Lubeluzole Repositioning as Chemosensitizing Agent on Multidrug-Resistant Human Ovarian A2780/DX3 Cancer Cells
Abstract
1. Introduction
2. Results
2.1. Inhibition of Cell Proliferation by Lube S, Lube R, and Lube S/R
2.2. Inhibition of A2780/DX3 Cell Proliferation by Dox Associated with Lube Enantiomers or the Racemate
2.3. Detection of Apoptosis
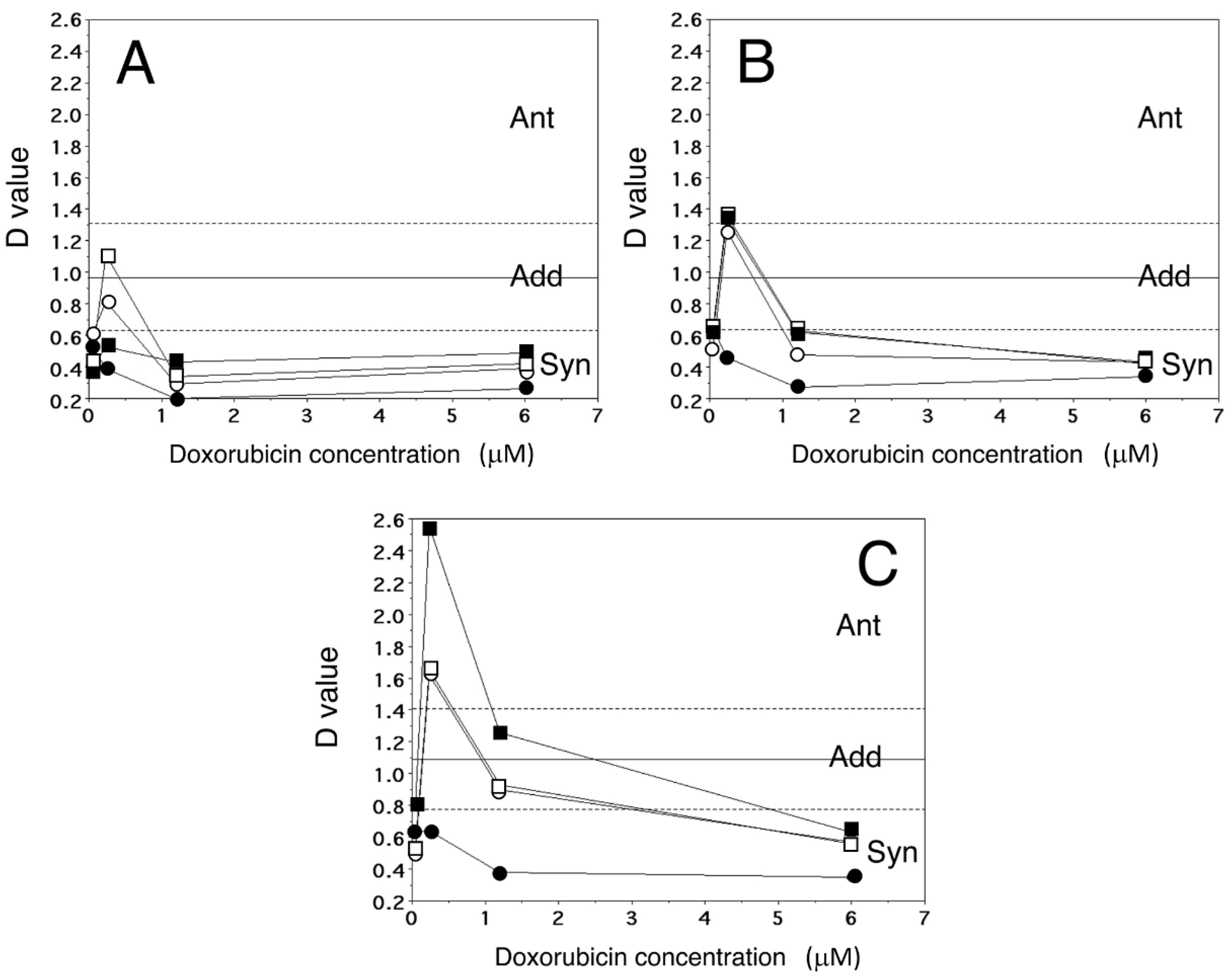
2.4. Analysis of Dox Accumulation by Flow Cytometry
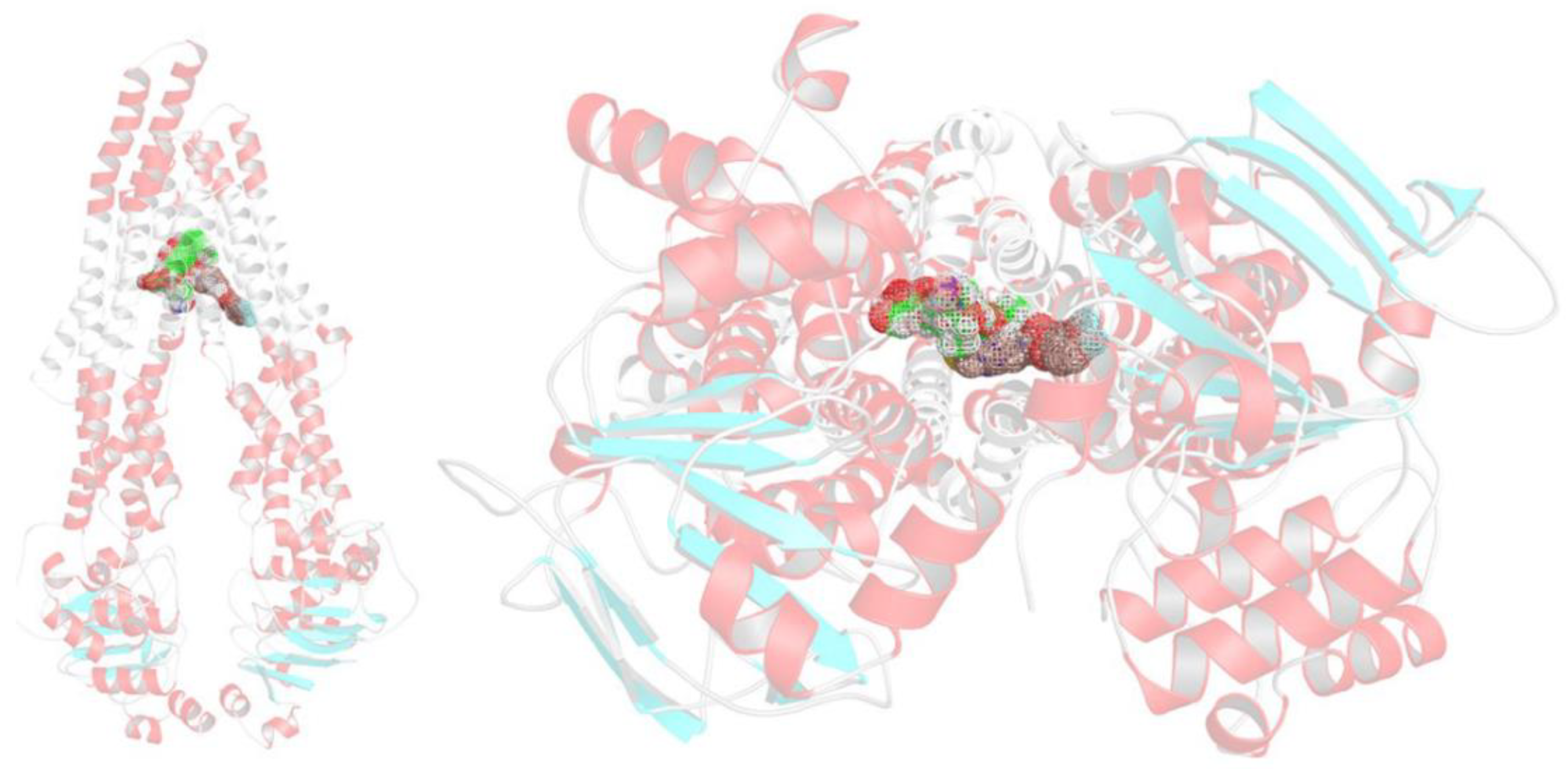
2.5. Docking Studies
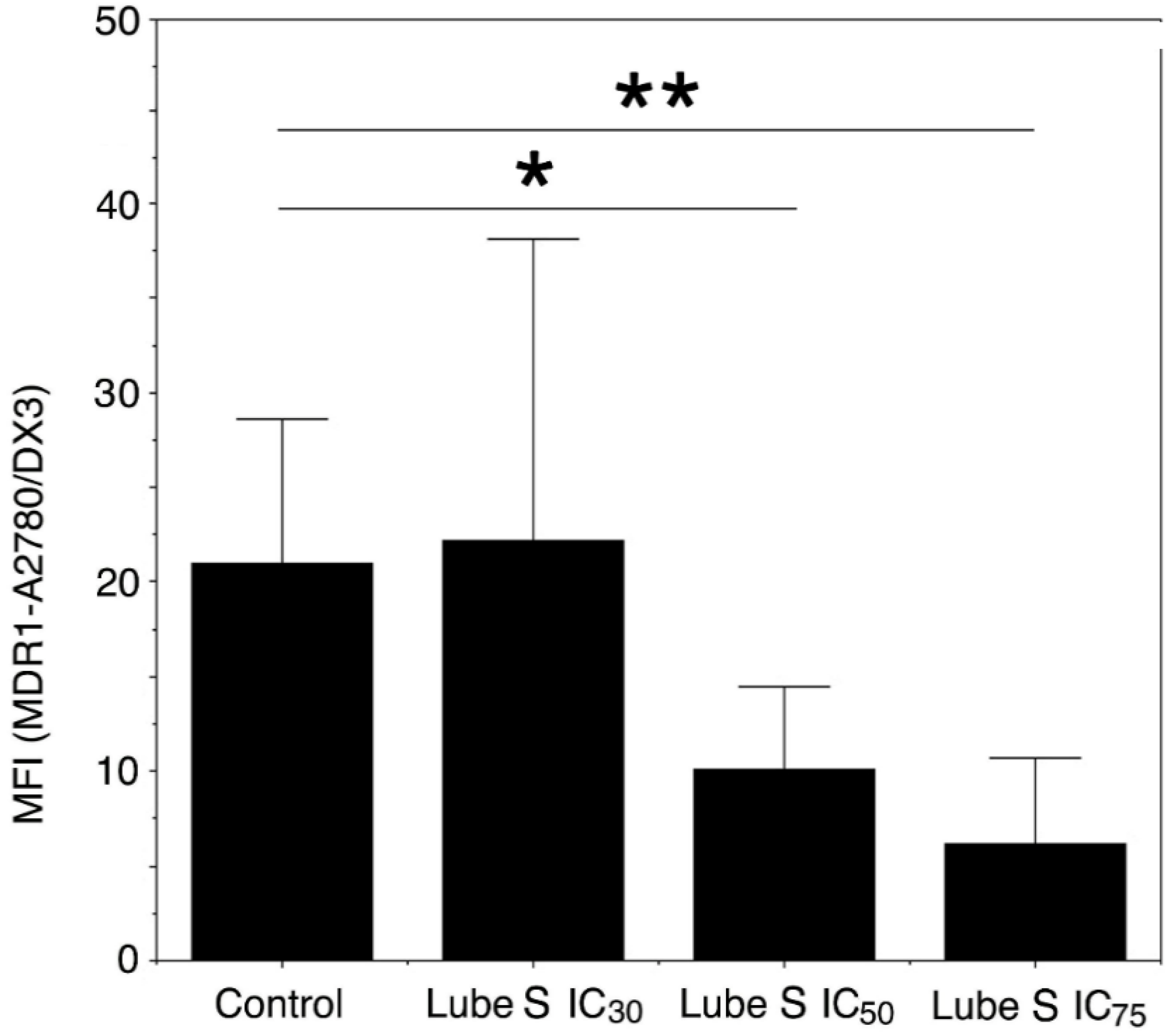
2.6. Influence of Lube S on MDR1 Expression
2.7. Effect of Lube S and Dox on the Intracellular Oxidative Stress Damages in A2780/DX3 Cells
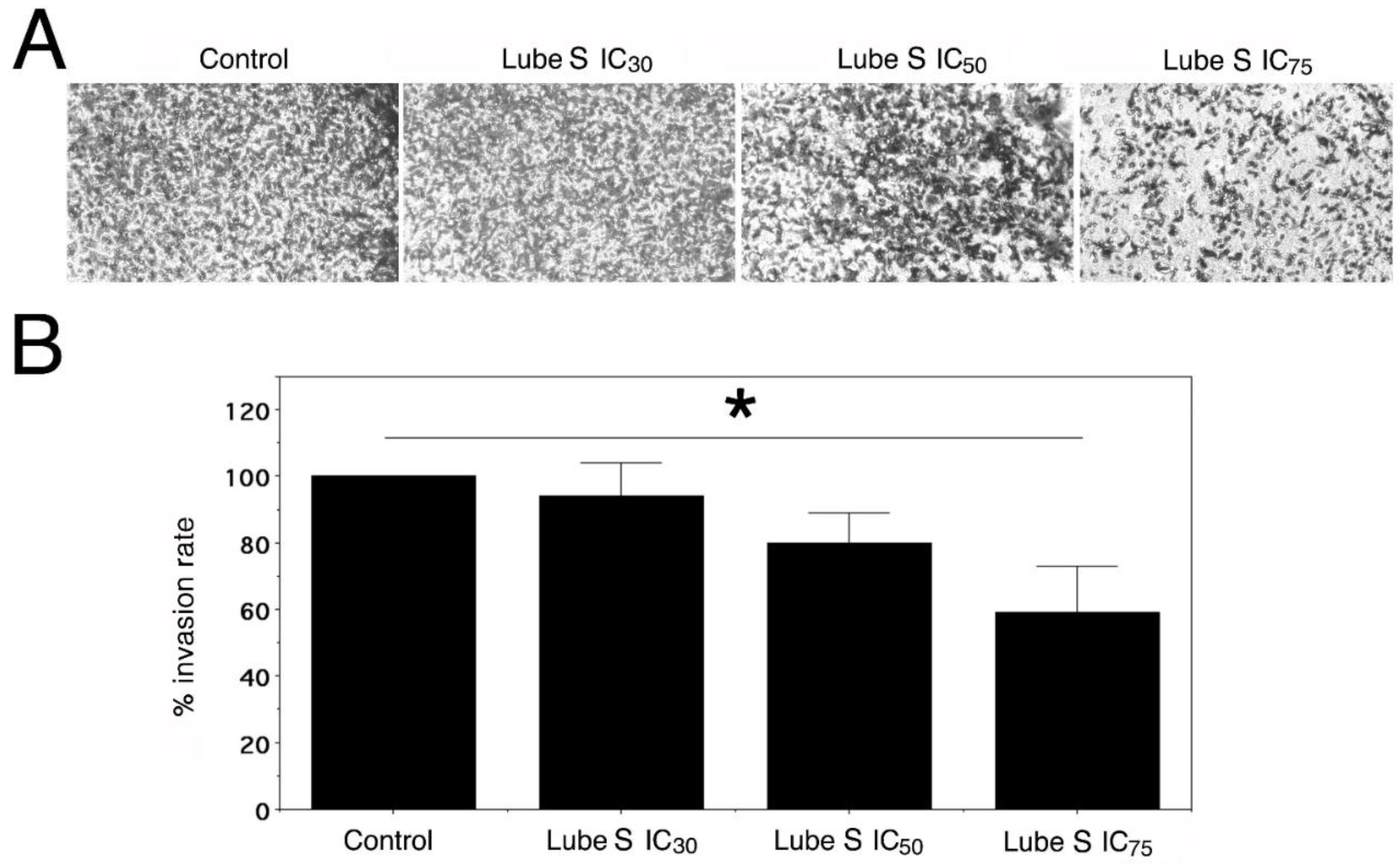
2.8. Effect of Lube S on the Invasion of MDA-MB-231
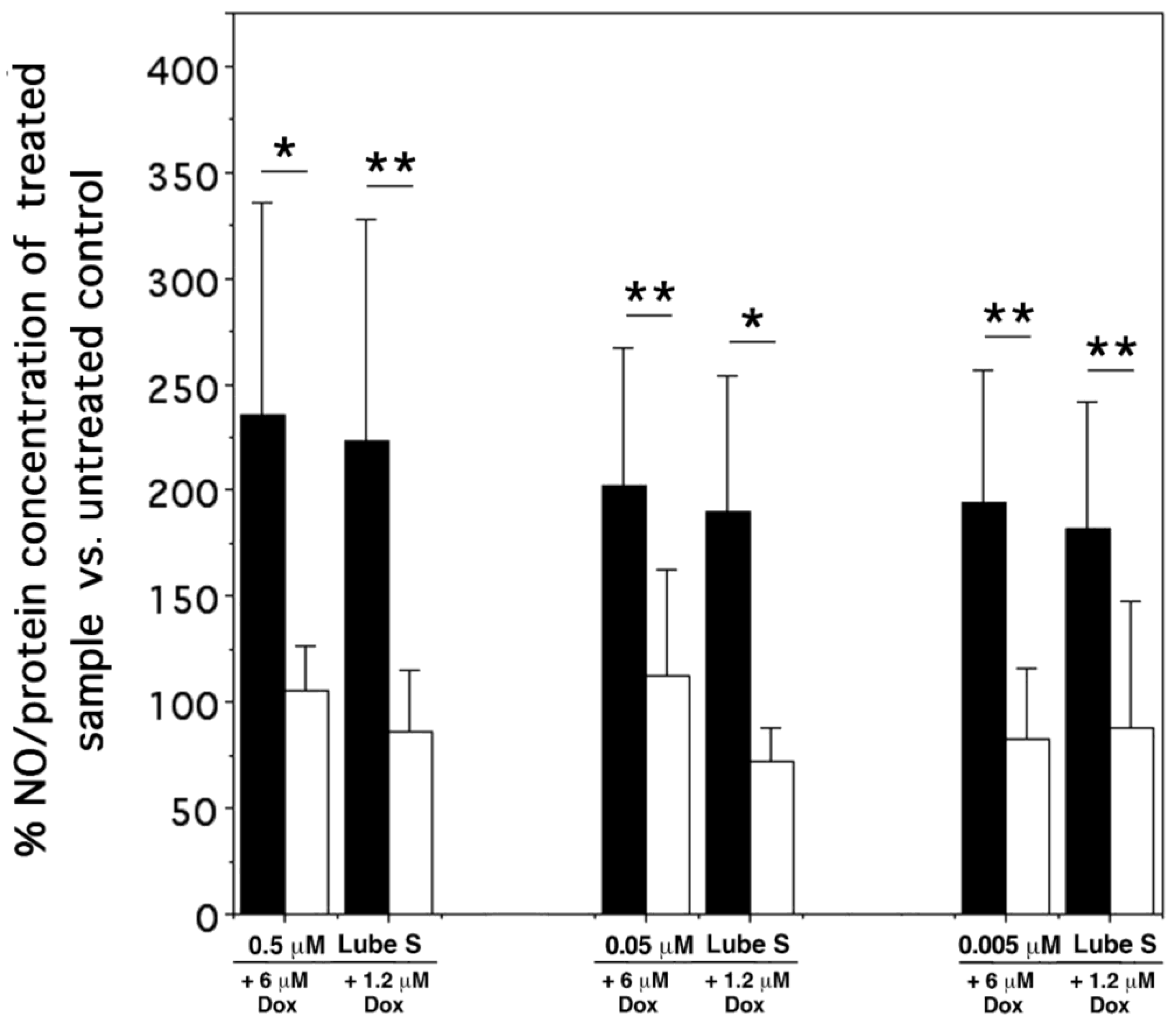
2.9. NO Detection
3. Discussion
4. Materials and Methods
4.1. Cells
4.2. Chemicals
4.3. In Vitro Studies of Inhibition of Cell Proliferation
4.4. Detection of Apoptosis by 4′,6-Diamidino-2-Phenylindole (DAPI) Test
4.5. Cytofluorimetric Study of Intracellular Accumulation of Dox
4.6. Study of MDR1 Expression
4.7. Cell Invasion Assay
4.8. Evaluation of Intracellular Malondialdehyde Level as a Marker of Oxidative Stress Damage
4.9. Molecular Modelling Methods
4.10. Detection of NO Intracellular Levels in A2780/DX3 Cells
4.11. Data Analysis and Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Ryck, M.; Keersmaekers, R.; Duytschaever, H.; Claes, C.; Clincke, G.; Janssen, M.; Van Reet, G.J. Lubeluzole Protects Sensorimotor Function and Reduces Infarct Size in a Photochemical Stroke Model in Rats. J. Pharmacol. Exp. Ther. 1996, 279, 748–758. [Google Scholar] [PubMed]
- Lesage, A.S.; Peeters, L.; Leysen, J.E. Lubeluzole, a Novel Long-Term Neuroprotectant, Inhibits the Glutamate-Activated Nitric Oxide Synthase Pathway. J. Pharmacol. Exp. Ther. 1996, 279, 759–766. [Google Scholar] [PubMed]
- Scheller, D.; Kolb, J.; Szathmary, S.; Zacharias, E.; Deryck, M.; Van Reempts, J.; Clinke, B.; Tegtmeier, F. Extracellular Changes of Glutamate in the Periinfarct Zone. Effect of Lubeluzole. J. Cereb. Blood Flow Metab. 1995, 15, S379. [Google Scholar]
- Mueller, R.N.; Deyo, D.J.; Brantley, D.R.; Disterhoft, J.F.; Zornow, M.H. Lubeluzole and Conditioned Learning after Cerebral Ischemia. Exp. Brain Res. 2003, 152, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Desaphy, J.-F.; Carbonara, R.; Costanza, T.; Lentini, G.; Cavalluzzi, M.M.; Bruno, C.; Franchini, C.; Camerino, D.C. Molecular Dissection of Lubeluzole Use-Dependent Block of Voltage-Gated Sodium Channels Discloses New Therapeutic Potentials. Mol. Pharmacol. 2013, 83, 406–415. [Google Scholar] [CrossRef]
- Bruno, C.; Cavalluzzi, M.M.; Rusciano, M.R.; Lovece, A.; Carriera, A.; Pracella, R.; Giannuzzi, G.; Polimeno, L.; Viale, M.; Illario, M.; et al. The Chemosensitizing Agent Lubeluzole Binds Calmodulin and Inhibits Ca2+/Calmodulin-Dependent Kinase II. Eur. J. Med. Chem. 2016, 116, 36–45. [Google Scholar] [CrossRef]
- Cavalluzzi, M.M.; Viale, M.; Bruno, C.; Carocci, A.; Catalano, A.; Carrieri, A.; Franchini, C.; Lentini, G. A Convenient Synthesis of Lubeluzole and Its Enantiomer: Evaluation as Chemosensitizing Agents on Human Ovarian Adenocarcinoma and Lung Carcinoma Cells. Bioorg. Med. Chem. Lett. 2013, 23, 4820–4823. [Google Scholar] [CrossRef]
- Ly, J.V.; Zavala, J.A.; Donnan, G.A. Neuroprotection and Thrombolysis: Combination Therapy in Acute Ischaemic Stroke. Expert Opin. Pharmacother. 2006, 7, 1571–1581. [Google Scholar] [CrossRef]
- Gandolfo, C.; Sandercock, P.; Conti, M. Lubeluzole for Acute Ischaemic Stroke. Cochrane Database Syst. Rev. 2002, CD001924. [Google Scholar] [CrossRef]
- Teodori, E.; Dei, S.; Scapecchi, S.; Gualtieri, F. The Medicinal Chemistry of Multidrug Resistance (MDR) Reversing Drugs. Farmaco 2002, 57, 385–415. [Google Scholar] [CrossRef]
- Nadkar, A.; Pungaliya, C.; Drake, K.; Zajac, E.; Singhal, S.S.; Awasthi, S. Therapeutic Resistance in Lung Cancer. Expert Opin. Drug Metab. Toxicol. 2006, 2, 753–777. [Google Scholar] [CrossRef] [PubMed]
- Gatti, L.; Zunino, F. Overview of Tumor Cell Chemoresistance Mechanisms. Methods Mol. Med. 2005, 111, 127–148. [Google Scholar] [CrossRef] [PubMed]
- Litman, T.; Druley, T.E.; Stein, W.D.; Bates, S.E. From MDR to MXR: New Understanding of Multidrug Resistance Systems, their Properties and Clinical Significance. Cell Mol. Life Sci. 2001, 58, 931–959. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.H.; Yu, A.M. ABC Transporters in Multidrug Resistance and Pharmacokinetics, and Strategies for Drug Development. Curr. Pharm. Des. 2014, 20, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Aszalos, A. Drug-Drug Interactions Affected by the Transporter Protein, P-Glycoprotein (ABCB1, MDR1) II. Clinical Aspects. Drug Discov. Today 2007, 12, 838–843. [Google Scholar] [CrossRef]
- Viale, M.; Cordazzo, C.; Cosimelli, B.; de Totero, D.; Castagnola, P.; Aiello, C.; Severi, E.; Petrillo, G.; Cianfriglia, M.; Spinelli, D. Inhibition of MDR1 Activity in vitro by a Novel Class of Diltiazem Analogues: Toward New Candidates. J. Med. Chem. 2009, 52, 259–266. [Google Scholar] [CrossRef]
- Micucci, M.; Viale, M.; Chiarini, A.; Spinelli, D.; Frosini, M.; Tavani, C.; Maccagno, M.; Bianchi, L.; Gangemi, R.; Budriesi, R. 3-Aryl-4-Nitrobenzothiochromans S,S-Dioxide: From Calcium-Channel Modulators Properties to Multidrug-Resistance Reverting Activity. Molecules 2020, 25, 1056. [Google Scholar] [CrossRef]
- Zhang, J.; Ming, C.; Zhang, W.; Okechukwu, P.N.; Morak-Młodawska, B.; Pluta, K.; Jeleń, M.; Akim, A.M.; Ang, K.-P.; Ooi, K.K. 10H-3,6-Diazaphenothiazine Induces G2/M Phase Cell Cycle Arrest and Caspase-Dependent Apoptosis and Inhibits Cell Invasion of A2780 Ovarian Carcinoma Cells through the Regulation of NF-Κb and (BIRC6-XIAP) Complexes. Drug Des. Devel. Ther. 2017, 11, 3045–3063. [Google Scholar] [CrossRef]
- Alam, A.; Kowal, J.; Broude, E.; Roninson, I.; Locher, K.P. Structural Insight into Substrate and Inhibitor Discrimination by Human P-Glycoprotein. Science 2019, 363, 753–756. [Google Scholar] [CrossRef]
- Guglielmo, S.; Lazzarato, L.; Contino, M.; Perrone, M.G.; Chegaev, K.; Carrieri, A.; Fruttero, R.; Colabufo, N.A.; Gasco, A. Structure-Activity Relationship Studies on Tetrahydroisoquinoline Derivatives: [4′-(6,7-Dimethoxy-3,4-Dihydro-1H-Isoquinolin-2-Ylmethyl)Biphenyl-4-Ol] (MC70) Conjugated through Flexible Alkyl Chains With Furazan Moieties Gives Rise to Potent and Selective Ligands of P-Glycoprotein. J. Med. Chem. 2016, 59, 6729–6738. [Google Scholar] [CrossRef]
- Contino, M.; Guglielmo, S.; Perrone, M.G.; Giampietro, R.; Rolando, B.; Carrieri, A.; Zaccaria, D.; Chegaev, K.; Borio, V.; Riganti, C.; et al. New Tetrahydroisoquinoline-Based P-Glycoprotein Modulators: Decoration of the Biphenyl Core Gives Selective Ligands. MedChemComm 2018, 9, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Nosol, K.; Romane, K.; Irobalieva, R.N.; Alam, A.; Kowal, J.; Fujita, N.; Locher, K.P. Cryo-EM Structures Reveal Distinct Mechanisms of Inhibition of the Human Multidrug Transporter ABCB1. Proc. Natl. Acad. Sci. USA 2020, 117, 26245–26253. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.; Küng, R.; Kowal, J.; McLeod, R.A.; Tremp, N.; Broude, E.V.; Roninson, I.B.; Stahlberg, H.; Locher, K.P. Structure of a Zosuquidar and UIC2-Bound Human-Mouse Chimeric ABCB1. Proc. Natl. Acad. Sci. USA 2018, 115, E1973–E1982. [Google Scholar] [CrossRef] [PubMed]
- Tsubaki, M.; Komai, M.; Itoh, T.; Imano, M.; Sakamoto, K.; Shimaoka, H.; Takeda, T.; Ogawa, N.; Mashimo, K.; Fujiwara, D.; et al. By Inhibiting Src, Verapamil and Dasatinib Overcome Multidrugresistance via Increased Expression of Bim and Decreased Expressions of MDR1 and Survivin in Human Multidrug-Resistant Myeloma Cells. Leuk. Res. 2014, 38, 121–130. [Google Scholar] [CrossRef]
- Tabata, M.; Tsubaki, M.; Takeda, T.; Tateishi, K.; Tsurushima, K.; Imano, M.; Satou, T.; Ishizaka, T.; Nishida, S. Dasatinib Reverses Drug Resistance by Downregulating MDR1 and Survivin in Burkitt Lymphoma Cells. BMC Complement Med. Ther. 2020, 20, 84. [Google Scholar] [CrossRef]
- Muller, C.; Goubin, F.; Ferrandis, E.; Cornil-Scharwtz, I.; Bailly, J.D.; Bordier, C.; Bénard, J.; Sikic, B.I.; Laurent, G. Evidence for Transcriptional Control of Human Mdr1 Gene Expression by Verapamil in Multidrug-Resistant Leukemic Cells. Mol. Pharmacol. 1995, 47, 51–56. [Google Scholar]
- James, P.; Vorherr, T.; Carafoli, E. Calmodulin-Binding Domains: Just Two Faced or Multi-Faceted? Trends Biochem. Sci. 1995, 20, 38–42. [Google Scholar] [CrossRef]
- Vertessy, B.G.; Harmat, V.; Böcskei, Z.; Náray-Szabó, G.; Orosz, F.; Ovádi, J. Simultaneous Binding of Drugs with Different Chemical Structures to Ca2+-calmodulin: Crystallographic and Spectroscopic Studies. Biochemistry 1998, 37, 15300–15310. [Google Scholar] [CrossRef]
- Mayur, Y.C.; Jagadeesh, S.; Thimmaiah, K.N. Targeting Calmodulin in Reversing Multi Drug Resistance in Cancer Cells. Mini Rev. Med. Chem. 2006, 6, 1383–1389. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, Y.; Chen, Y.; Qi, J.; Ren, S.; Xushi, M.Y.; Yang, C.; Zhu, H.; Xiong, D. A Novel Calmodulin Antagonist O-(4-Ethoxyl-Butyl)-Berbamine Overcomes Multidrug Resistance in Drug-Resistant MCF-7/ADR Breast Carcinoma Cells. J. Pharm. Sci. 2010, 99, 3266–3275. [Google Scholar] [CrossRef]
- Cavalluzzi, M.M.; Budriesi, R.; De Salvia, M.A.; Quintieri, L.; Piarulli, M.; Milani, G.; Gualdani, R.; Micucci, M.; Corazza, I.; Rosato, A.; et al. Lubeluzole: From Anti-Ischemic Drug to Preclinical Antidiarrheal Studies. Pharmacol. Rep. 2021, 73, 172–184. [Google Scholar] [CrossRef]
- Timm, K.N.; Tyler, D.J. The Role of AMPK Activation for Cardioprotection in Doxorubicin-Induced Cardiotoxicity. Cardiovasc. Drugs Ther. 2020, 34, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Mazevet, M.; Moulin, M.; Llach-Martinez, A.; Chargari, C.; Deutsch, E.; Gomez, A.M.; Morel, E. Complications of Chemotherapy, a Basic Science Update. Presse Med. 2013, 42, 352–361. [Google Scholar] [CrossRef] [PubMed]
- El-Sehemy, A.; Postovit, L.-M.; Fu, Y.X. Nitric Oxide Signaling in Human Ovarian Cancer: A Potential Therapeutic Target. Nitric Oxide 2016, 54, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Crowell, J.A.; Steele, V.E.; Sigman, C.C.; Fay, J.R. Is Inducible Nitric Oxide Synthase a Target for Chemoprevention? Mol. Cancer Ther. 2003, 2, 815–823. [Google Scholar] [PubMed]
- Juang, S.-H.; Xie, K.; Xu, L.; Shi, Q.; Wang, Y.; Yoneda, J.; Fidler, I.J. Suppression of Tumorigenicity and Metastasis of Human Renal Carcinoma Cells by Infection with Retroviral Vectors Harboring the Murine Inducible Nitric Oxide Synthase Gene. Hum. Gene Ther. 1998, 9, 845–854. [Google Scholar] [CrossRef]
- Bratasz, A.; Weir, N.M.; Parinandi, N.L.; Zweier, J.L.; Sridhar, R.; Ignarro, L.J.; Kuppusamy, P. Reversal to Cisplatin Sensitivity in Recurrent Human Ovarian Cancer Cells by NCX-4016, a Nitro Derivative of Aspirin. Proc. Natl. Acad. Sci. USA 2006, 103, 3914–3919. [Google Scholar] [CrossRef]
- Hays, E.; Bonavida, B. Nitric Oxide-Mediated Enhancement and Reversal of Resistance of Anticancer Therapies. Antioxidants 2019, 8, 407. [Google Scholar] [CrossRef]
- Raspollini, M.R.; Amunni, G.; Villanucci, A.; Boddi, V.; Baroni, G.; Taddei, A.; Taddei, G.L. Expression of Inducible Nitric Oxide Synthase and Cyclooxygenase-2 in Ovarian Cancer: Correlation with Clinical Outcome. Gynecol. Oncol. 2004, 92, 806–812. [Google Scholar] [CrossRef]
- Engels, K.; du Bois, A.; Harter, P.; Fisseler-Eckhoff, A.; Kommoss, F.; Stauber, R.; Kaufmann, M.; Nekljudova, V.; Loibl, S. VEGF-A and I-NOS Expression are Prognostic Factors in Serous Epithelial Ovarian Carcinomas after Complete Surgical Resection. J. Clin. Pathol. 2009, 62, 448–454. [Google Scholar] [CrossRef]
- Rizi, B.S.; Caneba, C.; Nowicka, A.; Nabiyar, A.W.; Liu, X.; Chen, K.; Klopp, A.; Nagrath, D. Nitric Oxide Mediates Metabolic Coupling of Omentum-Derived Adipose Stroma to Ovarian and Endometrial Cancer Cells. Cancer Res. 2015, 75, 456–471. [Google Scholar] [CrossRef] [PubMed]
- Saed, G.M.; Ali-Fehmi, R.; Jiang, Z.L.; Fletcher, N.M.; Diamond, M.P.; Abu-Soud, H.M.; Munkarah, A.R. Myeloperoxidase Serves as a Redox Switch that Regulates Apoptosis in Epithelial Ovarian Cancer. Gynecol. Oncol. 2010, 116, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Fraser, M.; Chan, S.L.; Chan, S.S.L.; Fiscus, R.R.; Tsang, B.K. Regulation of P53 and Suppression of Apoptosis by the Soluble Guanylyl Cyclase/Cgmp Pathway in Human Ovarian Cancer Cells. Oncogene 2006, 25, 2203–2221. [Google Scholar] [CrossRef]
- Bruno, C.; Carocci, A.; Catalano, A.; Cavalluzzi, M.M.; Corbo, F.; Franchini, C.; Lentini, G.; Tortorella, V. Facile, Alternative Route to Lubeluzole, its Enantiomer, and the Racemate. Chirality 2006, 18, 227–231. [Google Scholar] [CrossRef]
- Oliveri, V.; Lanza, V.; Milardi, D.; Viale, M.; Maric, I.; Sgarlata, C.; Vecchio, G. Amino- and Chloro-8-Hydroxyquinolines and Their Copper(II) Complexes as Proteasome Inhibitors and Antiproliferative Agents. Metallomics 2017, 9, 1439–1446. [Google Scholar] [CrossRef] [PubMed]
- Cianfriglia, M.; Willingham, M.C.; Tombesi, M.; Scagliotti, G.V.; Frasca, G.; Chersi, A. P-Glycoprotein Epitope Mapping. I. Identification of a Linear Human-Specific Epitope in The Fourth Loop of the P-Glycoprotein Extracellular Domain by MM4.17 Murine Monoclonal Antibody to Human Multidrug-Resistant Cells. Int. J. Cancer 1994, 56, 153–160. [Google Scholar] [CrossRef]
- Cappelli, E.; Cuccarolo, P.; Stroppiana, G.; Miano, M.; Bottega, R.; Cossu, V.; Degan, P.; Ravera, S. Defects in Mitochondrial Energetic Function Compels Fanconi Anaemia Cells to Glycolytic Metabolism. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1214–1221. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Peeters, O.M.; Blaton, N.M.; De Ranter, C.J. (+)-(S)-1-{4-[(2-Benzothiazolyl)(Methyl)Amino]Piperidyl}-3-(3,4-Difluorophenoxy)-2-Propanol (Lubeluzole). Acta Crystallogr. Sect. C Cryst. Struct. Commun. 1995, C51, 2129–2132. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 31703, Doxorubicin. 2021. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Doxorubicin (accessed on 15 September 2021).
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Schrödinger Release 2021-1, Maestro; Schrödinger, LLC: New York, NY, USA, 2021.
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Gould, I.R.; Merz, K.M.; Ferguson, D.M.; Spellmeyer, D.C.; Fox, T.; Caldwell, J.W.; Kollman, P.A. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995, 117, 5179–5193. [Google Scholar] [CrossRef]
- Forli, S.; Olson, A.J. A Force Field With Discrete Displaceable Waters and Desolvation Entropy for Hydrated Ligand Docking. J. Med. Chem. 2012, 55, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated Docking Using a Lamarckian Genetic Algorithm and Empirical Binding Free Energy Function. J. Comput. Chem. 1999, 19, 1639–1662. [Google Scholar] [CrossRef]
- El Khoury, L.; Santos-Martins, D.; Sasmal, S.; Eberhardt, J.; Bianco, G.; Ambrosio, F.A.; Solis-Vasquez, L.; Koch, A.; Forli, S.; Mobley, D.L. Comparison of Affinity Ranking Using Autodock-GPU And MM-GBSA Scores for BACE-1 Inhibitors in the D3R Grand Challenge 4. J. Comput. Aided Mol. Des. 2019, 33, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Berembaum, M.C. What is Synergy? Pharmacol. Rev. 1989, 41, 93–141. [Google Scholar]
- Berembaum, M.C. Criteria for Analyzing Interactions Between Biologically Active Agents. Adv. Cancer Res. 1981, 35, 269–335. [Google Scholar] [CrossRef]

| Drug/Compound | Cell Lines | ||||
|---|---|---|---|---|---|
| A2780 | A2780/DX3 | HepG2 | MDAMB231 | A549 | |
| Dox | 0.018 ± 0.004 a | 1.4 ± 0.5 | 0.127 ± 0.018 | 0.053 ± 0.020 | 0.058 ± 0.014 |
| Lube S | 4.7 ± 1.5 | 6.5 ± 2.9 | 7.4 ± 1.3 | 7.4 ± 2.0 | 28.8 ± 10.0 |
| Lube R | 12.0 ± 2.3 | 18.6 ± 0.8 | 6.1 ± 1.3 | 16.2 ± 2.1 | 40.5 ± 10.1 |
| Lube S/R | 8.6 ± 2.0 | 29.6 ± 4.0 | 6.3 ± 1.9 | 8.6 ± 1.5 | 29.8 ± 9.2 |
| Drug/Compound | A2780/DX3 | ||||
|---|---|---|---|---|---|
| IC5 a | IC10 | IC30 | IC50 | IC75 | |
| Lube S | 0.29 | 0.42 | 1.7 | 6.5 | 30.8 |
| Lube R | 0.65 | 1.5 | 7.3 | 18.6 | 39.2 |
| Lube S/R | 0.10 | 0.50 | 12.5 | 29.6 | 58.5 |
| LubeS (μM) | D Value | |
|---|---|---|
| MTT | Apoptosis | |
| Doxorubicin (1.2 μM) | Doxorubicin (1.2 μM) | |
| 50 | 0.94 ± 0.17 | 1.02 ± 0.01 |
| 5 | 0.20 ± 0.06 | 0.69 ± 0.26 b |
| 0.5 | 0.29 ± 0.11 | 0.39 ± 0.17 c |
| Dox + IC10 Lube S | Dox + IC30 Lube S | Dox + IC50 Lube S | Dox + IC75 Lube S |
|---|---|---|---|
| −12 ± 3% a | −4 ± 9% | −11 ± 8% | −13 ± 12% |
| Drug | FEB a | ΔE b | EFF c | POP d |
|---|---|---|---|---|
| Dox | −13.15 | 0.00 | −0.337 | 127/1000 |
| Lube S | −10.48 | 0.00 | −0.350 | 73/1000 |
| Dox Concentration (μM) | Lube S Concentration (μM) | ||
|---|---|---|---|
| 0.5 | 0.05 | 0.005 | |
| 6 | 0.26 ± 0.13 | 0.14 ± 0.02 | 1.7 ± 0.7 |
| p < 0.001 | p < 0.001 | p = 0.004 | |
| Syn | Syn | Ant | |
| 1.2 | 0.40 ± 0.26 | 0.4 ± 0.4 | 0.7 ± 0.4 |
| p = 0.001 | p = 0.02 | p = 0.036 | |
| Syn | Syn | Add | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viale, M.; Lentini, G.; Gangemi, R.; Castagnola, P.; Milani, G.; Ravera, S.; Bertola, N.; Carrieri, A.; Cavalluzzi, M.M. Lubeluzole Repositioning as Chemosensitizing Agent on Multidrug-Resistant Human Ovarian A2780/DX3 Cancer Cells. Molecules 2022, 27, 7870. https://doi.org/10.3390/molecules27227870
Viale M, Lentini G, Gangemi R, Castagnola P, Milani G, Ravera S, Bertola N, Carrieri A, Cavalluzzi MM. Lubeluzole Repositioning as Chemosensitizing Agent on Multidrug-Resistant Human Ovarian A2780/DX3 Cancer Cells. Molecules. 2022; 27(22):7870. https://doi.org/10.3390/molecules27227870
Chicago/Turabian StyleViale, Maurizio, Giovanni Lentini, Rosaria Gangemi, Patrizio Castagnola, Gualtiero Milani, Silvia Ravera, Nadia Bertola, Antonio Carrieri, and Maria Maddalena Cavalluzzi. 2022. "Lubeluzole Repositioning as Chemosensitizing Agent on Multidrug-Resistant Human Ovarian A2780/DX3 Cancer Cells" Molecules 27, no. 22: 7870. https://doi.org/10.3390/molecules27227870
APA StyleViale, M., Lentini, G., Gangemi, R., Castagnola, P., Milani, G., Ravera, S., Bertola, N., Carrieri, A., & Cavalluzzi, M. M. (2022). Lubeluzole Repositioning as Chemosensitizing Agent on Multidrug-Resistant Human Ovarian A2780/DX3 Cancer Cells. Molecules, 27(22), 7870. https://doi.org/10.3390/molecules27227870










