Dual Targeting of MDM4 and FTH1 by MMRi71 for Induced Protein Degradation and p53-Independent Apoptosis in Leukemia Cells
Abstract
1. Introduction
2. Results and Discussion
2.1. Structural Requirement for the Anti-Proliferative Activity of MMRi62 and MMRi67 Derivatives
2.2. MMRi67 Is a Bona Fide Inhibitor of MDM2-MDM4 E3 Ligase
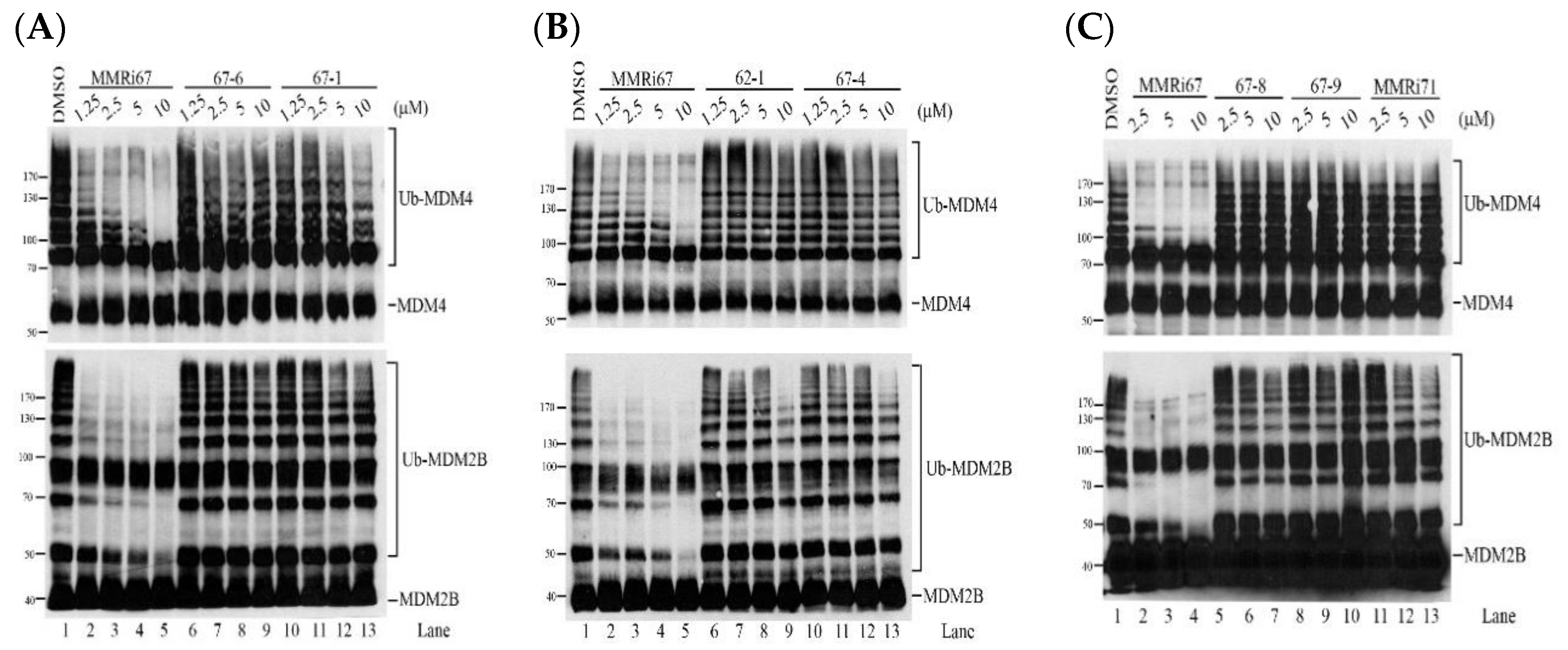
2.3. Structural Requirement of MMRi67 Derivatives for Inhibitory Activity toward MDM2-MDM4 E3 Ligase
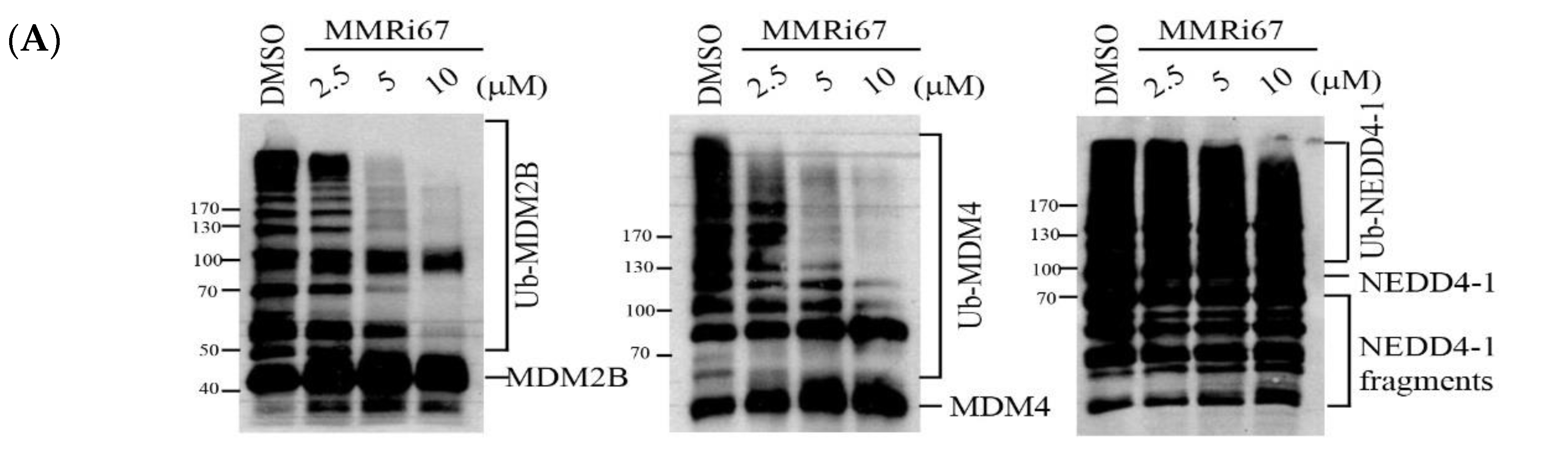
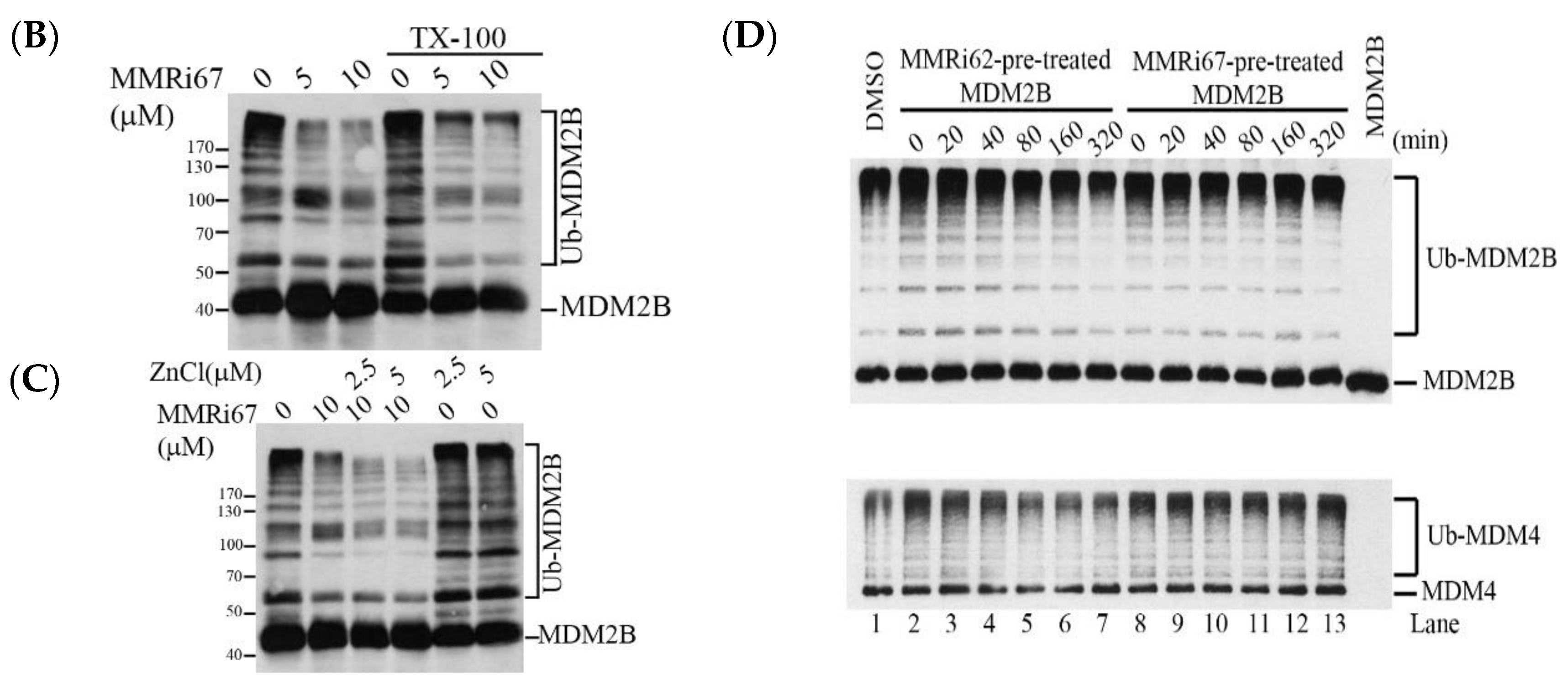
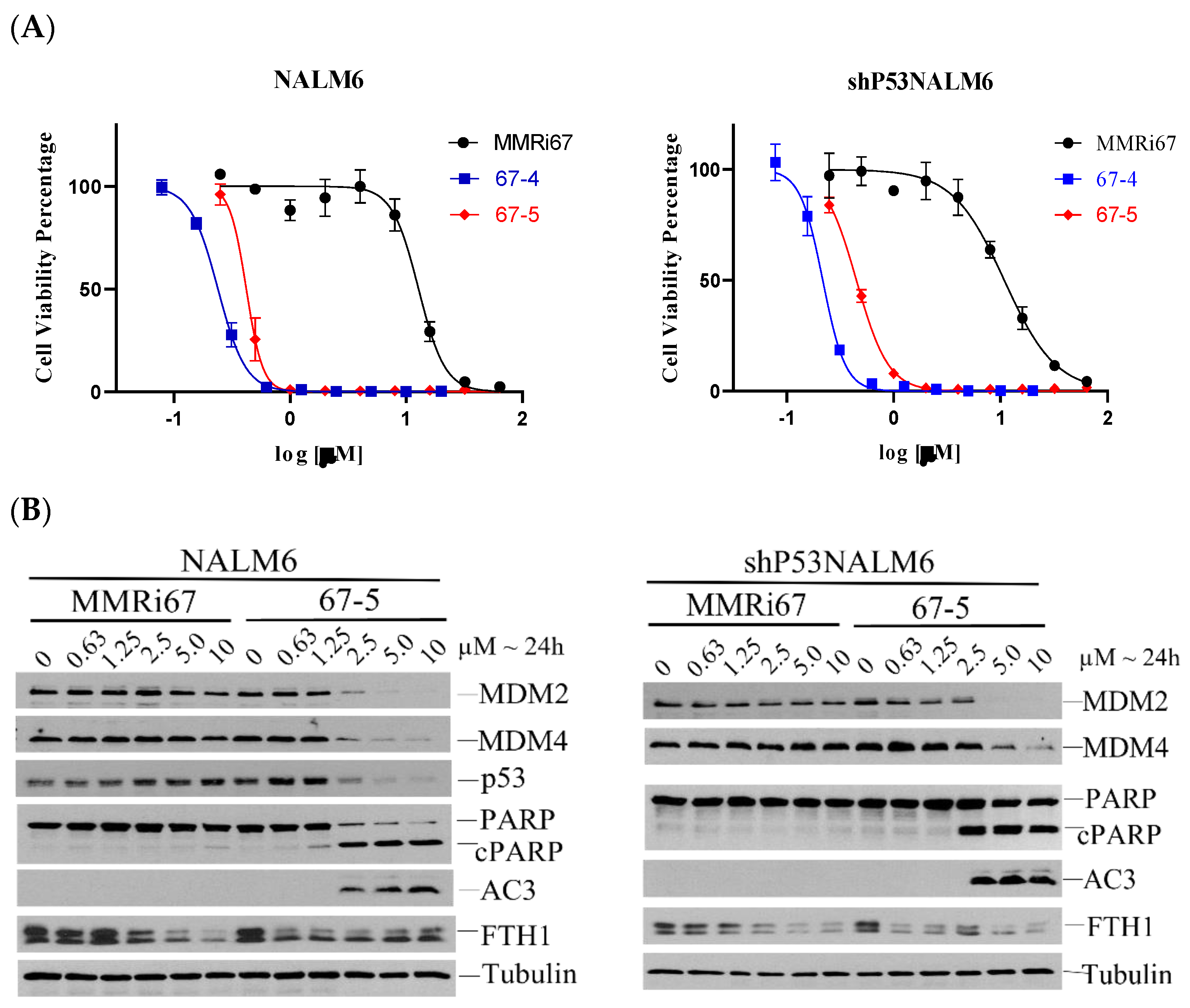
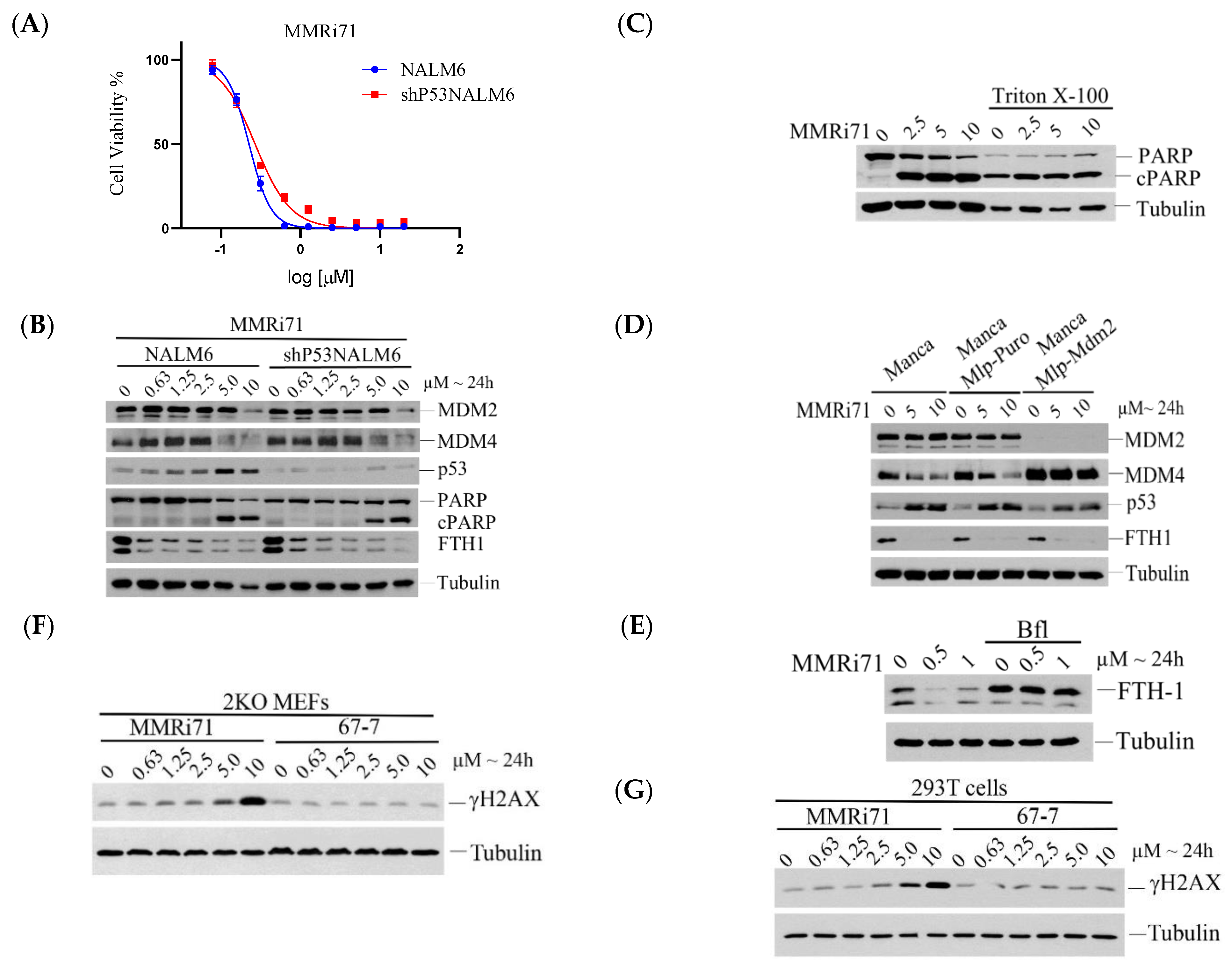
2.4. Induced MDM4 and FTH1 Degradation Is Associated with the Pro-Apoptotic Activity of MMRi67 Derivatives
3. Materials and Methods
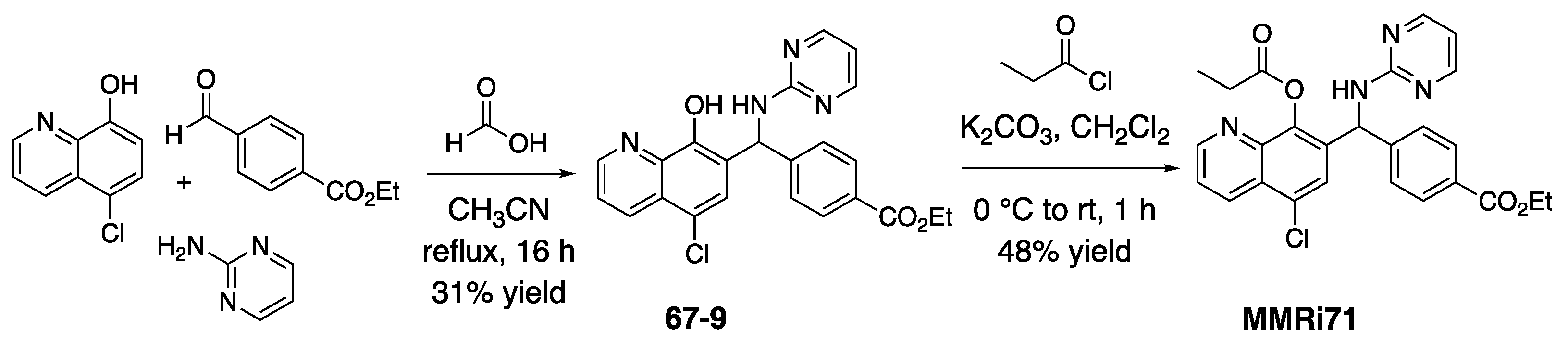
3.1. Representative Chemistry Methods
3.1.1. Ethyl 4-((5-Chloro-8-hydroxyquinolin-7-yl)(pyrimidin ylamino)methyl)benzoate (67-9) Synthesis
3.1.2. Ethyl 4-((8-(Propionyloxy)quinolin-7-yl)(pyrimidin-2-ylamino)methyl)benzoate (MMRi71) Synthesis
3.2. Biological Assays and Methods
3.2.1. Cell Culture and Small-Molecule Compounds
3.2.2. Western Blotting, In Vitro Ubiquitination, and Apoptosis Analysis
3.2.3. IC50 Measurement and Analysis
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martins, C.P.; Brown-Swigart, L.; Evan, G.I. Modeling the Therapeutic Efficacy of p53 Restoration in Tumors. Cell 2006, 127, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Ventura, A.; Kirsch, D.G.; McLaughlin, M.E.; Tuveson, D.A.; Grimm, J.; Lintault, L.; Newman, J.; Reczek, E.E.; Weissleder, R.; Jacks, T. Restoration of p53 function leads to tumour regression in vivo. Nature 2007, 445, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Zender, L.; Miething, C.; Dickins, R.A.; Hernando, E.; Krizhanovsky, V.; Cordon-Cardo, C.; Lowe, S.W. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007, 445, 656–660. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, X. Mdm2 and MdmX partner to regulate p53. FEBS Lett. 2012, 586, 1390–1396. [Google Scholar] [CrossRef] [PubMed]
- Wade, M.; Li, Y.-C.; Wahl, G.M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 2013, 13, 83–96. [Google Scholar] [CrossRef]
- Marine, J.-C.; Francoz, S.; Maetens, M.; Wahl, G.M.; Toledo, F.; Lozano, G. Keeping p53 in check: Essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006, 13, 927–934. [Google Scholar] [CrossRef]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef]
- Konopleva, M.; Martinelli, G.; Daver, N.; Papayannidis, C.; Wei, A.; Higgins, B.; Ott, M.; Mascarenhas, J.; Andreeff, M. MDM2 inhibition: An important step forward in cancer therapy. Leukemia 2020, 34, 2858–2874. [Google Scholar] [CrossRef]
- Wang, S.; Chen, F.-E. Small-molecule MDM2 inhibitors in clinical trials for cancer therapy. Eur. J. Med. Chem. 2022, 236, 114334. [Google Scholar] [CrossRef]
- Patton, J.T.; Mayo, L.D.; Singhi, A.D.; Gudkov, A.V.; Stark, G.R.; Jackson, M.W. Levels of HdmX Expression Dictate the Sensitivity of Normal and Transformed Cells to Nutlin-3. Cancer Res. 2006, 66, 3169–3176. [Google Scholar] [CrossRef]
- Hu, B.; Gilkes, D.M.; Farooqi, B.; Sebti, S.M.; Chen, J. MDMX Overexpression Prevents p53 Activation by the MDM2 Inhibitor Nutlin. J. Biol. Chem. 2006, 281, 33030–33035. [Google Scholar] [CrossRef] [PubMed]
- Gembarska, A.; Luciani, F.; Fedele, C.; Russell, E.A.; Dewaele, M.; Villar, S.; Zwolinska, A.; Haupt, S.; de Lange, J.; Yip, D.; et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nat. Med. 2012, 18, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Linares, L.K.; Hengstermann, A.; Ciechanover, A.; Müller, S.; Scheffner, M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc. Natl. Acad. Sci. USA 2003, 100, 12009–12014. [Google Scholar] [CrossRef] [PubMed]
- Kawai, H.; Lopez-Pajares, V.; Kim, M.M.; Wiederschain, D.; Yuan, Z.-M. RING Domain–Mediated Interaction Is a Requirement for MDM2’s E3 Ligase Activity. Cancer Res. 2007, 67, 6026–6030. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, J.; Jiang, X. MdmX Protein Is Essential for Mdm2 Protein-mediated p53 Polyubiquitination. J. Biol. Chem. 2011, 286, 23725–23734. [Google Scholar] [CrossRef] [PubMed]
- Itahana, K.; Mao, H.; Jin, A.; Itahana, Y.; Clegg, H.V.; Lindström, M.S.; Bhat, K.P.; Godfrey, V.L.; Evan, G.I.; Zhang, Y. Targeted Inactivation of Mdm2 RING Finger E3 Ubiquitin Ligase Activity in the Mouse Reveals Mechanistic Insights into p53 Regulation. Cancer Cell 2007, 12, 355–366. [Google Scholar] [CrossRef]
- Huang, L.; Yan, Z.; Liao, X.; Li, Y.; Yang, J.; Wang, Z.; Zuo, Y.; Kawai, H.; Shadfan, M.; Ganapathy, S.; et al. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 12001–12006. [Google Scholar] [CrossRef]
- Pant, V.; Xiong, S.; Iwakuma, T.; Quintás-Cardama, A.; Lozano, G. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc. Natl. Acad. Sci. USA 2011, 108, 11995–12000. [Google Scholar] [CrossRef]
- Linke, K.; Mace, P.D.; Smith, C.A.; Vaux, D.L.; Silke, J.; Day, C.L. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008, 15, 841–848. [Google Scholar] [CrossRef]
- Chinnam, M.; Xu, C.; Lama, R.; Zhang, X.; Cedeno, C.D.; Wang, Y.; Stablewski, A.B.; Goodrich, D.W.; Wang, X. MDM2 E3 ligase activity is essential for p53 regulation and cell cycle integrity. PLoS Genet. 2022, 18, e1010171. [Google Scholar] [CrossRef]
- Wu, W.; Xu, C.; Ling, X.; Fan, C.; Buckley, B.P.; Chernov, M.V.; Ellis, L.; Li, F.; Muñoz, I.G.; Wang, X. Targeting RING domains of Mdm2–MdmX E3 complex activates apoptotic arm of the p53 pathway in leukemia/lymphoma cells. Cell Death Dis. 2015, 6, e2035. [Google Scholar] [CrossRef] [PubMed]
- Lama, R.; Xu, C.; Galster, S.L.; Querol-García, J.; Portwood, S.; Mavis, C.K.; Ruiz, F.M.; Martin, D.; Wu, J.; Giorgi, M.C.; et al. Small molecule MMRi62 targets MDM4 for degradation and induces leukemic cell apoptosis regardless of p53 status. Front. Oncol. 2022, 12, 933446. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lama, R.; Galster, S.L.; Inigo, J.R.; Wu, J.; Chandra, D.; Chemler, S.R.; Wang, X. Small-Molecule MMRi62 Induces Ferroptosis and Inhibits Metastasis in Pancreatic Cancer via Degradation of Ferritin Heavy Chain and Mutant p53. Mol. Cancer Ther. 2022, 21, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, M.; Pastuch-Gawołek, G.; Pluta, A.; Erfurt, K.; Domiński, A.; Kurcok, P. 8-Hydroxyquinoline Glycoconjugates: Modifications in the Linker Structure and Their Effect on the Cytotoxicity of the Obtained Compounds. Molecules 2019, 24, 4181. [Google Scholar] [CrossRef] [PubMed]
- Oliveri, V.; Giuffrida, M.L.; Vecchio, G.; Aiello, C.; Viale, M. Gluconjugates of 8-hydroxyquinolines as potential anti-cancer prodrugs. Dalton Trans. 2012, 41, 4530–4535. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef]
- Aldrich, C.; Bertozzi, C.; Georg, G.I.; Kiessling, L.; Lindsley, C.; Liotta, D.; Merz, J.K.M.; Schepartz, A.; Wang, S. The Ecstasy and Agony of Assay Interference Compounds. ACS Med. Chem. Lett. 2017, 8, 379–382. [Google Scholar] [CrossRef]
- Kenny, P.W. Comment on The Ecstasy and Agony of Assay Interference Compounds. J. Chem. Inf. Model. 2017, 57, 2640–2645. [Google Scholar] [CrossRef]
- Baell, J.B.; Nissink, J.W.M. Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017—Utility and Limitations. ACS Chem. Biol. 2017, 13, 36–44. [Google Scholar] [CrossRef]
- McLean, L.R.; Zhang, Y.; Li, H.; Li, Z.; Lukasczyk, U.; Choi, Y.-M.; Han, Z.; Prisco, J.; Fordham, J.; Tsay, J.T.; et al. Discovery of covalent inhibitors for MIF tautomerase via cocrystal structures with phantom hits from virtual screening. Bioorganic Med. Chem. Lett. 2009, 19, 6717–6720. [Google Scholar] [CrossRef]
- Mumbauer, S.; Pascual, J.; Kolotuev, I.; Hamaratoglu, F. Ferritin heavy chain protects the developing wing from reactive oxygen species and ferroptosis. PLoS Genet. 2019, 15, e1008396. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Chen, J. MDM2 Promotes Ubiquitination and Degradation of MDMX. Mol. Cell Biol. 2003, 23, 5113–5121. [Google Scholar] [CrossRef] [PubMed]
- Prachayasittikul, V.; Prachayasittikul, V.; Prachayasittikul, S.; Ruchirawat, S. 8-Hydroxyquinolines: A review of their metal chelating properties and medicinal applications. Drug Des. Dev. Ther. 2013, 7, 1157–1178. [Google Scholar] [CrossRef] [PubMed]
- Mah, L.J.; El-Osta, A.; Karagiannis, T.C. gammaH2AX: A sensitive molecular marker of DNA damage and repair. Leukemia 2010, 24, 679–686. [Google Scholar] [CrossRef]
- Heylmann, D.; Rödel, F.; Kindler, T.; Kaina, B. Radiation sensitivity of human and murine peripheral blood lymphocytes, stem and progenitor cells. Biochim. Et Biophys. Acta 2014, 1846, 121–129. [Google Scholar] [CrossRef]
- Godar, S.; Ince, T.A.; Bell, G.W.; Feldser, D.; Donaher, J.L.; Bergh, J.; Liu, A.; Miu, K.; Watnick, R.S.; Reinhardt, F.; et al. Growth-Inhibitory and Tumor- Suppressive Functions of p53 Depend on Its Repression of CD44 Expression. Cell 2008, 134, 62–73. [Google Scholar] [CrossRef]
- Rosso, M.; Polotskaia, A.; Bargonetti, J. Homozygous mdm2 SNP309 cancer cells with compromised transcriptional elongation at p53 target genes are sensitive to induction of p53-independent cell death. Oncotarget 2015, 6, 34573–34591. [Google Scholar] [CrossRef]
- Chou, T.-C. Theoretical Basis, Experimental Design, and Computerized Simulation of Synergism and Antagonism in Drug Combination Studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development setting. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Shergalis, A.; Xue, D.; Gharbia, F.Z.; Driks, H.; Shrestha, B.; Tanweer, A.; Cromer, K.; Ljungman, M.; Neamati, N. Characterization of Aminobenzylphenols as Protein Disulfide Isomerase Inhibitors in Glioblastoma Cell Lines. J. Med. Chem. 2020, 63, 10263–10286. [Google Scholar] [CrossRef] [PubMed]
- Weinert, E.E.; Dondi, R.; Colloredo-Melz, S.; Frankenfield, K.N.; Mitchell, C.H.; Freccero, M.; Rokita, S.E. Substituents on Quinone Methides Strongly Modulate Formation and Stability of Their Nucleophilic Adducts. J. Am. Chem. Soc. 2006, 128, 11940–11947. [Google Scholar] [CrossRef] [PubMed]
- Kanizsai, I.; Madácsi, R.; Hackler, L., Jr.; Gyuris, M.; Szebeni, G.J.; Huzián, O.; Puskás, L.G. Synthesis and Cytoprotective Characterization of 8-Hydroxyquinoline Betti Products. Molecules 2018, 23, 1934. [Google Scholar] [CrossRef]
- Pobbati, A.V.; Mejuch, T.; Chakraborty, S.; Karatas, H.; Bharath, S.R.; Guéret, S.M.; Goy, P.-A.; Hahne, G.; Pahl, A.; Sievers, S.; et al. Identification of Quinolinols as Activators of TEAD-Dependent Transcription. ACS Chem. Biol. 2019, 14, 2909–2921. [Google Scholar] [CrossRef] [PubMed]
- Olyaei, A.; Sadeghpour, M. Recent advances in the synthesis and synthetic applications of Betti base (aminoalkylnaphthol) and bis-Betti base derivatives. RSC Adv. 2019, 9, 18467–18497. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Popov, A.A.; Tselikov, G.; Heo, J.; Pliss, A.; Kim, S.; Kabashin, A.V.; Prasad, P.N. Organic Solvent and Surfactant Free Fluorescent Organic Nanoparticles by Laser Ablation of Aggregation-Induced Enhanced Emission Dyes. Adv. Opt. Mater. 2018, 6, 1800164. [Google Scholar] [CrossRef]
- Pauli, G.F.; Chen, S.-N.; Simmler, C.; Lankin, D.C.; Gödecke, T.; Jaki, B.U.; Friesen, J.B.; McAlpine, J.B.; Napolitano, J.G. Importance of Purity Evaluation and the Potential of Quantitative 1H NMR as a Purity Assay. J. Med. Chem. 2014, 57, 9220–9231. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.-Y.; He, Y.-B.; Yang, Z.-Y. A new method for reduction of phenyl fluoroalkanesulphonates to arenes catalysed by palladium. J. Chem. Soc. Chem. Commun. 1986, 1452–1453. [Google Scholar] [CrossRef]
- Andreu, R.; Ronda, J.C. Synthesis of 3,4-Dihydro-2H-1,3-benzoxazines by Condensation of 2-Hydroxyaldehydes and Primary Amines: Application to the Synthesis of Hydroxy-Substituted and Deuterium-Labeled Compounds. Synth. Commun. 2008, 38, 2316–2329. [Google Scholar] [CrossRef]
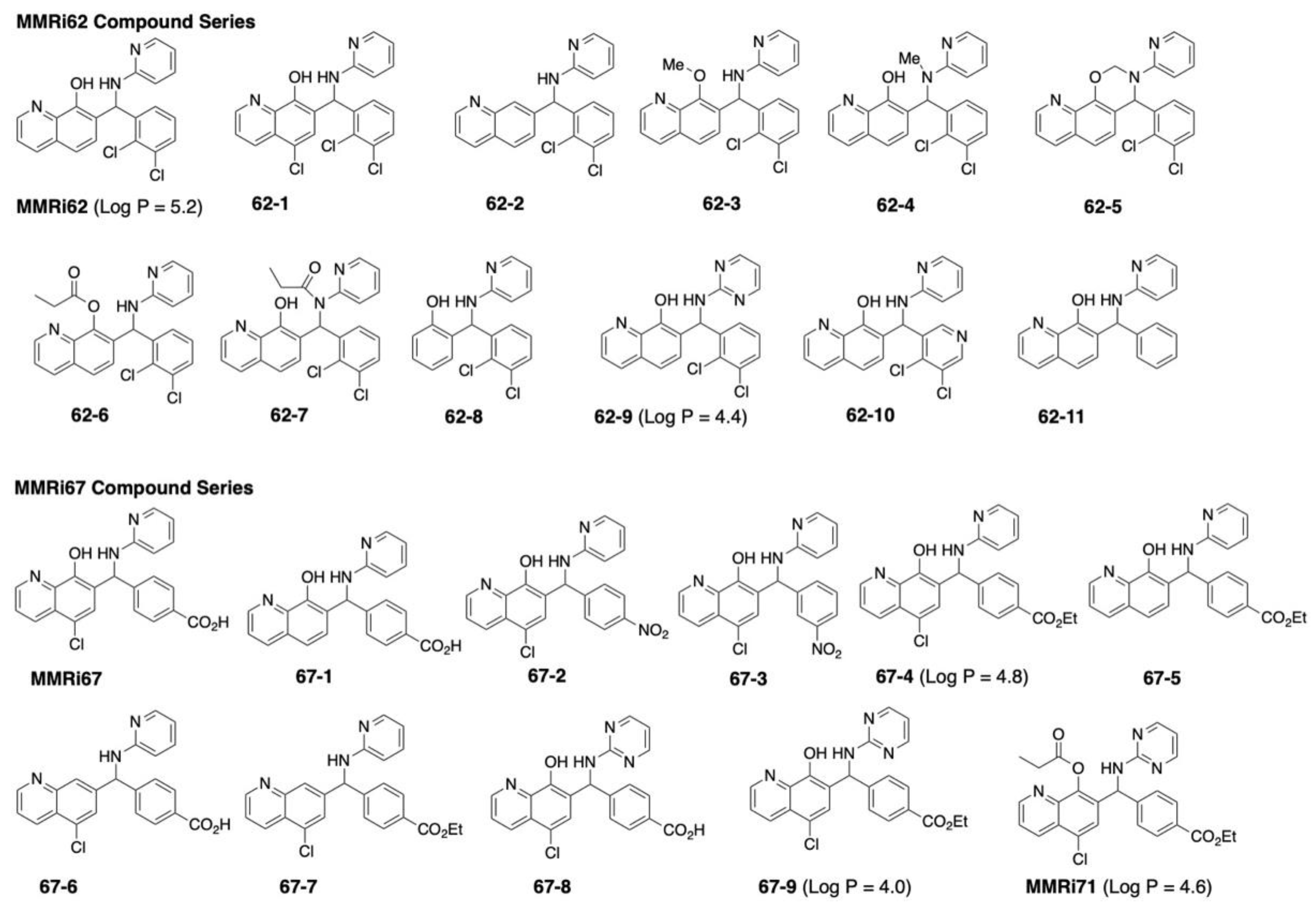
| Entry | NALM6 IC50 (μM) | shP53NALM6 IC50 (μM) |
|---|---|---|
| MMRi62 | 0.12 ± 0.001 | 0.14 ± 0.001 |
| 62-1 | 0.29 ± 0.01 | 0.46 ± 0.04 |
| 62-2 | 32.4 ± 2.25 | 35.5 ± 2.62 |
| 62-3 | 8.58 ± 0.83 | 24.2 ± 4.38 |
| 62-4 | 4.84 ± 0.48 | 4.68 ± 0.52 |
| 62-5 | 2.62 ± 0.36 | 1.51 ± 0.12 |
| 62-6 | 1.89 ± 0.53 | 1.08 ± 0.08 |
| 62-7 | 0.35 ± 0.01 | 0.26 ± 0.02 |
| 62-8 | 28.6 ± 2.87 | 28.5 ± 3.82 |
| 62-9 | 0.30 ± 0.02 | 0.36 ± 0.03 |
| 62-10 62-11 MMRi67 67-1 67-2 67-3 67-4 67-5 67-6 67-7 67-8 67-9 MMRi71 | 12.0 ± 1.79 2.10 ± 0.22 12.80 ± 1.57 8.93 ± 0.41 0.40 ± 0.01 0.38 ± 0.03 0.53 ± 0.03 0.38 ± 0.03 >100 18.2 ± 6.54 2.13 ± 0.18 0.27 ± 0.02 0.23 ± 0.01 | 27.1 ± 5.58 2.66 ± 0.17 10.89 ± 1.56 10.1 ± 0.51 0.47 ± 0.01 0.48 ± 0.02 0.57 ± 0.04 0.45 ± 0.01 >100 31.7 ± 3.52 2.36 ± 0.15 0.37 ± 0.003 0.29 ± 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lama, R.; Galster, S.L.; Xu, C.; Davison, L.W.; Chemler, S.R.; Wang, X. Dual Targeting of MDM4 and FTH1 by MMRi71 for Induced Protein Degradation and p53-Independent Apoptosis in Leukemia Cells. Molecules 2022, 27, 7665. https://doi.org/10.3390/molecules27227665
Lama R, Galster SL, Xu C, Davison LW, Chemler SR, Wang X. Dual Targeting of MDM4 and FTH1 by MMRi71 for Induced Protein Degradation and p53-Independent Apoptosis in Leukemia Cells. Molecules. 2022; 27(22):7665. https://doi.org/10.3390/molecules27227665
Chicago/Turabian StyleLama, Rati, Samuel L. Galster, Chao Xu, Luke W. Davison, Sherry R. Chemler, and Xinjiang Wang. 2022. "Dual Targeting of MDM4 and FTH1 by MMRi71 for Induced Protein Degradation and p53-Independent Apoptosis in Leukemia Cells" Molecules 27, no. 22: 7665. https://doi.org/10.3390/molecules27227665
APA StyleLama, R., Galster, S. L., Xu, C., Davison, L. W., Chemler, S. R., & Wang, X. (2022). Dual Targeting of MDM4 and FTH1 by MMRi71 for Induced Protein Degradation and p53-Independent Apoptosis in Leukemia Cells. Molecules, 27(22), 7665. https://doi.org/10.3390/molecules27227665






