensor for Listeria Monocytogenes Detection. Biosens. Bioelectron. 2021, 179, 113073. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Zheng, B.; Li, J.T.; Gao, J.L.; Liu, Y.H.; Pang, D.W.; Tang, H.W. Holographic Optical Tweezers and Boosting Upconversion Luminescent Resonance Energy Transfer Combined Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas12a Biosensors. ACS Nano 2021, 15, 8142–8154. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Tian, W.; Liu, X.; Ren, W.; Liu, C. New CRISPR-Derived microRNA Sensing Mechanism Based on Cas12a Self-Powered and Rolling Circle Transcription-Unleashed Real-Time crRNA Recruiting. Anal. Chem. 2020, 92, 6702–6708. [Google Scholar] [CrossRef]
- Tian, W.M.; Liu, X.L.; Wang, G.T.; Liu, C.H. A Hyperbranched Transcription-Activated CRISPR-Cas12a Dignal Amplification Strategy for Sensitive MicroRNA Sensing. Chem. Commun. 2020, 56, 13445–13448. [Google Scholar] [CrossRef]
- Peng, L.; Zhou, J.; Liu, G.; Yin, L.; Ren, S.; Man, S.; Ma, L. CRISPR-Cas12a based Aptasensor for Sensitive and Selective ATP Detection. Sens. Actuators B Chem. 2020, 320, 128164. [Google Scholar] [CrossRef]
- Chen, P.; Wang, L.; Qin, P.; Yin, B.-C.; Ye, B.-C. An RNA-based Catalytic Hairpin Assembly Circuit Coupled with CRISPR-Cas12a for One-step Detection of MicroRNAs. Biosens. Bioelectron. 2022, 207, 114152. [Google Scholar] [CrossRef]
- Wang, D.-X.; Wang, J.; Du, Y.-C.; Ma, J.-Y.; Wang, S.-Y.; Tang, A.-N.; Kong, D.-M. CRISPR/Cas12a-based Dual Amplified BioSensing System for Sensitive and Rapid Detection of Polynucleotide Kinase/Phosphatase. Biosens. Bioelectron. 2020, 168, 112556. [Google Scholar] [CrossRef]
- Zhou, M.; Teng, X.; Li, Y.; Deng, R.; Li, J. Cascade Transcription Amplification of RNA Aptamer for Ultrasensitive MicroRNA Detection. Anal. Chem. 2019, 91, 5295–5302. [Google Scholar] [CrossRef]
- Athanasios, D.; Kanchana, R.; Elissa, M.; Melissa, J.; Amy, E. An Engineered T7 RNA Polymerase That Produces mRNA Free of Immunostimulatory Byproducts. Nat. Biotechnol. 2022, 6, 1525. [Google Scholar]
- Elvan, C.; Luis, E.; Craig, T. High-salt Transcription of DNA Cotethered with T7 RNA Polymerase to Beads Generates Increased Yields of Highly Pure RNA. J. Biolo. Chem. 2021, 297, 100999. [Google Scholar]
- Huang, C.; Shen, G.; Ding, S.; Kan, A.; Jiang, D.; Jiang, W. Primer-Template Conversion-based Cascade Signal Amplification Strategy for Sensitive and Accurate Detection of Polynucleotide Kinase Activity. Anal. Chim. Acta 2021, 1187, 339139. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Lian, G.; Li, H.; Han, H.; Zhou, D.; Gong, X. Ultrasensitive Sensing of T4 PNKP Phosphatase Activity Through Establishing a Novel Transcription-based Signal Amplification Platform. Sens. Actuators B Chem. 2022, 15, 132269. [Google Scholar] [CrossRef]
- Xie, Z.; Wang, X.; Chen, S.; Zhao, Z.; Zhao, S.; Zhang, W.; Luo, L.; Yi, G. Construction of A Simple, Localized and Homogeneous Fluorescence Detection Platform for T4 PNKP Activity Based on Tetrahedral DNA Nanostructure-Mediated Primer Exchange Reaction. Microchem. J. 2022, 183, 107989. [Google Scholar] [CrossRef]
- Cui, W.; Fan, X.; Zhao, W.; Liu, J.; Zheng, L.; Zhou, L.; Zhang, J.; Zhang, X.; Wang, X. A Label-Free Fuorescent Biosensor for Amplified Detection of T4 Polynucleotide Kinase Activity based on Rolling Circle Amplification and Catalytic Hairpin Assembly. Spectrochem. Acta A. 2023, 285, 121938. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; He, G.; Wu, W.; Deng, W.; Tan, Y.; Xie, Q. Sensitive Photoelectrochemical Determination of T4 Polynucleotide Kinase Using AuNPs/SnS2/ZnIn2S4 Photoactive Material and Enzymatic Reaction-induced DNA Structure Switch Strategy. Talanta 2022, 249, 123660. [Google Scholar] [CrossRef]


| Method | Linear Range | LOD | Ref. |
|---|---|---|---|
| Photoelectrochemical | 0.1~20 mU/mL | 0.069 mU/mL | [12] |
| Fluorescence | 50~1500 mU/mL | 5 mU/mL | [14] |
| Electrochemical | 10~5000 mU/mL | 8.9 mU/mL | [15] |
| Electrochemical | 0.00001~20 U/mL | 3.0 × 10−6 U/mL | [17] |
| Fluorescence | 1~100 mU/mL | 0. 34 mU/mL | [19] |
| Fluorescence | 0.01~25 mU/mL | 0.0033 mU/mL | [28] |
| Fluorescence | 0.001~0.5 mU/μL | 0.0002 mU/μL | [32] |
| Fluorescence | 0.00001~0.01 U/mL | 8.1×10−6 U/mL | [33] |
| Fluorescence | 0.00008 to 0.1 U/m | 1.8 × 10−5 U/mL | [34] |
| Fluorescence | 0.001 to 0.05 U/mL | 6.63 × 10−4 U/mL | [35] |
| Photoelectrochemical | 10−4~1 U/mL | 6 × 10−5 U/mL | [36] |
| Fluorescence | 0.005~1 mU/mL | 0.0017 mU/mL | This work |
| Name | Sequence (5′-3′) |
|---|---|
| msg | CGC CTT ATT AGA TGA CTT CTC ATC TAC ACT TAG TAG AAA TTA CCC TAT AGT GAG TCG TAT TA |
| sg | TAA TAC GAC TCA CTA TA GGG TAA TTT CTA CTA AGT GTA GAT GAG AAG TCA TCT AAT AAG GCG |
| PR-30 | TGA GGC GCC TTA TTA GAT GAC TTC TCT AAA |
| F-Q | 6-FAM-TTA TT-BHQ1 |
| SS1 | TAMRA-TTT TCT CAT ACC ACT GCT CAT CCA TGC CTA GAC TGG CGA TAA GTA GCC AGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, X.; Yuan, H.; Liu, B.; Liu, Y. Self-Supplying Guide RNA-Mediated CRISPR/Cas12a Fluorescence System for Sensitive Detection of T4 PNKP. Molecules 2022, 27, 9019. https://doi.org/10.3390/molecules27249019
Yuan X, Yuan H, Liu B, Liu Y. Self-Supplying Guide RNA-Mediated CRISPR/Cas12a Fluorescence System for Sensitive Detection of T4 PNKP. Molecules. 2022; 27(24):9019. https://doi.org/10.3390/molecules27249019
Chicago/Turabian StyleYuan, Xiuhua, Hui Yuan, Bingxin Liu, and Yeling Liu. 2022. "Self-Supplying Guide RNA-Mediated CRISPR/Cas12a Fluorescence System for Sensitive Detection of T4 PNKP" Molecules 27, no. 24: 9019. https://doi.org/10.3390/molecules27249019
APA StyleYuan, X., Yuan, H., Liu, B., & Liu, Y. (2022). Self-Supplying Guide RNA-Mediated CRISPR/Cas12a Fluorescence System for Sensitive Detection of T4 PNKP. Molecules, 27(24), 9019. https://doi.org/10.3390/molecules27249019





