Identification of Novel Antifungal Skeleton of Hydroxyethyl Naphthalimides with Synergistic Potential for Chemical and Dynamic Treatments
Abstract
1. Introduction
2. Results and Discussion
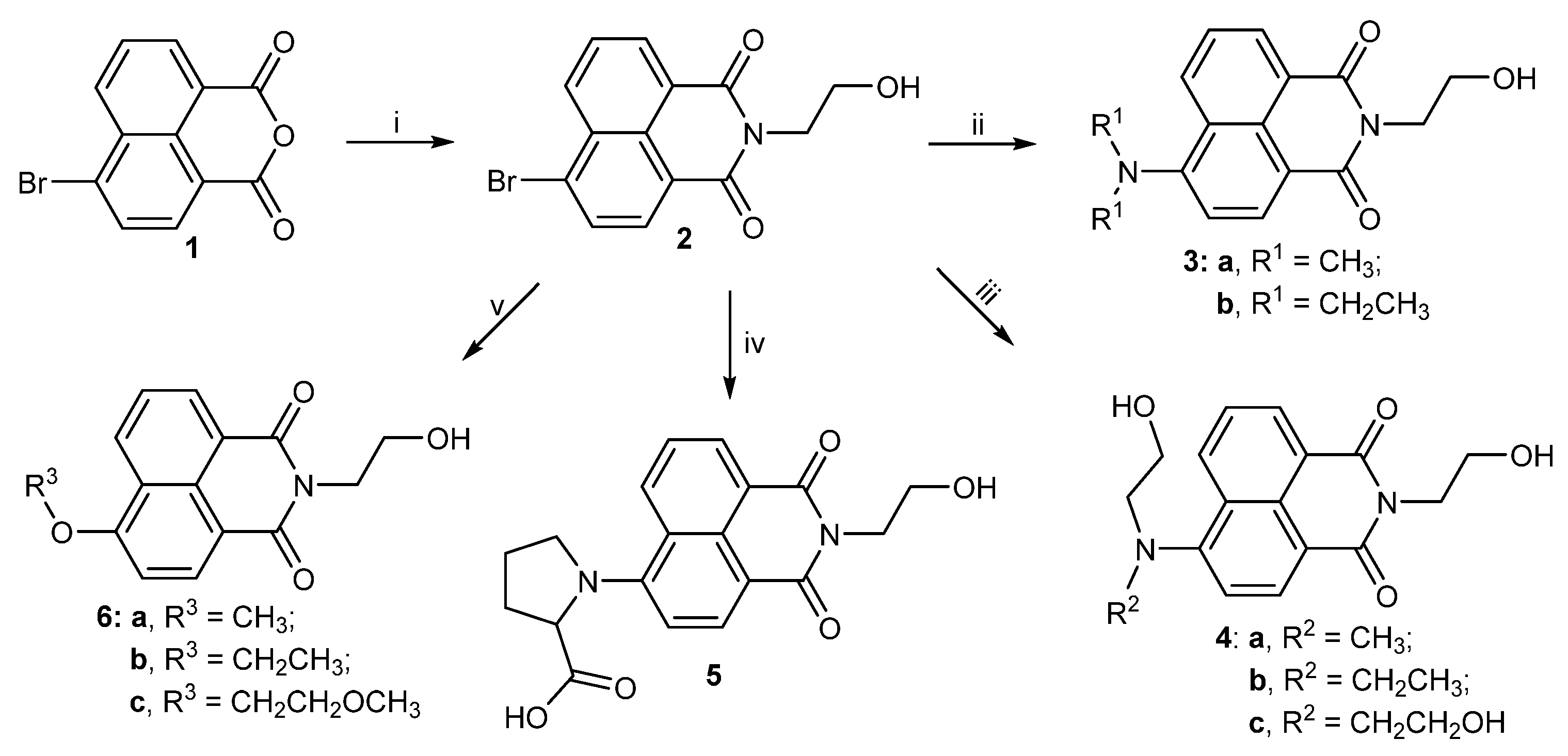
2.1. Chemistry
2.2. Relationship between DNA Binding and Antifungal Assay
2.3. Supramolecular Interaction of Thioether Benzimidazole 7f with DNA
2.4. Cytotoxicity, Hemolysis Assays and Resistance Development Assay
2.5. Pharmacokinetic Properties
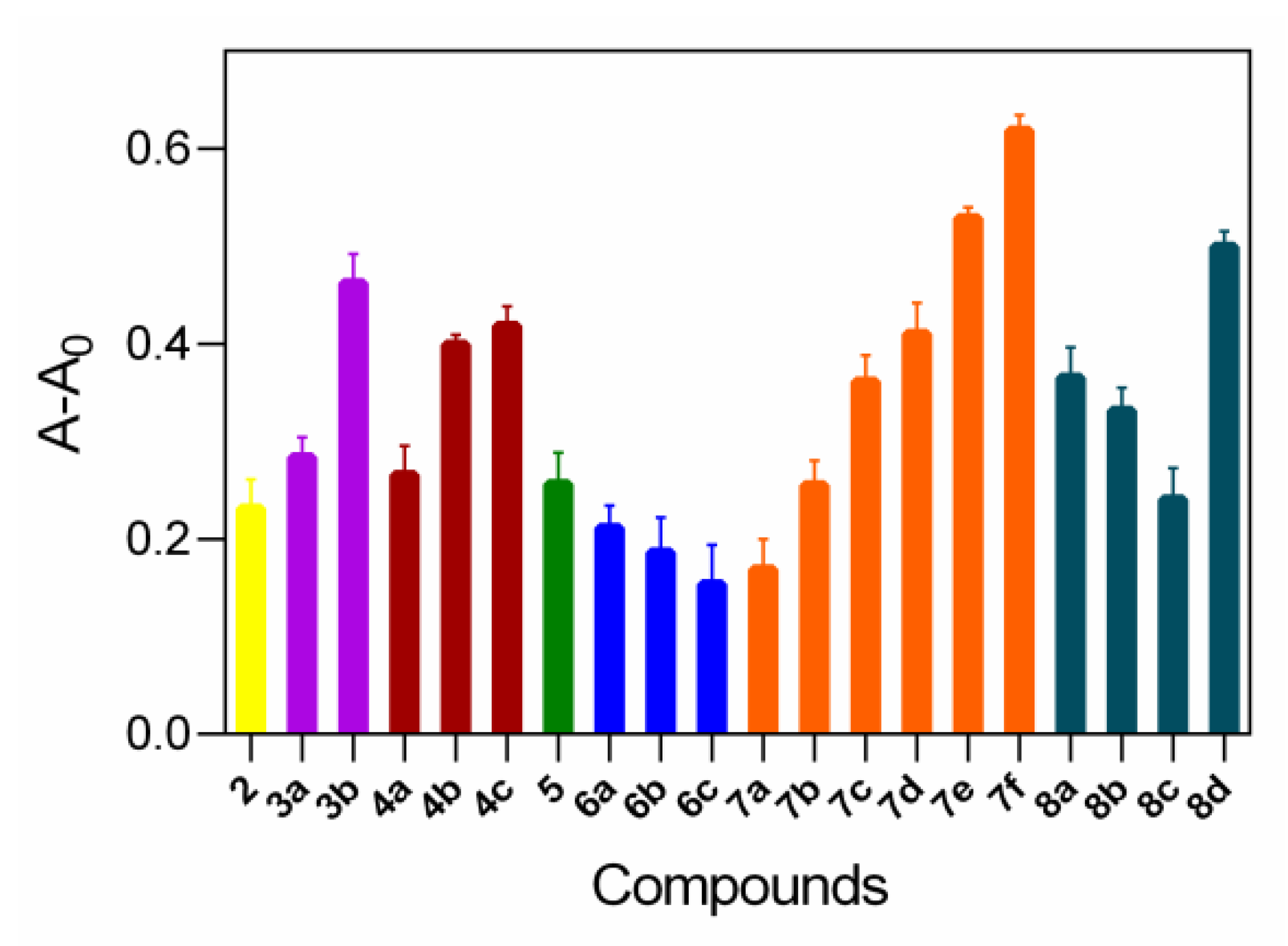
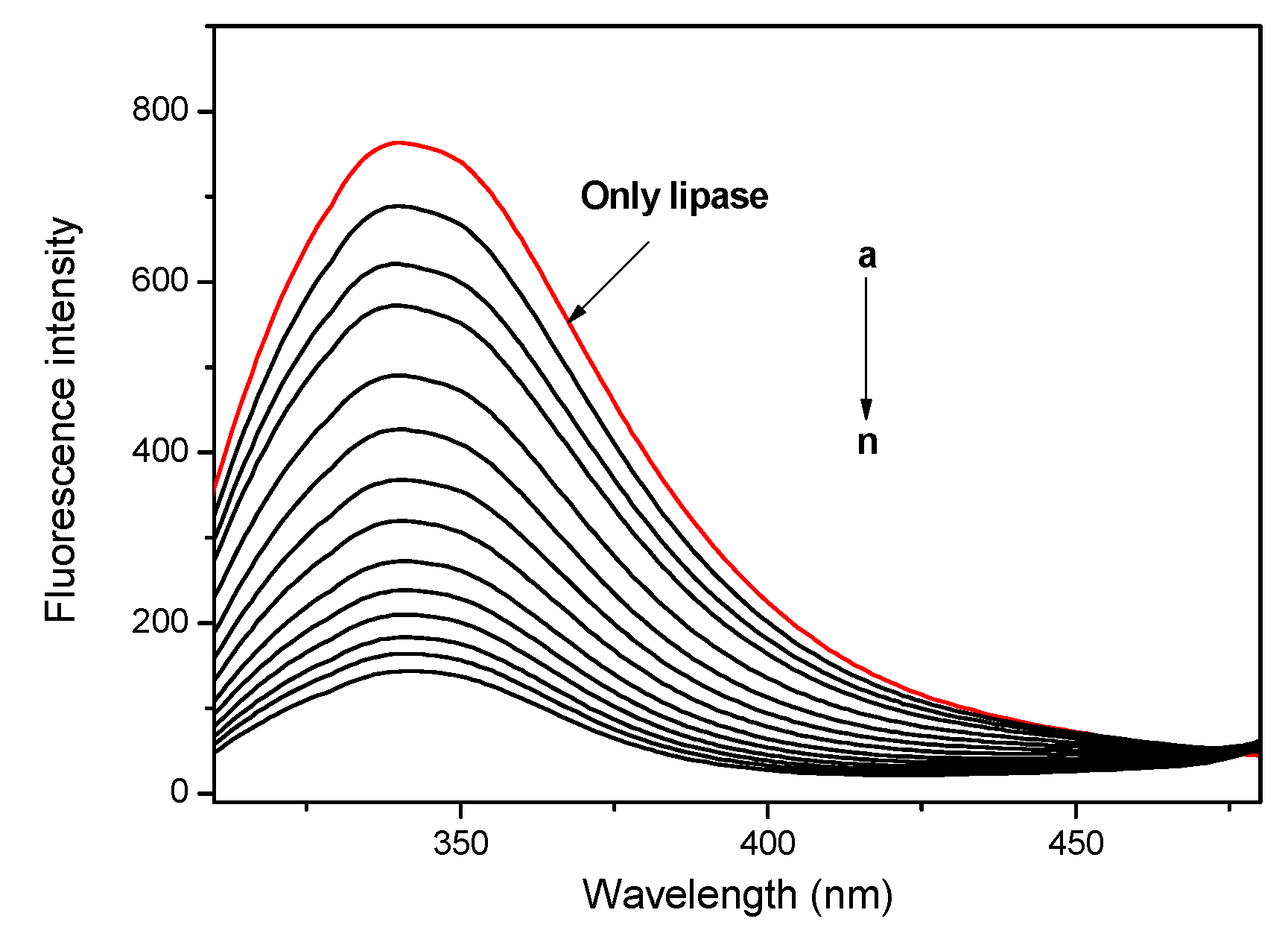
2.6. Lipase Affinity of Thioether Benzimidazole 7f
2.7. Membrane Damage Assay
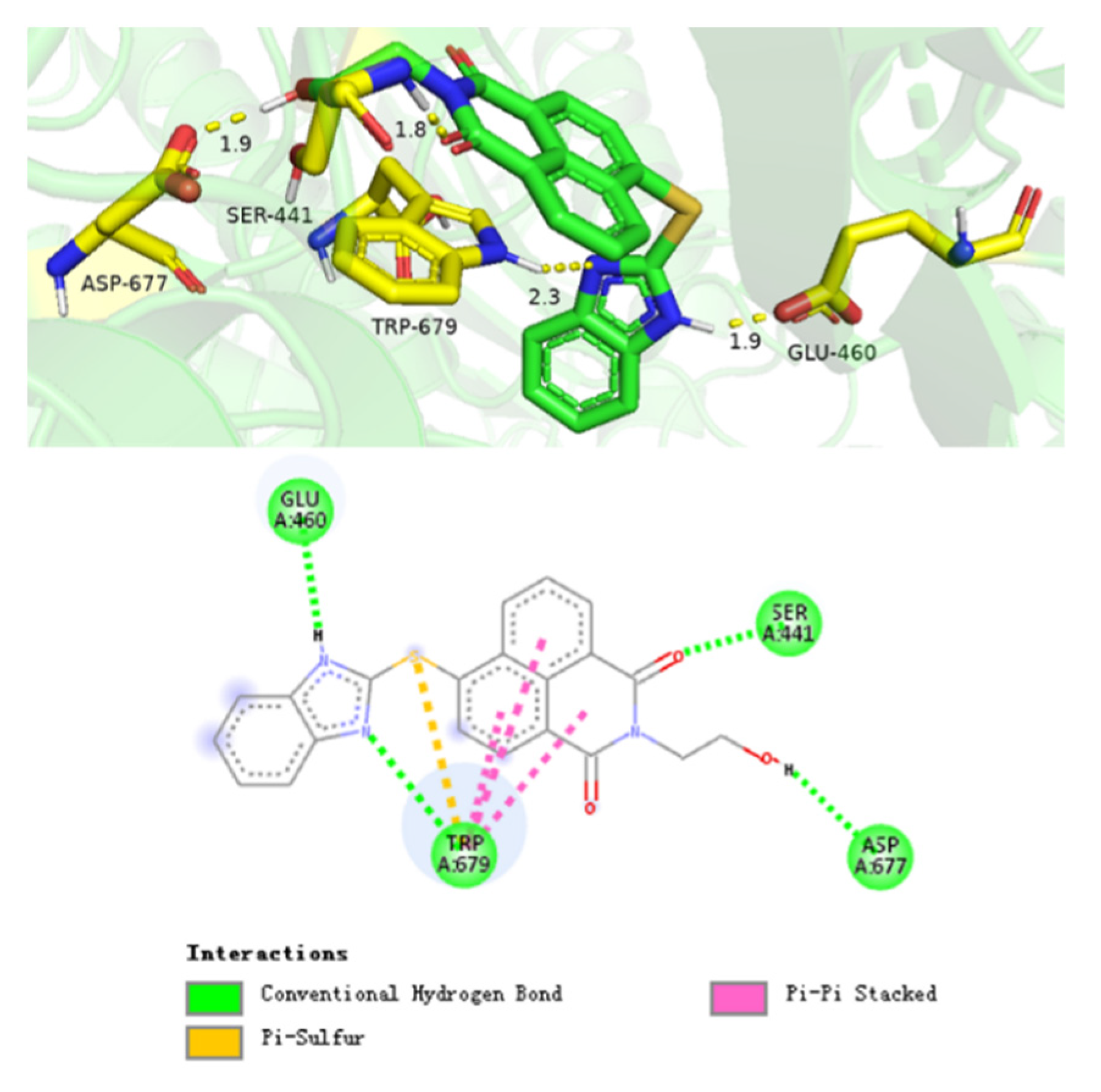
2.8. Supramolecular Interaction of Compound 7f with Cytochrome P450 Reductase
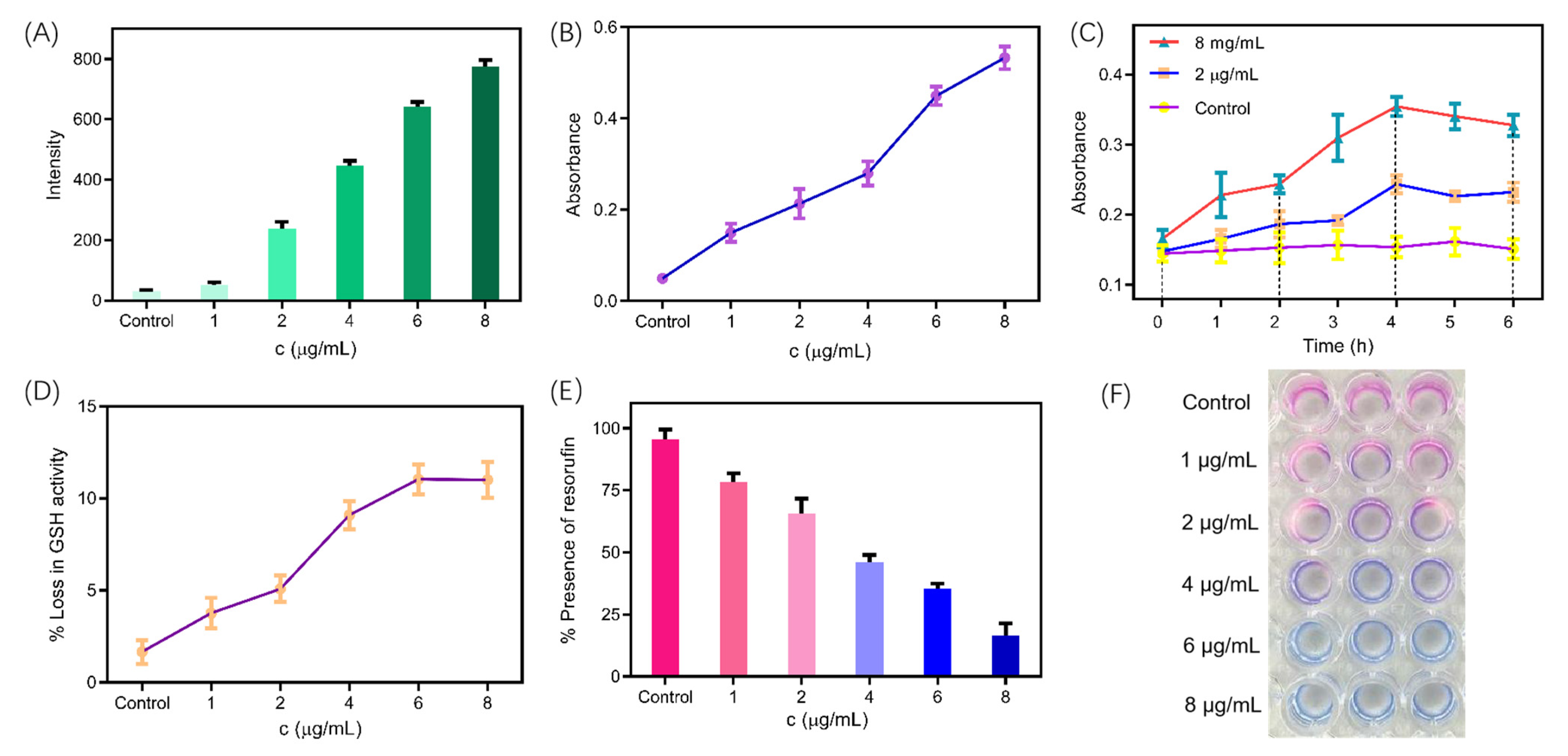
2.9. ROS-Mediated Dynamic Treatment
2.10. Measurement of Metabolic Activity
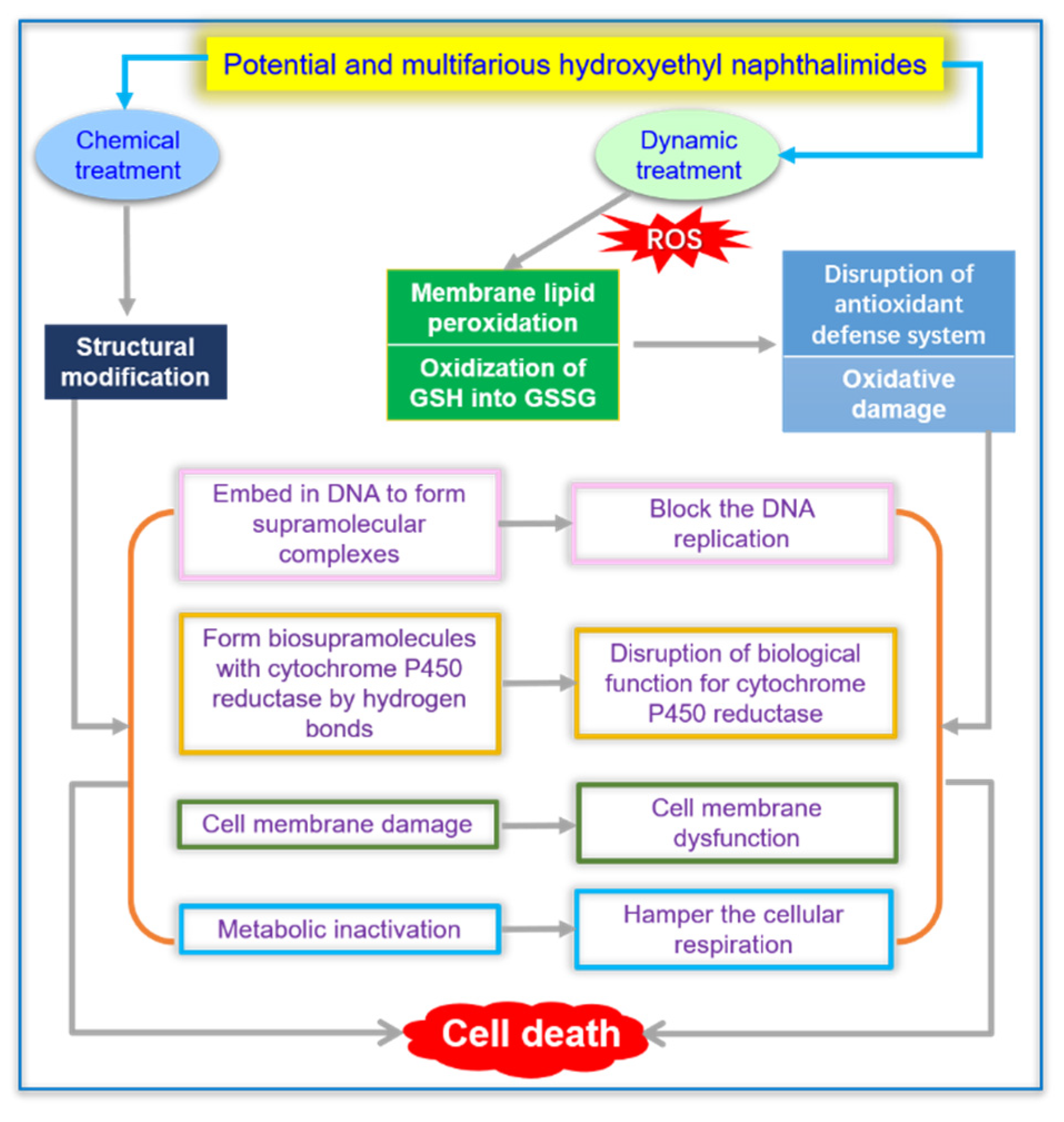
2.11. Synergistic Effect of Chemical and Dynamic Antifungal Treatment for Hydroxyethyl Naphthalimide Antifungals
3. Materials and Methods
3.1. Instruments and Chemicals
3.2. Synthesis of Hydroxyethyl Naphthalimides
3.2.1. Synthesis of 6-Bromo-2-(2-hydroxyethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione (2)
3.2.2. Synthesis of 6-(Dimethylamino)-2-(2-hydroxyethyl)-1H-benzo[de]isoquinoline-1,3 (2H)-dione (3a)
3.2.3. Synthesis of 6-(Diethylamino)-2-(2-hydroxyethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione (3b)
3.2.4. Synthesis of 2-(2-Hydroxyethyl)-6-((2-hydroxyethyl)(methyl)amino)-1H-benzo[de] isoquinoline-1,3(2H)-dione (4a)
3.2.5. Synthesis of 6-(Ethyl(2-hydroxyethyl)amino)-2-(2-hydroxyethyl)-1H-benzo[de] isoquinoline-1,3(2H)-dione (4b)
3.2.6. Synthesis of 6-(Bis(2-hydroxyethyl)amino)-2-(2-hydroxyethyl)-1H-benzo[de] isoquinoline-1,3(2H)-dione (4c)
3.2.7. Synthesis of (2-(2-Hydroxyethyl)-1,3-dioxo-2,3-dihydro-1H-benzo[de]isoquinolin-6-yl)proline (5)
3.2.8. Synthesis of 2-(2-Hydroxyethyl)-6-methoxy-1H-benzo[de]isoquinoline-1,3(2H)-dione (6a)
3.2.9. Synthesis of 6-Ethoxy-2-(2-hydroxyethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione (6b)
3.2.10. Synthesis of 2-(2-Hydroxyethyl)-6-(2-methoxyethoxy)-1H-benzo[de]isoquinoline-1,3(2H)-dione (6c)
3.2.11. Synthesis of 2-(2-Hydroxyethyl)-6-((1-methyl-1H-imidazol-2-yl)thio)-1H-benzo[de] isoquinoline-1,3(2H)-dione (7a)
3.2.12. Synthesis of 2-(2-Hydroxyethyl)-6-((1-methyl-1H-tetrazol-5-yl)thio)-1H-benzo[de] isoquinoline-1,3(2H)-dione (7b)
3.2.13. Synthesis of 6-((1H-1,2,4-Triazol-5-yl)thio)-2-(2-hydroxyethyl)-1H-benzo[de] isoquinoline-1,3(2H)-dione (7c)
3.2.14. Synthesis of 2-(2-Hydroxyethyl)-6-((5-methyl-1,3,4-thiadiazol-2-yl)thio)-1H-benzo [de]isoquinoline-1,3(2H)- dione (7d)
3.2.15. Synthesis of 6-(Benzo[d]thiazol-2-ylthio)-2-(2-hydroxyethyl)-1H-benzo[de] isoquinoline-1,3(2H)-dione (7e)
3.2.16. Synthesis of 6-((1H-Benzo[d]imidazol-2-yl)thio)-2-(2-hydroxyethyl)-1H-benzo [de]isoquinoline-1,3(2H)-dione (7f)
3.2.17. Synthesis of 2-(2-Hydroxyethyl)-6-(pyrimidin-2-ylthio)-1H-Benzo[de]isoquinoline-1,3(2H)-dione (8a)
3.2.18. Synthesis of 2-(2-Hydroxyethyl)-6-((4-methylpyrimidin-2-yl)thio)-1H-benzo[de] isoquinoline-1,3(2H)-dione (8b)
3.2.19. Synthesis of 6-((4,6-Dimethylpyrimidin-2-yl)thio)-2-(2-hydroxyethyl)-1H-benzo[de] isoquinoline-1,3(2H)-dione (8c)
3.2.20. Synthesis of 6-((4-Hydroxy-6-methylpyrimidin-2-yl)thio)-2-(2-hydroxyethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione (8d)
3.3. Biological Assay
3.3.1. Antifungal Assay
3.3.2. UV Absorption Spectra of Fluorophores with DNA
3.3.3. Competitive Reaction of Compound 7f and AO or DAPI with DNA
3.3.4. Measurement of Intracellular ROS Production
3.3.5. Measurement of RNIs by Griess’s Reaction
3.3.6. Measurement of MDA
3.3.7. Measurement of Intracellular Glutathione (GSH) Activity
3.3.8. Measurement of Alamar Blue Assay
3.3.9. Drug Resistance Development Assay
3.3.10. Hemolysis Assay
3.3.11. In Vitro Cytotoxicity
3.3.12. Membrane Depolarization Assay
3.3.13. Protein Leakage Assay
3.3.14. Measurement of Metabolic Activity
3.3.15. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Janbon, G.; Quintin, J.; Lanternier, F.; Enfert, C. Studying fungal pathogens of humans and fungal infections: Fungal diversity and diversity of approaches. Genes. Immun. 2019, 20, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Burnham, C.A.D.; Jennifer, L.; Patrice, N.; Justin, O.G.; Jean, P. Diagnosing antimicrobial resistance. Nat. Rev. Microbiol. 2017, 15, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.P.; Ansari, M.F.; Zhang, S.L.; Zhou, C.H. Novel carbazole-oxadiazoles as potential Staphylococcus aureus germicides. Pestic. Biochem. Phys. 2021, 175, 104849. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef]
- West, A.P.; Igor, E.B.; Christoph, R.; Dong, K.W.; Hediye, E.B.; Paul, T.; Matthew, C.W.; Yongwon, C.; Gerald, S.S.; Sankar, G. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 2011, 472, 476–480. [Google Scholar] [CrossRef]
- Zhou, Y.; Sun, H.J.; Wang, F.M.; Ren, J.S.; Qu, X.G. How functional groups influence the ROS generation and cytotoxicity of graphene quantum dots. Chem. Commun. 2017, 53, 10588–10591. [Google Scholar] [CrossRef]
- Judith, A.; Brian, D.B. Silence of the ROS. Immunity 2016, 44, 520–522. [Google Scholar]
- Tan, Y.M.; Li, D.; Li, F.F.; Ansari, M.F.; Fang, B.; Zhou, C.H. Pyrimidine-conjugated fluoroquinolones as new potential broad-spectrum antibacterial agents. Bioorg. Med. Chem. Lett. 2022, 73, 128885. [Google Scholar] [CrossRef]
- Cui, S.F.; Addla, D.; Zhou, C.H. Novel 3-aminothiazol quinolones: Design, synthesis, bioactive evaluation, SARs, and preliminary antibacterial mechanism. J. Med. Chem. 2016, 59, 4488–4510. [Google Scholar] [CrossRef]
- Tandon, R.; Luxami, V.; Kaur, H.; Tandon, N.; Paul, K. 1,8-Naphthalimide: A potent DNA intercalator and target for cancer therapy. Chem. Rec. 2017, 17, 956–993. [Google Scholar] [CrossRef]
- Gong, H.H.; Addla, D.; Lv, J.S.; Zhou, C.H. Heterocyclic naphthalimides as new skeleton structure of compounds with increasingly expanding relational medicinal applications. Curr. Topics Med. Chem. 2016, 16, 3303–3364. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Gopala, L.; Bheemanaboina, R.R.Y.; Liu, H.B.; Cheng, Y.; Geng, R.X.; Zhou, C.H. Novel naphthalimide aminothiazoles as potential multitargeting antimicrobial agents. ACS Med. Chem. Lett. 2017, 8, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Gopala, L.; Tangadanchu, V.K.R.; Gao, W.W.; Zhou, C.H. Novel naphthalimide nitroimidazoles as multitargeting antibacterial agents against resistant Acinetobacter baumannii. Future Med. Chem. 2018, 10, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Tangadanchu, V.K.R.; Gopala, L.; Gao, W.W.; Cheng, Y.; Liu, H.B.; Geng, R.X.; Li, S.; Zhou, C.H. Novel potentially antibacterial naphthalimide-derived metronidazoles: Design, synthesis, biological evaluation and supramolecular interactions with DNA, human serum albumin and topoisomerase II. Chin. Chem. Lett. 2017, 28, 1369–1374. [Google Scholar] [CrossRef]
- Zhang, P.L.; Gopala, L.; Yu, Y.; Fang, B.; Zhou, C.H. Identification of a novel antifungal backbone of naphthalimide thiazoles with synergistic potential for chemical and dynamic treatment. Future Med. Chem. 2021, 13, 2047–2067. [Google Scholar] [CrossRef]
- Luo, Y.L.; Baathulaa, K.; Kannekanti, V.K.; Zhou, C.H.; Cai, G.X. Novel benzimidazole derived naphthalimide triazoles: Synthesis, antimicrobial activity and interactions with calf thymus DNA. Sci. China Chem. 2015, 58, 483–494. [Google Scholar] [CrossRef]
- Yordanova, S.; Temiz, H.T.; Boyaci, I.H.; Stoyanov, S.; Vasileva-Tonkova, E.; Asiri, A.; Grabchev, I. Synthesis, characterization and in vitro antimicrobial activity of a new blue fluorescent Cu(II) metal complex of bis-1,8-naphthalimide. J. Mol. Struct. 2015, 1101, 50–56. [Google Scholar] [CrossRef]
- Damu, G.L.V.; Wang, Q.P.; Zhang, H.Z.; Zhang, Y.Y.; Lv, J.S.; Zhou, C.H. A series of naphthalimide azoles: Design, synthesis and bioactive evaluation as potential antimicrobial agents. Sci. China Chem. 2013, 56, 952–969. [Google Scholar] [CrossRef]
- Zhang, P.L.; Gopala, L.; Zhang, S.L.; Cai, G.X.; Zhou, C.H. An unanticipated discovery towards novel naphthalimide corbelled aminothiazoximes as potential anti-MRSA agents and allosteric modulators for PBP2a. Eur. J. Med. Chem. 2022, 229, 114050. [Google Scholar] [CrossRef]
- Luo, X.; Yang, Y.J.; Qian, X.H. Recent progresses on the development of thioxo-naphthalimides. Chin. Chem. Lett. 2020, 31, 2877–2883. [Google Scholar] [CrossRef]
- Shinde, R.G.; Khan, A.A.; Barik, A. Formation of two centre three electron bond by hydroxyl radical induced reaction of thiocoumarin: Evidence from experimental and theoretical studies. Free Radic. Res. 2019, 53, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Jennifer, N.M.; Thomas, G.C. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018, 80, 50–64. [Google Scholar]
- Zhang, P.L.; Laiche, M.H.; Li, Y.L.; Gao, W.W.; Lin, J.M.; Zhou, C.H. An unanticipated discovery of novel naphthalimidopropanediols as potential broad-spectrum antibacterial members. Eur. J. Med. Chem. 2022, 241, 114657. [Google Scholar] [CrossRef] [PubMed]
- Kubo, I.; Cespedes, C.L. Antifungal activity of alkanols: Inhibition of growth of spoilage yeasts. Phytochem. Rev. 2013, 12, 961–977. [Google Scholar] [CrossRef]
- Zhang, Y.; Tangadanchu, V.K.R.; Bheemanaboina, R.R.Y.; Cheng, Y.; Zhou, C.H. Novel carbazole-triazole conjugates as DNA-targeting membrane active potentiators against clinical isolated fungi. Eur. J. Med. Chem. 2018, 155, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tangadanchu, V.K.R.; Cheng, Y.; Yang, R.G.; Lin, J.M.; Zhou, C.H. Potential antimicrobial isopropanol-conjugated carbazole azoles as dual targeting inhibitors of Enterococcus faecalis. ACS Med. Chem. Lett. 2018, 9, 244–249. [Google Scholar] [CrossRef]
- Hu, Y.Y.; Bheemanaboina, R.R.Y.; Battini, N.; Zhou, C.H. Sulfonamide-derived four-component molecular hybrids as novel DNA-targeting membrane active potentiators against clinical Escherichia coli. Mol. Pharmaceutics 2019, 16, 1036–1052. [Google Scholar] [CrossRef]
- Yang, Z.H.; Ge, G.B.; Liu, F.Y.; Chen, S.S.; Sun, S.G. Study of a series of fluorophore-labeled artesunate derivatives-Design, analysis and application. Dyes Pigments 2017, 146, 414–419. [Google Scholar] [CrossRef]
- Krasnovskaya, O.O.; Malinnikov, V.M.; Dashkova, N.S.; Gerasimov, V.M.; Grishina, I.V.; Kireev, I.I.; Lavrushkina, S.V.; Panchenko, P.A.; Zakharko, M.A.; Ignatov, P.A.; et al. Thiourea modified doxorubicin: A perspective pH-sensitive prodrug. Bioconjugate Chem. 2019, 30, 741–750. [Google Scholar] [CrossRef]
- Sun, H.; Ansari, M.F.; Fang, B.; Zhou, C.H. Natural berberine-hybridized benzimidazoles as novel unique bactericides against Staphylococcus aureus. J. Agric. Food Chem. 2021, 69, 7831–7840. [Google Scholar] [CrossRef]
- Nanna, H.L.; Jérémie, K.; Jenifer, R.M.; Julien, I.; David, B.; Patrick, N.O.; Mathieu, S.; Mathieu, L. Origin of DNA-induced circular dichroism in a minor-groove binder. J. Am. Chem. Soc. 2017, 139, 14947–14953. [Google Scholar]
- Sun, H.; Ansari, M.F.; Battini, N.; Bheemanaboina, R.R.Y.; Zhou, C.H. Novel potential artificial MRSA DNA intercalators: Synthesis and biological evaluation of berberine-derived thiazolidinediones. Org. Chem. Front. 2019, 6, 319–334. [Google Scholar] [CrossRef]
- Lin, S.M.; Wade, J.D.; Liu, S.P. De Novo design of flavonoid-based mimetics of cationic antimicrobial peptides: Discovery, development, and applications. Acc. Chem. Res. 2021, 54, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.M.; Li, H.X.; Tao, Y.W.; Liu, J.Y.; Yuan, W.C.; Chen, Y.Z.; Liu, Y.; Liu, S.P. In vitro and in vivo evaluation of membrane-active flavone amphiphiles: Semisynthetic kaempferol-derived antimicrobials against drug-resistant gram-positive bacteria. J. Med. Chem. 2020, 63, 5797–5815. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.C.; Wang, Y.; Liu, N.; Dong, G.Q.; Sheng, C.Q. Tackling fungal resistance by biofilm inhibitors. J. Med. Chem. 2017, 60, 2193–2211. [Google Scholar] [CrossRef]
- Wang, L.L.; Battini, N.; Bheemanaboina, R.R.Y.; Zhang, S.L.; Zhou, C.H. Design and synthesis of aminothiazolyl norfloxacin analogues as potential antimicrobial agents and their biological evaluation. Eur. J. Med. Chem. 2019, 167, 105–123. [Google Scholar] [CrossRef]
- Zhang, L.; Peng, X.M.; Damu, G.L.V.; Geng, R.X.; Zhou, C.H. Comprehensive review in current developments of imidazole-based medicinal chemistry. Med. Res. Rev. 2014, 34, 340–437. [Google Scholar]
- Zhang, H.Z.; Zhao, Z.L.; Zhou, C.H. Recent advance in oxazole-based medicinal chemistry. Eur. J. Med. Chem. 2018, 144, 444–492. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, P.L.; Ansari, M.F.; Li, S.; Zhou, C.H. Molecular design and preparation of 2-aminothiazole sulfanilamide oximes as membrane active antibacterial agents for drug resistant Acinetobacter baumannii. Bioorg. Chem. 2021, 113, 105039. [Google Scholar] [CrossRef]
- Nunzio, C.; Vera, M.; Luana, P.; Corrado, T. Natural isoflavones and semisynthetic derivatives as pancreatic lipase inhibitors. J. Nat. Prod. 2021, 84, 654–665. [Google Scholar]
- Yang, X.; Sun, H.; Maddili, S.K.; Li, S.; Yang, R.G.; Zhou, C.H. Dihydropyrimidinone imidazoles as unique structural antibacterial agents for drug-resistant gram-negative pathogens. Eur. J. Med. Chem. 2022, 232, 114188. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Z.; Tangadanchu, V.K.R.; Battini, N.; Bheemanaboina, R.R.Y.; Zang, Z.L.; Zhang, S.L.; Zhou, C.H. Indole-nitroimidazole conjugates as efficient manipulators to decrease the genes expression of methicillin-resistant Staphylococcus aureus. Eur. J. Med. Chem. 2019, 179, 723–735. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.P.; Battini, N.; Ansari, M.F.; Zhou, C.H. Membrane active 7-thiazoxime quinolones as novel DNA binding agents to decrease the genes expression and exert potent antimethicillin-resistant Staphylococcus aureus activity. Eur. J. Med. Chem. 2021, 217, 113340. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Catucci, G.; Nardo, G.D.; Gilardi, G. Effector role of cytochrome P450 reductase for androstenedione binding to human aromatase. Int. J. Biol. Macromol. 2020, 164, 510–517. [Google Scholar] [CrossRef]
- Wang, J.; Battini, N.; Ansari, M.F.; Zhou, C.H. Synthesis and biological evaluation of quinazolonethiazoles as new potential conquerors towards Pseudomonas aeruginosa. Chin. J. Chem. 2021, 39, 1093–1103. [Google Scholar] [CrossRef]
- Sun, H.; Huang, S.Y.; Jeyakkumar, P.; Cai, G.X.; Fang, B.; Zhou, C.H. Natural berberine-derived azolyl ethanols as new structural antibacterial agents against drug-resistant Escherichia coli. J. Med. Chem. 2022, 65, 436–459. [Google Scholar] [CrossRef]
- Xie, Y.P.; Sangaraiah, N.; Meng, J.P.; Zhou, C.H. Unique Carbazole-oxadiazole derivatives as new potential antibiotics for combating gram-positive and -negative bacteria. J. Med. Chem. 2022, 65, 6171–6190. [Google Scholar] [CrossRef]
- Sui, Y.F.; Ansari, M.F.; Fang, B.; Zhang, S.L.; Zhou, C.H. Discovery of novel purinylthiazolylethanone derivatives as anti-Candida albicans agents through possible multifaceted mechanisms. Eur. J. Med. Chem. 2021, 221, 113557. [Google Scholar] [CrossRef]
- Wang, J.; Ansari, M.F.; Lin, J.M.; Zhou, C.H. Design and synthesis of sulfanilamide aminophosphonates as novel antibacterial agents towards Escherichia coli. Chin. J. Chem. 2021, 39, 2251–2263. [Google Scholar] [CrossRef]
- Sui, Y.F.; Ansari, M.F.; Zhou, C.H. Pyrimidinetrione-imidazoles as a unique structural type of potential agents towards Candida albicans: Design, synthesis and biological evaluation. Chem. Asian J. 2021, 16, 1417–1429. [Google Scholar] [CrossRef]
- Li, F.F.; Zhang, P.L.; Tangadanchu, V.K.R.; Li, S.; Zhou, C.H. Novel metronidazole-derived three-component hybrids as promising broad-spectrum agents to combat oppressive bacterial resistance. Bioorgan. Chem. 2022, 122, 105718. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.C.; Zhang, P.L.; Kumar, K.V.; Li, S.; Geng, R.X.; Zhou, C.H. Discovery of unique thiazolidinone-conjugated coumarins as novel broad spectrum antibacterial agents. Eur. J. Med. Chem. 2022, 232, 114192. [Google Scholar] [CrossRef]
- Zhang, P.L.; Lv, J.S.; Ansari, M.F.; Battini, N.; Cai, G.X.; Zhou, C.H. Synthesis of naphthalimide triazoles with a novel structural framework and their anti-Aspergillus fumigatus effects. Sci. Sin. Chim. 2021, 51, 1094–1103. [Google Scholar] [CrossRef]
- Liu, Y.; Niu, L.Y.; Chen, Y.Z.; Yang, Q.Z. A self-assembled fluorescent nanoprobe for detection of GSH and dual-channel imaging. J. Photochem. Photobiol. A Chem. 2018, 355, 311–317. [Google Scholar] [CrossRef]
- Sun, H.; Li, Z.Z.; Jeyakkumar, P.; Zang, Z.L.; Fang, B.; Zhou, C.H. A new discovery of unique 13-(benzimidazolylmethyl)berberines as promising broad-spectrum antibacterial agents. J. Agric. Food Chem. 2022, 70, 12320–12329. [Google Scholar] [CrossRef]
- Wang, J.; Ansari, M.F.; Zhou, C.H. Identification of unique quinazolone thiazoles as novel structural scaffolds for potential gram-negative bacterial conquerors. J. Med. Chem. 2021, 64, 7630–7645. [Google Scholar] [CrossRef]
- Yang, X.C.; Hu, C.F.; Zhang, P.L.; Li, S.; Hu, C.S.; Geng, R.X.; Zhou, C.-H. Coumarin thiazoles as unique structural skeleton of potential antimicrobial agents. Bioorgan. Chem. 2022, 124, 105855. [Google Scholar] [CrossRef]
- Deng, Z.; Bheemanaboina, R.R.Y.; Luo, Y.; Zhou, C.H. Aloe emodin-conjugated sulfonyl hydrazones as novel type of antibacterial modulators against S. aureus 25923 through multifaceted synergistic effects. Bioorgan. Chem. 2022, 127, 106035. [Google Scholar] [CrossRef]
- Li, F.F.; Zhao, W.H.; Tangadanchu, V.K.R.; Meng, J.P.; Zhou, C.H. Discovery of novel phenylhydrazone-based oxindole-thiolazoles as potent antibacterial agents toward Pseudomonas aeruginosa. Eur. J. Med. Chem. 2022, 239, 114521. [Google Scholar] [CrossRef]
- Yang, X.; Syed, R.; Fang, B.; Zhou, C.H. A new discovery towards novel skeleton of benzimidazole-conjugated pyrimidinones as unique effective antibacterial agents. Chin. J. Chem. 2022, 40, 2642–2654. [Google Scholar] [CrossRef]
- Yang, X.C.; Zeng, C.M.; Avula, S.R.; Peng, X.M.; Geng, R.X.; Zhou, C.H. Novel coumarin aminophosphonates as potential multitargeting antibacterial agents against Staphylococcus aureus. Eur. J. Med. Chem. 2023, 245, 114891. [Google Scholar] [CrossRef] [PubMed]






| Compounds | Fungi | ||||
|---|---|---|---|---|---|
| Candida albicans | Candida albicans 90023 | Aspergillus fumigatus | Candida tropicalis | Candida parapsilosis 22019 | |
| 2 | 128 | 256 | 128 | 64 | 64 |
| 3a | 128 | 64 | 128 | 32 | 128 |
| 3b | 128 | 64 | 128 | 32 | 128 |
| 4a | 128 | 64 | 128 | 16 | 32 |
| 4b | 128 | 128 | 128 | 64 | 64 |
| 4c | 256 | 128 | 64 | 64 | 128 |
| 5 | 128 | 64 | 128 | 128 | 128 |
| 6a | 256 | 64 | 64 | 32 | 64 |
| 6b | 256 | 128 | 256 | 128 | 128 |
| 6c | 256 | 64 | 128 | 32 | 16 |
| 7a | 256 | 64 | 128 | 8 | 32 |
| 7b | 256 | 128 | 256 | 64 | 256 |
| 7c | 256 | 256 | 256 | 128 | 256 |
| 7d | 128 | 64 | 64 | 32 | 32 |
| 7e | 128 | 64 | 64 | 32 | 32 |
| 7f | 128 | 128 | 32 | 4 | 64 |
| 8a | 128 | 64 | 128 | 32 | 64 |
| 8b | 128 | 64 | 128 | 32 | 64 |
| 8c | 128 | 64 | 128 | 16 | 32 |
| 8d | 32 | 32 | 64 | 16 | 32 |
| Fluconazole | 4 | 4 | 512 | 256 | 128 |
| Parameters | 7f | Fluconazole |
|---|---|---|
| MW (g/mol) < 500 | 389.43 | 306.27 |
| MLog P ≤ 4.15 | 2.93 | 1.47 |
| H-bond acceptors ≤ 10 | 4 | 7 |
| H-bond donors ≤ 5 | 2 | 1 |
| Lipinski violations | 0 | 0 |
| Skin permeation (cm/s) | −6.24 | −7.92 |
| Human intestinal absorption (HIA, %) | 94.42 (+) | 98.83 (+) |
| Acute oral toxicity | III | III |
| BBB permeant | No | No |
| Bioavailability Score | 0.55 | 0.55 |
| PAINS | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.; Tangadanchu, V.K.R.; Zhou, C. Identification of Novel Antifungal Skeleton of Hydroxyethyl Naphthalimides with Synergistic Potential for Chemical and Dynamic Treatments. Molecules 2022, 27, 8453. https://doi.org/10.3390/molecules27238453
Zhang P, Tangadanchu VKR, Zhou C. Identification of Novel Antifungal Skeleton of Hydroxyethyl Naphthalimides with Synergistic Potential for Chemical and Dynamic Treatments. Molecules. 2022; 27(23):8453. https://doi.org/10.3390/molecules27238453
Chicago/Turabian StyleZhang, Pengli, Vijai Kumar Reddy Tangadanchu, and Chenghe Zhou. 2022. "Identification of Novel Antifungal Skeleton of Hydroxyethyl Naphthalimides with Synergistic Potential for Chemical and Dynamic Treatments" Molecules 27, no. 23: 8453. https://doi.org/10.3390/molecules27238453
APA StyleZhang, P., Tangadanchu, V. K. R., & Zhou, C. (2022). Identification of Novel Antifungal Skeleton of Hydroxyethyl Naphthalimides with Synergistic Potential for Chemical and Dynamic Treatments. Molecules, 27(23), 8453. https://doi.org/10.3390/molecules27238453







