HPLC with Fluorescence and Photodiode Array Detection for Quantifying Capmatinib in Biological Samples: Application to In Vivo and In Vitro Studies
Abstract
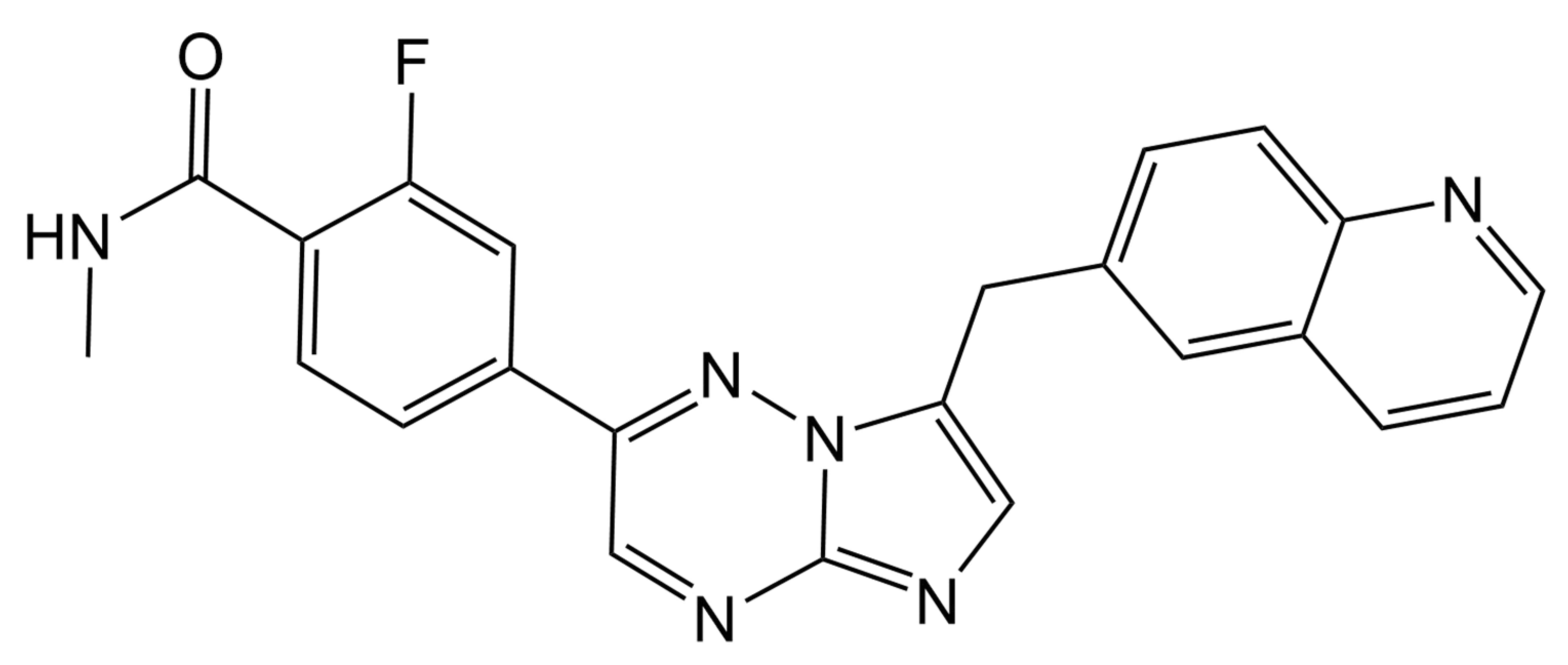
1. Introduction
2. Results and Discussion
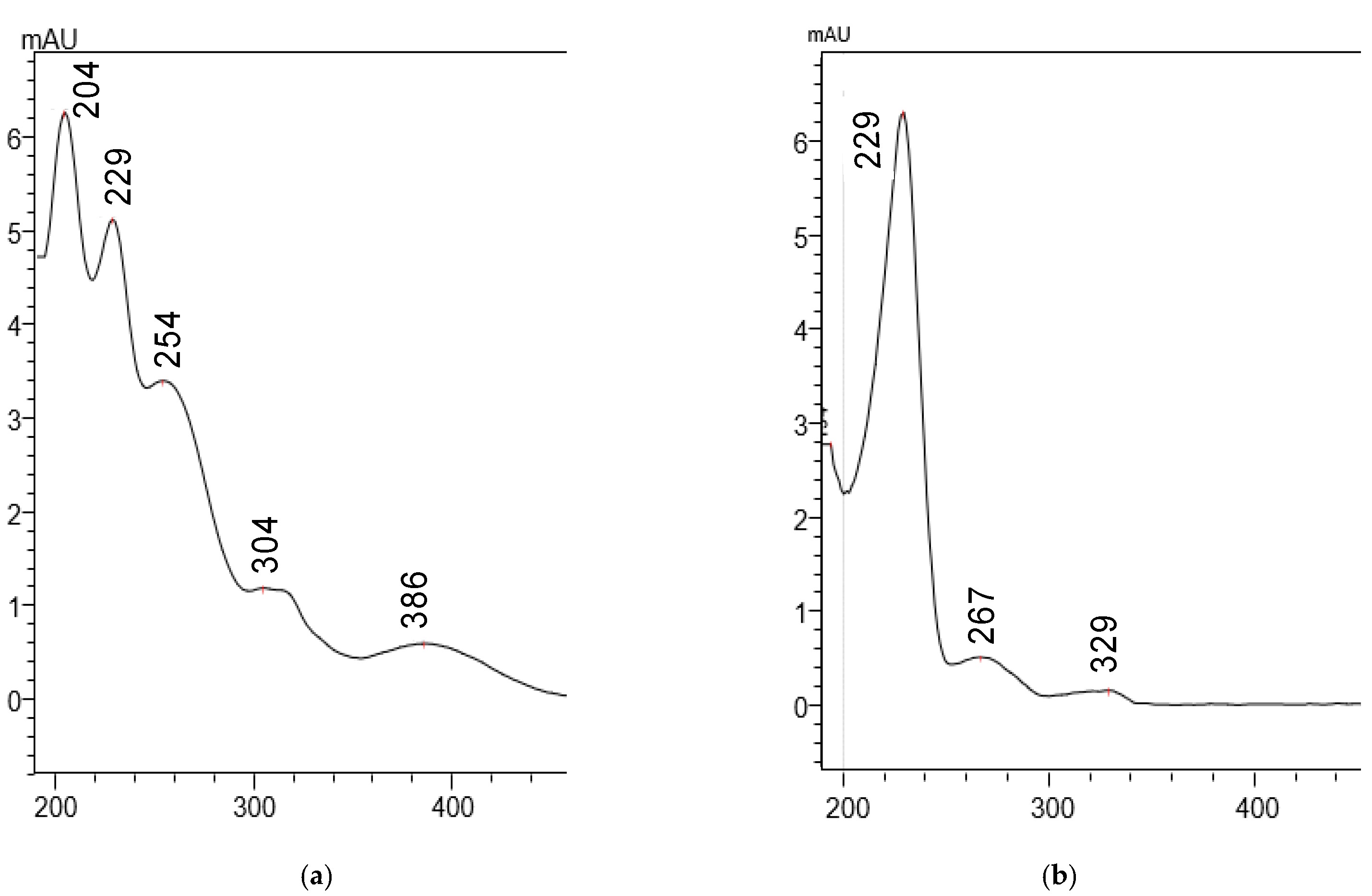
2.1. Method Development
2.2. Method Validation
2.2.1. HPLC-FLD Method Validation
Linearity and LLOQ
Accuracy and Precision
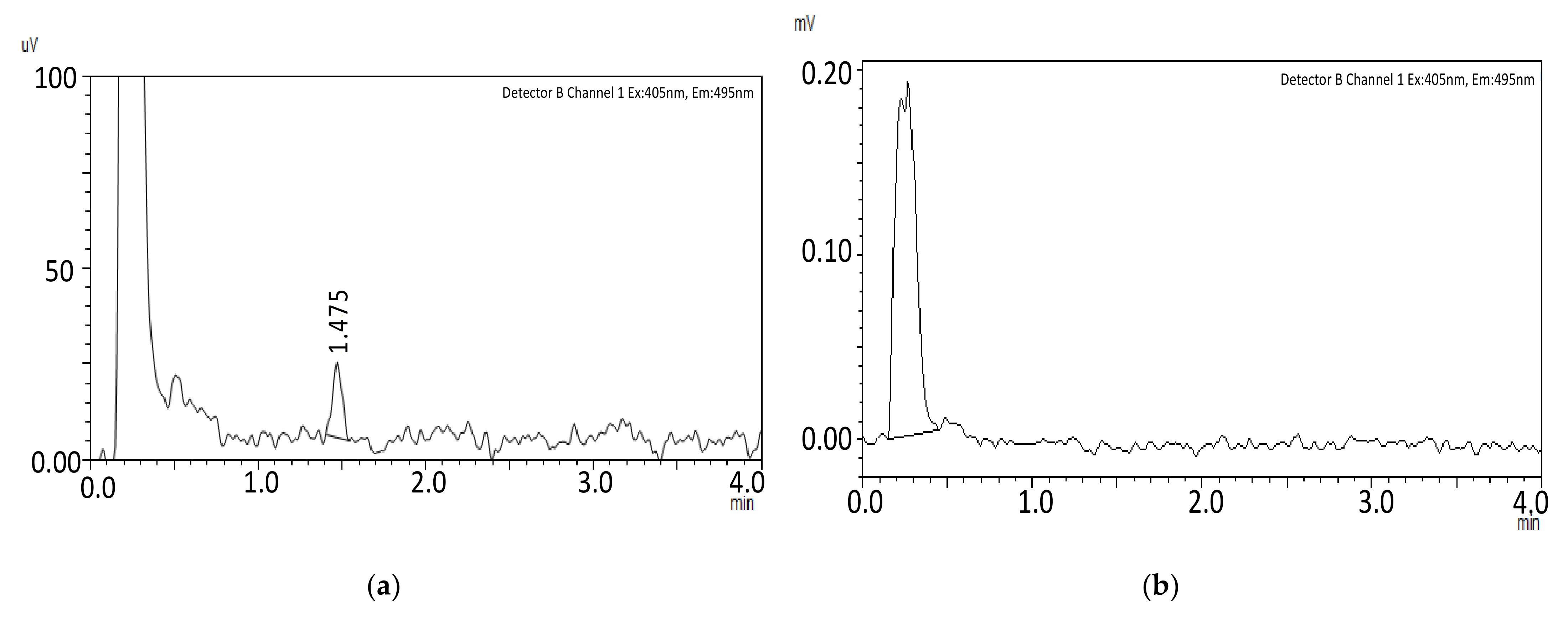
Selectivity
Carryover
2.2.2. HPLC-PDA Method Validation
Linearity and LLOQ
Accuracy and Precision
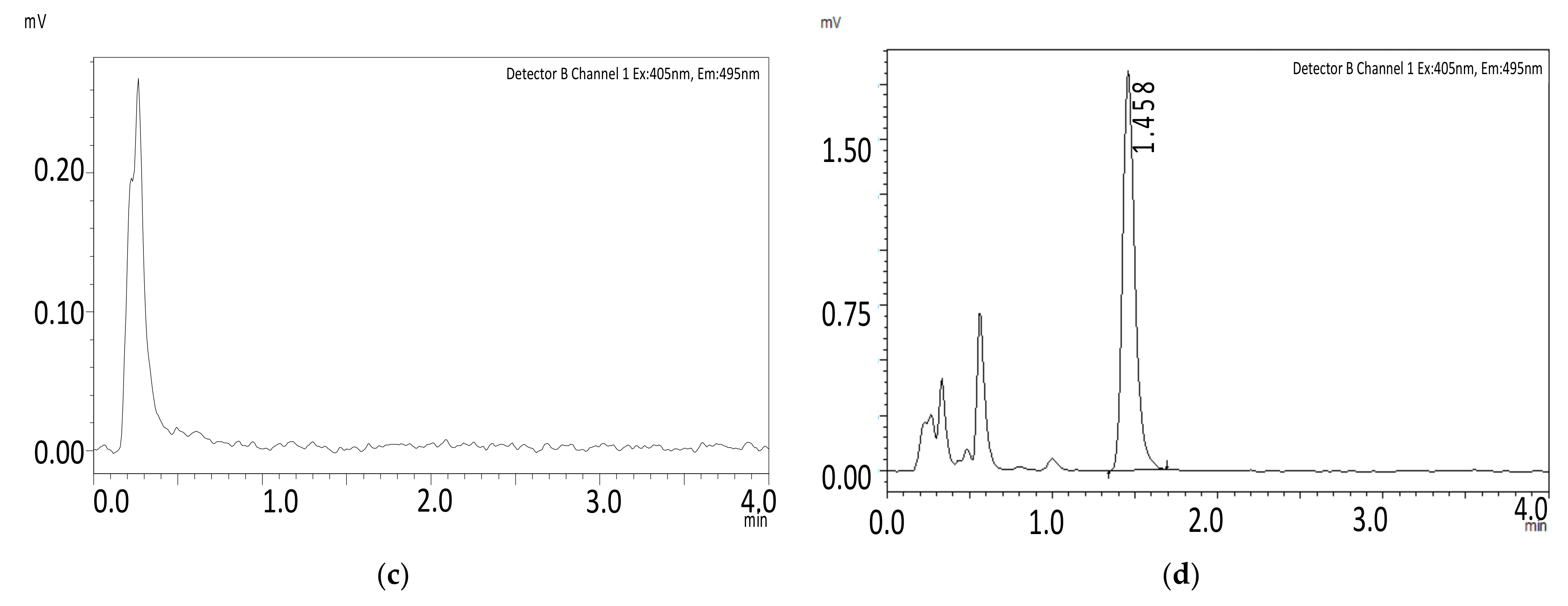
Selectivity
Carryover
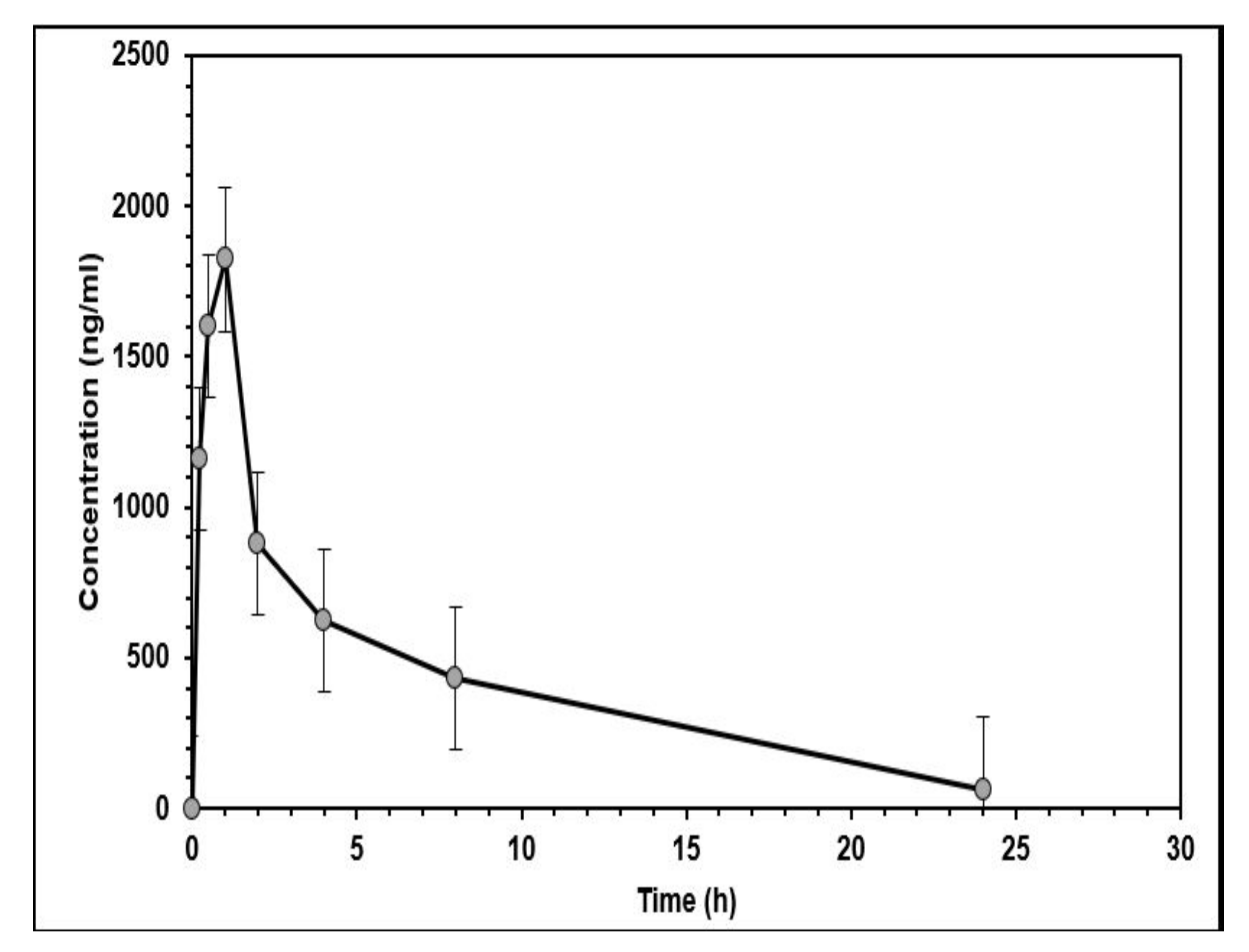
2.3. In Vivo Pharmacokinetic Study
2.4. In Vitro Metabolic Study
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Equipment
3.3. Chromatographic Conditions
3.4. Preparation of Standard Solutions, Calibration, and Quality Control Samples
3.5. Sample Preparation
3.6. Method Validation
3.6.1. Linearity and LLOQ
3.6.2. Accuracy and Precision
3.6.3. Selectivity
3.6.4. Carryover
3.7. In Vivo Pharmacokinetic Study
3.7.1. Animal Handling
3.7.2. In Vivo Experimental Design
3.7.3. Data Analysis
3.8. In Vitro Metabolic Stability Study in Human Liver Microsomes
3.8.1. Microsomal Incubation
3.8.2. Data Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Wang, X.; Zhang, Z.; Tang, C.; Ye, H.; Jones, L.; Lou, F.; Zhang, D.; Jiang, S.; Sun, H.; et al. Identification of genetic mutations in human lung cancer by targeted sequencing. Cancer Inform. 2015, 14, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Garon, E.B.; Brodrick, P. Targeted Therapy Approaches for MET Abnormalities in Non-Small Cell Lung Cancer. Drugs 2021, 81, 547–554. [Google Scholar] [CrossRef] [PubMed]
- FDA Approves Capmatinib for Metastatic Non-Small Cell Lung Cancer. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-capmatinib-metastatic-non-small-cell-lung-cancer (accessed on 15 August 2022).
- Baltschukat, S.; Engstler, B.S.; Huang, A.; Hao, H.-X.; Tam, A.; Wang, H.Q.; Liang, J.; DiMare, M.T.; Bhang, H.-E.C.; Wang, Y.; et al. Capmatinib (INC280) is active against models of non–small cell lung cancer and other cancer types with defined mechanisms of MET activation. Clin. Cancer Res. 2019, 25, 3164–3175. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Park, S.-Y.; Lee, Y.; Lim, K.Y.; Park, M.; Lee, G.K.; Han, J.-Y. The Clinical Impact of Capmatinib in the Treatment of Advanced Non–Small Cell Lung Cancer with MET Exon 14 Skipping Mutation or Gene Amplification. Cancer Res. Treat. 2021, 53, 1024–1032. [Google Scholar] [CrossRef]
- Bang, Y.; Su, W.; Schuler, M.; Nam, D.; Lim, W.T.; Bauer, T.M.; Azaro, A.; Poon, R.T.P.; Hong, D.; Lin, C.; et al. Phase 1 study of capmatinib in MET-positive solid tumor patients: Dose escalation and expansion of selected cohorts. Cancer Sci. 2020, 111, 536–547. [Google Scholar] [CrossRef]
- Glaenzel, U.; Jin, Y.; Hansen, R.; Schroer, K.; Rahmanzadeh, G.; Pfaar, U.; Van Lier, J.J.; Borell, H.; Meissner, A.; Camenisch, G.; et al. Absorption, distribution, metabolism, and excretion of capmatinib (inc280) in healthy male volunteers and in vitro aldehyde oxidase phenotyping of the major metabolite. Drug Metab. Dispos. 2020, 48, 873–885. [Google Scholar] [CrossRef]
- Chen, X.; Cui, X.; Pognan, N.; Quinlan, M.; Kapoor, S.; Rahmanzadeh, G.; Giovannini, M.; Marbury, T.C. Pharmacokinetics of capmatinib in participants with hepatic impairment: A phase 1, open-label, single-dose, parallel-group study. Br. J. Clin. Pharmacol. 2022, 88, 91–102. [Google Scholar] [CrossRef]
- Fan, X.; Yang, G.; Cui, W.; Liu, Q.; Zhang, Z.; Zhang, Z. Development and full validation of an LC–MS/MS methodology to quantify capmatinib (INC280) following intragastric administration to rats. Biomed. Chromatogr. 2020, 34, e4768. [Google Scholar] [CrossRef]
- Zhou, C.; Tian, J.; Lin, P.; Liu, T.; He, A.; Fang, L.; Sun, L. Quantitation of capmatinib, a mesenchymal-epithelial transition factor inhibitor by UPLC-MS/MS in rat plasma and its application to a pharmacokinetic study. Bioanalysis 2020, 12, 285–293. [Google Scholar] [CrossRef]
- Ali, H.M.; Essawy, A.A.; Alsohaimi, I.H.; Nayl, A.; Ibrahim, H.; Essawy, A.E.-N.I.; Elmowafy, M.; Gamal, M. Tailoring the photoluminescence of capmatinib towards a novel ultrasensitive spectrofluorimetric and HPLC-DAD monitoring in human serum ; investigation of the greenness characteristics. Microchem. J. 2022, 181, 107838. [Google Scholar] [CrossRef]
- Wu, D.Y.; Xie, L.X.; Zhang, C.L.; Duan, C.Y.; Zhao, Y.G.; Guo, Z.J. Quinoline-based molecular clips for selective fluorescent detection of Zn2+. J. Chem. Soc. Dalt. Trans. 2006, 29, 3528–3533. [Google Scholar] [CrossRef]
- Meng, X.; Wang, S.; Zhu, M. Quinoline-Based Fluorescence Sensors. Mol. Photochem. Var. Asp. 2012, 16. [Google Scholar] [CrossRef]
- Zayed, A.; Babaresh, W.M.; Darweesh, R.S.; El-Elimat, T. Simultaneous determination of warfarin and 7-hydroxywarfarin in rat plasma by HPLC-FLD. Acta Pharm. 2020, 70, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Corr. Guideline on Bioanalytical Method Validation; EMEA: London, UK, 1922; pp. 1–23. [Google Scholar]
- Esaki, T.; Hirai, F.; Makiyama, A.; Seto, T.; Bando, H.; Naito, Y.; Yoh, K.; Ishihara, K.; Kakizume, T.; Natsume, K.; et al. Phase I dose-escalation study of capmatinib (INC280) in Japanese patients with advanced solid tumors. Cancer Sci. 2019, 110, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Full Prescribing Information for Tabrecta. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/213591s004lbl.pdf (accessed on 26 August 2022).
- Savjani, K.T.; Gajjar, A.K.; Savjani, J.K. Drug Solubility: Importance and Enhancement Techniques. ISRN Pharm. 2012, 112, 1395–1401. [Google Scholar] [CrossRef]
- McNaney, C.A.; Drexler, D.; Hnatyshyn, S.Y.; Zvyaga, T.A.; Knipe, J.O.; Belcastro, J.V.; Sanders, M. An automated liquid chromatography-mass spectrometry process to determine metabolic stability half-life and intrinsic clearance of drug candidates by substrate depletion. Assay. Drug Dev. Technol. 2008, 6, 121–129. [Google Scholar] [CrossRef]
- Houston, J.B. Utility of in vitro drug metabolism data in predicting in vivo metabolic clearance. Biochem. Pharmacol. 1994, 47, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Chao, P.; Uss, A.S.; Cheng, K. Use of intrinsic clearance for prediction of human hepatic clearance. Expert Opin. Drug Metab. Toxicol. 2010, 6, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Hosea, N.A.; Collard, W.T.; Cole, S.; Maurer, T.S.; Fang, R.X.; Jones, H.; Kakar, S.M.; Nakai, Y.; Smith, B.J.; Webster, R.; et al. Prediction of human pharmacokinetics from preclinical information: Comparative accuracy of quantitative prediction approaches. J. Clin. Pharmacol. 2009, 49, 513–533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huo, M.; Zhou, J.; Xie, S.; Solver, P.K. An add-in program for pharmacokinetic and pharmacodynamic data analysis in Microsoft Excel. Comput. Methods Progr. Biomed. 2010, 99, 306–314. [Google Scholar] [CrossRef] [PubMed]
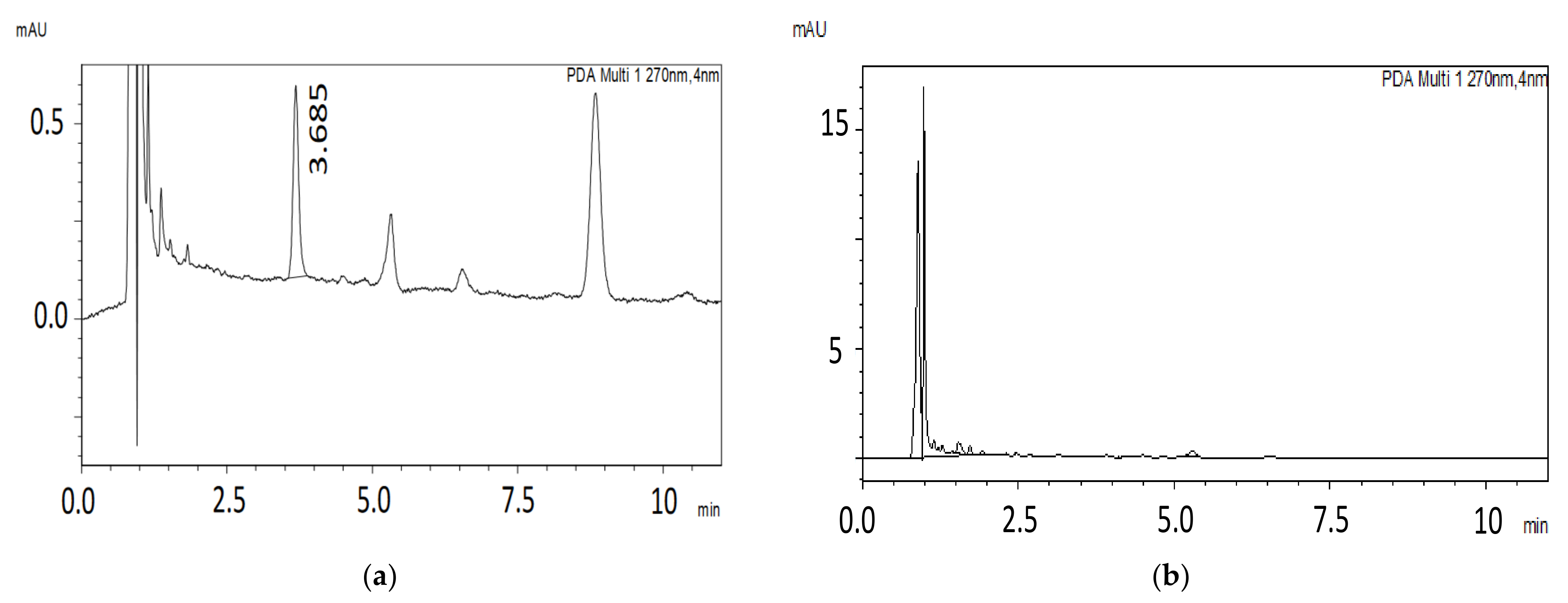

| Analyte | Tailing Factor | Retention Time (tR) min | Number of Theoretical Plates (N) | Capacity Factor | Selectivity Factor | Resolution |
|---|---|---|---|---|---|---|
| Capmatinib | 1.26 | 1.45 | 1644 | 4.61 | 2.27 | 7.87 |
| Warfarin | 1.20 | 2.73 | 1486 | 10.67 | 7.99 |
| Analyte | Tailing Factor | Retention Time (tR) min | Number of Theoretical Plates (N) | Capacity Factor | Selectivity Factor | Resolution |
|---|---|---|---|---|---|---|
| Capmatinib | 1.36 | 3.68 | 5996 | 2.82 | 2.84 | 5.05 |
| Naproxen | 1.03 | 8.88 | 11,447 | 8.02 | 19.88 |
| Linear Range (ng/mL) | Slope Mean ± SD | Intercept Mean ± SD | Correlation Coefficients Mean ± SD |
|---|---|---|---|
| 19.5–3920 | 6.76 ± 4.40 | 0.0066 ± 0.0028 | 0.9933 ± 0.0022 |
| Nominal Concentration (ng/mL) | Intra-Day (Single Batch) a | Inter-Day (Three Batches) b | |||||
|---|---|---|---|---|---|---|---|
| Measured Concentration | er (%) | RSD (%) | Measured Mean Concentration | er (%) | RSD (%) | ||
| LLOQ | 19.5 | 20.45 | 6.41 | 6.66 | 18.77 | 9.78 | 11.83 |
| QCL | 58.5 | 59.51 | 8.06 | 9.57 | 55.71 | 10.96 | 10.54 |
| QCM | 1470 | 1394.25 | 5.15 | 4.15 | 1469.00 | 5.55 | 7.25 |
| QCH | 3136.5 | 2893.19 | 7.76 | 4.60 | 3,082.44 | 7.10 | 8.55 |
| Linear Range (ng/mL) | Slope Mean ± SD | Intercept Mean ± SD | Correlation Coefficients Mean ± SD |
|---|---|---|---|
| 206–4124 | 1.026 ± 0.073 | 0.03 ± 0.019 | 0.992 ± 0.0012 |
| Nominal Concentration (ng/mL) | Intra-Day (Single Batch) a | Inter-Day (Three Batches) b | |||||
|---|---|---|---|---|---|---|---|
| Measured Concentration | er (%) | RSD (%) | Measured Mean Concentration | er (%) | RSD (%) | ||
| LLOQ | 206 | 210 | 2.0 | 3.8 | 202 | 4 | 8.9 |
| QCL | 618.6 | 660 | 6.6 | 2.2 | 635.1 | 2.7 | 7.7 |
| QCM | 2000 | 2316 | 12.4 | 1.9 | 2185 | 6 | 8.7 |
| QCH | 3093 | 3500 | 14 | 0.9 | 3270 | 6.4 | 9.0 |
| Parameter | Unit | Value (Mean ± SD) |
|---|---|---|
| t1/2 | h | 3.57 ± 1.16 |
| Tmax | h | 0.67 ± 0.26 |
| Cmax | ng/mL | 1636.55 ± 509.62 |
| ng/mL.h | 8150.59 ± 3754.15 | |
| ng/mL.h | 8383.64 ± 3639.34 | |
| h | 4.83 ± 1.4 | |
| Vz/F | L | 1.83 ± 1.22 |
| Cl/F | L/min/kg | 0.03 ± 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zayed, A.; Jaber, S.A.; Al Hroot, J.; Hawamdeh, S.; Ayoub, N.M.; Qinna, N.A. HPLC with Fluorescence and Photodiode Array Detection for Quantifying Capmatinib in Biological Samples: Application to In Vivo and In Vitro Studies. Molecules 2022, 27, 8582. https://doi.org/10.3390/molecules27238582
Zayed A, Jaber SA, Al Hroot J, Hawamdeh S, Ayoub NM, Qinna NA. HPLC with Fluorescence and Photodiode Array Detection for Quantifying Capmatinib in Biological Samples: Application to In Vivo and In Vitro Studies. Molecules. 2022; 27(23):8582. https://doi.org/10.3390/molecules27238582
Chicago/Turabian StyleZayed, Aref, Sana’a A. Jaber, Jomana Al Hroot, Sahar Hawamdeh, Nehad M. Ayoub, and Nidal A. Qinna. 2022. "HPLC with Fluorescence and Photodiode Array Detection for Quantifying Capmatinib in Biological Samples: Application to In Vivo and In Vitro Studies" Molecules 27, no. 23: 8582. https://doi.org/10.3390/molecules27238582
APA StyleZayed, A., Jaber, S. A., Al Hroot, J., Hawamdeh, S., Ayoub, N. M., & Qinna, N. A. (2022). HPLC with Fluorescence and Photodiode Array Detection for Quantifying Capmatinib in Biological Samples: Application to In Vivo and In Vitro Studies. Molecules, 27(23), 8582. https://doi.org/10.3390/molecules27238582






