Abstract
Peucedanum japonicum (Umbelliferae) is widely distributed throughout Southeast Asian countries. The root of this plant is used in traditional medicine to treat colds and pain, whereas the young leaves are considered an edible vegetable. In this study, the differences in coumarin profiles for different parts of P. japonicum including the flowers, roots, leaves, and stems were compared using ultra-performance liquid chromatography time-of-flight mass spectrometry. Twenty-eight compounds were tentatively identified, including three compounds found in the genus Peucedanum for the first time. Principal component analysis using the data set of the measured mass values and intensities of the compounds exhibited distinct clustering of the flower, leaf, stem, and root samples. In addition, their anticancer activities were screened using an Aldo–keto reductase (AKR)1C1 assay on A549 human non-small-cell lung cancer cells and the flower extract inhibited AKR1C1 activity. Based on these results, seven compounds were selected as potential markers to distinguish between the flower part versus the root, stem, and leaf parts using an orthogonal partial least-squares discriminant analysis. This study is the first to provide information on the comparison of coumarin profiles from different parts of P. japonicum as well as their AKR1C1 inhibitory activities. Taken together, the flowers of P. japonicum offer a new use related to the efficacy of overcoming anticancer drug resistance, and may be a promising source for the isolation of active lead compounds.
1. Introduction
Peucedanum japonicum Thunb. (Umbelliferae) is widely distributed in Korea, Japan, China, and Taiwan [1]. In the records of the traditional medicinal uses of P. japonicum in Southeast Asia, the stem and root have been used to treat sore throat [2], whereas the root has been used to treat cough, colds, and headache, and exhibits antipyretic and anti-inflammatory activity [3,4,5]. Coumarins are characteristic compounds of P. japonicum and several types of coumarins have been identified in this plant, such as simple coumarins, linear furanocoumarins, linear dihydro furanocoumarins, angular furanocoumarins, angular dihydrofuranocoumarins, linear dihydropyranocoumarins, and angular dihydropyranocoumarins [1,6,7]. Other constituents include chrome, phenolic compounds, and flavonoids [1]. The essential oil extracted from the aerial parts of P. japonicum contains α-pinene (24.68%) and β-pinene (66.07%) [5]. Various biological activities reported from the extracts and constituents of P. japonicum, such as antioxidant, anti-tyrosinase, anti-microbial, anti-allergic, neuroprotective, antidiabetic, and antiplatelet aggregative effects, have been summarized in Sarkhail et al., 2014 [1]. Other recent reports of in vitro and in vivo activities include anticancer [8], anti-adipogenic [9], antidiabetic [9], anti-osteoporosis [10], anti-allergic lung inflammation [11], antinociceptive [12], prostate contraction inhibitory [13], anti-osteoarthritis [14], antioxidant [15], anti-obesity [16], endothelial dysfunction improvement [17], anti-Alzheimer’s disease [18], corneal damage treatment [19], anti-atopic dermatitis [20], anti-inflammatory [21,22], and anti-influenza virus [23] effects.
Drug resistance is a major obstacle to the overall effectiveness of chemotherapy. Aldo–keto reductases (AKRs), which consist of the AKR1C1-AKR1C4 members, play a pivotal role in nicotinamide-adenine dinucleotide phosphate (NADPH)-dependent reduction and are involved in biosynthesis, intermediate metabolism, and detoxification [24,25]. With respect to the catalytic-dependent function of AKR1C isoforms in cancer chemotherapy, AKRs mediate the reduction of carbonyl groups in anticancer drugs (e.g., daunorubicin, doxorubicin, and oracin) to their inactive metabolites, thus contributing to drug resistance [26,27,28,29]. In addition, AKR1C1 is associated with cisplatin-resistance in colon cancer cell, head and neck squamous cell carcinoma, nasopharyngeal carcinoma, and paediatric T-cell acute lymphoblastic leukemia [30,31,32,33]. It has been recently reported that AKR1C3 mediates chemotherapy resistance in breast cancer and esophageal adenocarcinoma [34,35]. Thus, AKR1C isoforms may be a potential target for preventing the development of anticancer drug resistance. Moreover, AKR1C1 plays a role in regulating the cellular concentration of progesterone by catalyzing the reduction of progesterone to its inactive form, 20-alphahydroxy-progesterone, thus it is also considered a drug target for the prevention of pre-term birth throughout the inhibition of progesterone metabolism mediated by AKR1C1 [36].
In our work to identify AKR1C1 inhibitors from natural products to overcome cisplatin-resistance in human non-small lung cancer cells (NSCLC), the chemical composition of the extracts from four parts of P. japonicum and their AKR1C1 inhibitory activities in NSCLC (A549) were examined. Ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UPLC-QTof MS) and multivariate analysis was used to characterize the metabolomic differences between the four extracts.
2. Results and Discussion
2.1. Identification of Coumarins in the Different Parts of P. japonicum Using UPLC-QTof MS
Four parts of P. japonicum (flowers, roots, leaves, and stems) were prepared with six samples from each source that were designated as follows: S1–S6 for flower, S13–S18 for root, S7–S12 for leaf, S19–S24 for stem parts. Using UPLC-QTof MS with a gradient solvent system of acetonitrile (containing 0.1% formic acid) and water (containing 0.1% formic acid), the metabolites of the 24 P. japonicum samples were separated at high resolution within 22 min in the base peak ion (BPI) chromatogram. The representative BPI chromatograms of the methanol extract of flower, root, leaf, and stem of P. japonicum are shown in Figure 1. In these BPI chromatograms, the flower extract exhibited the most peaks and was fractioned using dichloromethane to obtain a chromatogram with increased intensity of coumarins (Figure S1). The mass spectrum of each peak in the BPI chromatogram of the dichloromethane fraction of the P. japonicum flowers (Figures S2–S29) was carefully interpreted by analyzing its experimental and theoretical high-resolution MS (the positive mode), error ppm, molecular formula, and MS/MS fragmentation (Table 1), as well as by comparing with existing data for coumarins of the Peucedanum species reported in the literature [3,6,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53]. In addition, UPLC-PDA chromatograms of all the peaks present in the methanol extract of four parts of P. japonicum and their UV-Vis spectra between 210–500 nm were shown in Figures S30 and S31. The coumarin structures, which were tentatively identified from the analysis of their MS data, are shown in Figure 2.
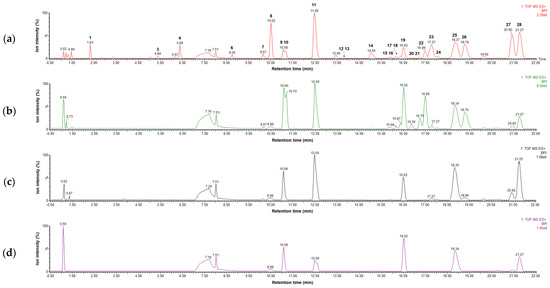
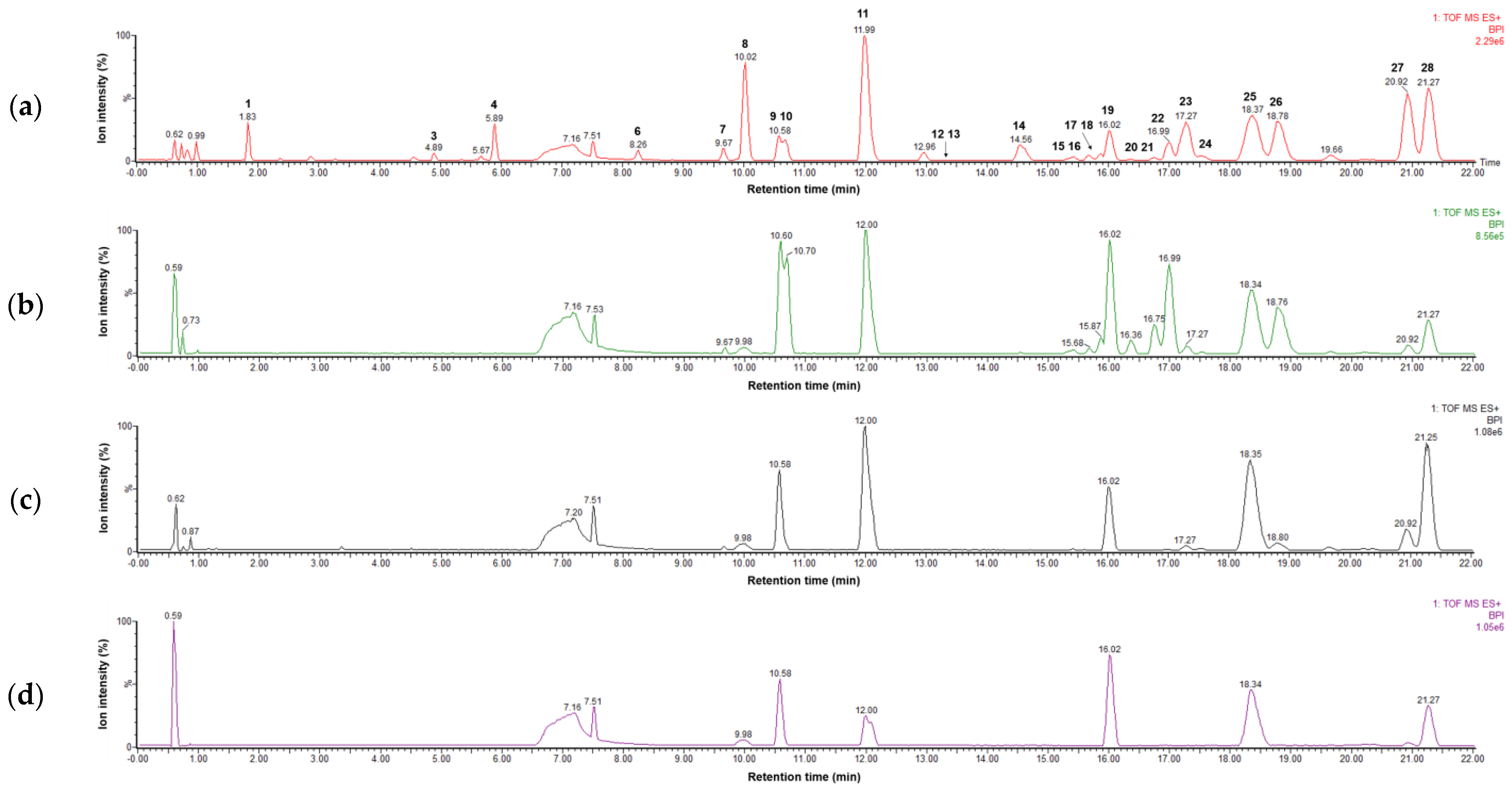
Figure 1.
LC–MS base peak ion chromatograms of four extracts representing (a) flowers (S3), (b) roots (S15), (c) leaves (S9), and (d) stems (S21) of Peucedanum japonicum in positive ion mode (6 eV, ESI+).

Table 1.
Characterization and tentative identification of coumarins found in Peucedanum japonicum using UPLC-QTof MS.
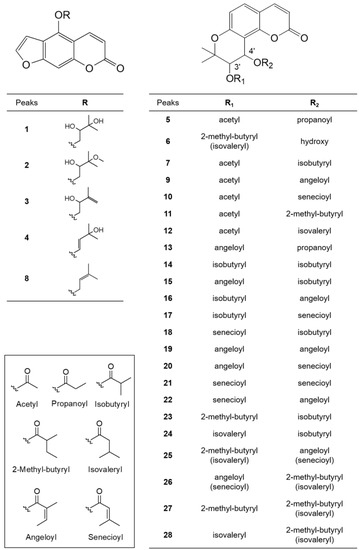
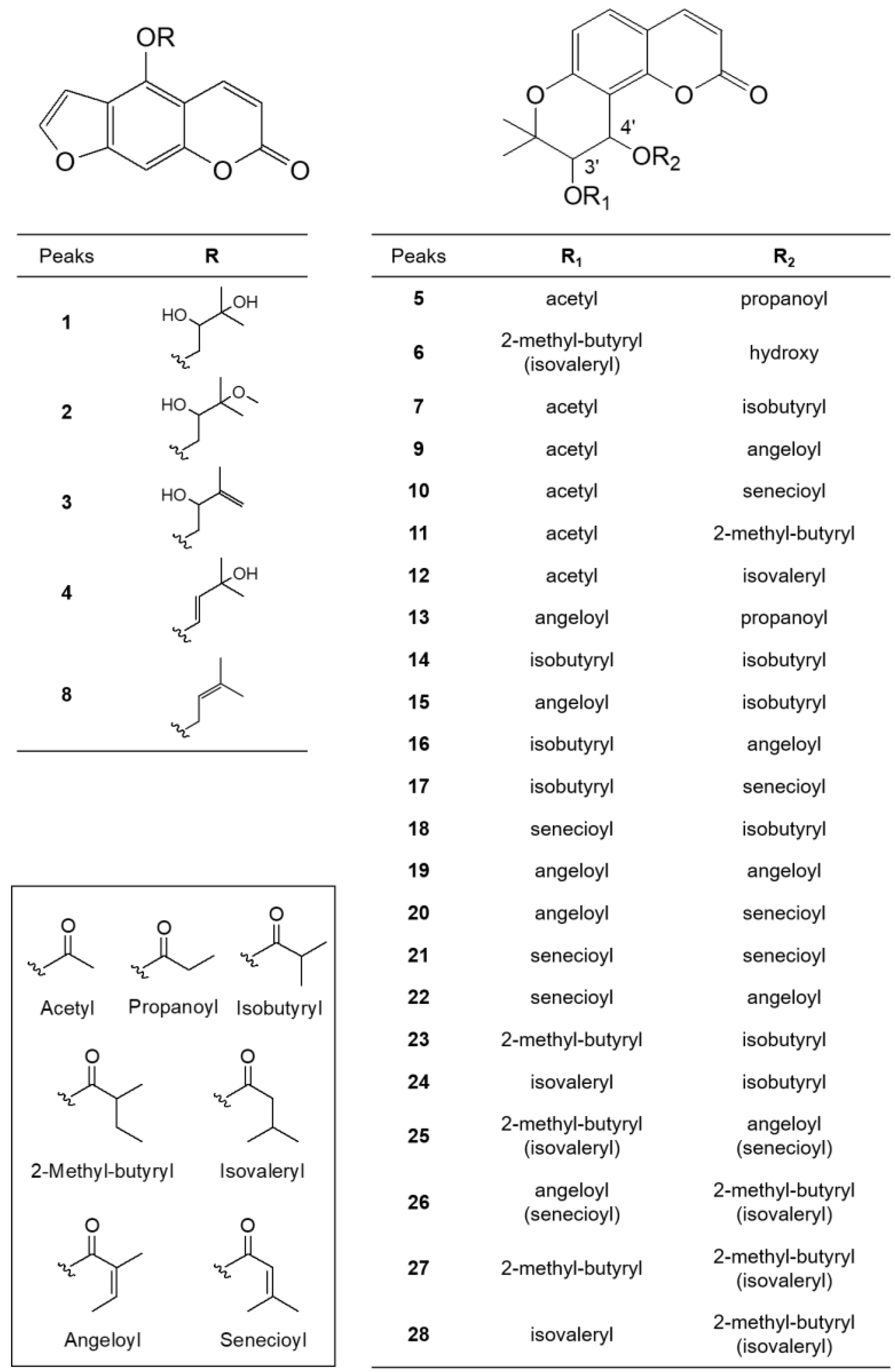
Figure 2.
Structures of identified coumarins (Peaks 1–28).
Peaks 1–4 and 8 had a common fragment ion for 4-hydroxy-7H-furo [3,2-g][1]benzopyran-7-one (5-hydroxypsoralen) or 9-hydroxy-7H-furo [3,2-g][1]benzopyran-7-one (8-hydroxypsoralen) at m/z 203.0344 [M − R + H]+, which resulted from the loss of different substituents at the C-5 oxygen or the C-8 oxygen [37]. In a review of coumarins isolated from Peucedanum species [1], the isolation of linear furanocoumarins, such as bergapten, cnidilin, isoimperatorin, and oxypeucetanin hydrate from P. japonicum was reported. Therefore, the linear furanocoumarins in this study were tentatively identified by giving preference to the structure based on the 5-hydroxypsoralen backbone. Peak 1 (tR 1.831 min) produced a major molecular ion at m/z 305.1027 [M + H]+ (calculated for C16H17O6+, 305.1025) and yielded a fragment ion at m/z m/z 203.0344 [M − R + H]+, indicating that a 2,3-dihydroxy-3-methylbutyl group was substituted at the 5-OH of 5-hydroxypsoralen. Therefore, peak 1 was tentatively identified as oxypeucedanin hydrate, which has been previously reported in P. japonicum [38]. Peak 2 (tR 4.712 min) produced a molecular ion at m/z 319.1223 [M + H]+ (calculated for C17H18O6+, 319.1223), which was 14 Da less than that of peak 1, indicating the presence of a methoxy group instead of a hydroxyl group. The fragment ions at m/z 203.0366 [M − R + H]+ represent a 2-hydroxy-3-methoxy-3-methylbutyl at the 5-OH of 5-hydroxypsoralen. Thus, it tentatively was identified as oxypeucedanin methanolate, which has been previously reported in P. ostruthium [39], although it was first detected in P. japonicum. Peaks 3 (tR 4.895 min) and 4 (tR 5.889 min) produced a molecular ion at m/z 287.0919 [M + H]+ (calculated for C16H15O5+, 287.0920), which is 18 Da less than that of peak 1, indicating the loss of a hydroxy group. The fragment ion at m/z 203.0340 [M − R + H]+ indicated the presence of a 2-hydroxy-3methyl-3-butenyl group or a 33-hydroxy-3-methyl-1-butenyl group at the 5-OH of 5-hydroxypsoralen. Therefore, these two peaks were tentatively identified as 5-[(2-hydroxy-3methyl-3-butenyl)oxy]psoralen (pabulenol) and 5-[(3-hydroxy-3-methyl-1-butenyl)oxy]psoralen, respectively, but they are interchangeable. 5-[(2-Hydroxy-3methyl-3-butenyl)oxy]psoralen (pabulenol) has been identified in various species of Peucedanum [40], but not in P. japonicum, whereas 5-[(3-Hydroxy-3-methyl-1-butenyl)oxy]psoralen has not been reported previously. Peak 8 (tR 10.015 min) produced a molecular ion at m/z 293.0759 [M + Na]+ (calculated for C16H14O4Na+, 293.0790), which is 16 Da less than that of peak 4, indicating the absence of a hydroxyl group. The fragment ions at m/z 203.0344 [M − R + H]+ represent the presence of a 2-hydroxy-3-methoxy-3-methylbutyl group at the 5-OH of 5-hydroxypsoralen. Thus, it was tentatively identified as isoimperatorin, which has been reported previously in P. japonicum [41].
In the ion fragmentation pathways of angular dihydropyranocoumarins, such as khellactone (3,4,5-trihydroxy-2,2-dimethyl-6-chromanacrylic acid δ-lactone), the MS fragment peak of the molecules resulting from a loss of the C-4′ substituent was observed as the main peak and the molecular ion peak due to fragmentation of the C-3′ substituent was small or undetectable [6]. Based on this phenomenon, we concluded that the position of the C-3′ and C-4′ substituents could be determined by MS fragmentation. In this study, molecular ions for angular dihydropyranocoumarins also exhibited characteristic fragment ions of [M-R2OH+H]+ by the loss of the C-4′ substituent, and [M-R2OH-(R1-H)+H]+ and [M-R2OH-(OR1-H)+H]+ by the loss of the C-3′ substituent. Therefore, in the ion fragmentation pathways of angular dihydropyranocoumarins, fragment ions at m/z 245.08 [M-R2OH-(R1-H)+H]+ for 5-hydroxy-2,2-dimethyl-3-oxo-6-chromanacrylic acid δ-lactone (jatamasinone) and 227.07 [M-R2OH-(OR1-H)+H]+ for 5-hydroxy-2,2-dimethyl-3,4-didehydro-6-chromanacrylic acid δ-lactone were frequently observed. Peak 5 (tR 7.878 min) showed a molecular ion at m/z 383.1159 [M + Na]+ (calculated for C19H20O7Na+, 383.1107) and fragment ions at m/z 287.0963 [M-R2OH+H]+, 245.0847 [M-R2OH-(R1-H)+H]+ and 227.0740 [M-R2OH-(OR1-H)+H]+, indicating the presence of a propionate group at C-4′ and an acetate group at C-3′. Thus, peak 5 was identified as 3′-O-acetyl-4′-O-propanoylkhellactone, which was first detected in P. japonicum; however, it has also been isolated from P. prearuptorum [41]. Peak 6 (tR 8.261 min) produced a major molecular ion at m/z 369.1314 [M + Na]+ (calculated for C19H22O6Na+, 369.1314) and yielded fragment ions at m/z 329.1384 [M-R2OH+H]+ and 245.0814 [M-R2OH-(R1-H)+H]+, indicating the presence of a hydroxyl group at C-4′ and a 2-methyl butanoate or isovalerate group at C-3′. Therefore, peak 6 was tentatively identified as 3′-O-(2-methyl-butyryl)-4′-hydroxy khellactone, which was isolated from P. japonicum [6] or 3′-O-(isovaleryl)-4′-hydroxy khellactone, which was isolated from P. turgeniifolium [42], but not P. japonicum. Peak 7 (tR 9.667 min) showed a molecular ion at m/z 397.1269 [M + Na]+ (calculated for C20H22O7Na+, 397.1263), which is 14 Da greater than that of peak 5, with fragment ions at m/z 287.0924 [M-R2OH+H]+, showing the presence of the isobutyrate group at C-4′. The fragment ions at m/z 245.0802 [M-R2OH-(R1-H)+H]+ and 227.0709 [M-R2OH-(OR1-H)+H]+ indicate the presence of the acetate group at C-3′. Thus, peak 7 was identified as 3′-O-acetyl-4′-O-isobutyryl khellactone, which was first detected in P. japonicum; however, it has also been isolated from P. prearuptorum [43]. Peaks 9 (tR 10.581 min) and 10 (tR 10.678 min) yielded identical molecular ions at m/z 409.1266 [M + Na]+ (calculated for C21H22O7Na+, 409.1263) and identical fragment ions to one another at m/z 287.0916 [M-R2OH+H]+, 245.0814 [M-R2OH-(R1-H)+H]+, and 227.0707 [M-R2OH-(OR1-H)+H]+, indicating the presence of an angeloate or a senecioate group at C-4′ and an acetate group at C-3′. Therefore, peaks 9 and 10 were tentatively identified as 3′-O-acetyl-4′-O-angeloylkhellactone and 3′-O-acetyl-4′-O-senecioylkhellactone, respectively, but they are interchangeable. In addition, both compounds were isolated from P. japonicum [38]. Peak 11 (tR 11.987 min) yielded a major molecular ion at m/z 411.1419 [M + Na]+ (calculated for C21H24O7Na+, 411.1420) and a fragment ion at m/z 287.0921 [M-R2OH+H]+, suggesting the presence of the 2-methyl butanoate or isovalerate groups at C-4′. The fragment ions at m/z 245.0820 [M-R2OH-(R1-H)+H]+ and 227.0715 [M-R2OH-(OR1-H)+H]+ indicated that the acetate group was substituted at C-3′. Therefore, peak 11 was tentatively identified as 3′-O-acetyl-4′-O-(2-methyl butanoate)khellactone [6] or 3′-O-acetyl-4′-O-isovalerylkhellactone [44], both of which have been isolated from P. japonicum [6,44]. Peak 12 (tR 13.181 min) produced a major molecular ion at m/z 423.1532 [M + Na]+ (calculated for C22H24O7Na+, 423.1420) and yielded fragment ions at m/z 301.1155 [M-R2OH+H]+, indicating the presence of the angeloate or a senecioate group at C-4′. The fragment ions at m/z 245.0878 [M-R2OH-(R1-H)+H]+ and 227.0769 [M-R2OH-(OR1-H)+H]+ suggested a propionate group at C-3′. Thus, peak 12 was tentatively identified as 3′-O-propanoyl-4′-O-angeloyl khellactone or 3′-O-propanoyl-4′-O-senecioylkhellactone, both of which have not been previously reported. Peak 13 (tR 13.433 min) produced a major molecular ion at m/z 423.1539 [M + Na]+ (calculated for C22H24O7Na+, 423.1420) and yielded fragment ions at m/z 327.1321 [M-R2OH+H]+, indicating the presence of a propionate group at C-4′. The fragment ions at m/z 245.0879 [M-R2OH-(R1-H)+H]+ and 227.0769 [M-R2OH-(OR1-H)+H]+ suggested the presence of the angeloate or the senecioate group at C-3′. Thus, peak 13 was tentatively identified as 3′-O-angeloyl-4′-O-propanoylkhellactone or 3′-O-senecioyl-4′-O-propanoylkhellactone, both of which have been previously reported in a profiling study of pyranocoumarins in P. prearuptorum [45]; however, these molecules were first detected in P. japonicum. Peak 14 (tR 14.542 min) produced a molecular ion at m/z 425.1577 [M + Na]+, corresponding to the molecular formula C22H26O7Na+. The fragment ions at m/z 315.1229 [M-R2OH+H]+, 245.0810 [M-R2OH-(R1-H)+H]+, and 227.0710 [M-R2OH-(OR1-H)+H]+ indicated the presence of two isobutyrate groups at C-3′ and C-4′. As a result, peak 14 was tentatively identified as 3′,4′-O-diisobutyrylkhellactone. 3′,4′-O-Diisobutyrylkhellactone has not been detected in P. japonicum; however, it has been isolated from Glehnia littoralis [46] and Phlojodicarpus sibiricus [47]. Peaks 15 (tR 15.319 min) and 18 (tR 15.885 min) produced identical molecular ions at m/z 437.1574 [M + Na]+ (calculated for C23H26O7Na+, 437.1576) and identical fragmentation patterns. The fragment ion at m/z 327.1227 [M-R2OH+H]+ indicated the presence of an isobutyrate group at C-4′. The fragment ions at m/z 245.0810 [M-R2OH-(R1-H)+H]+ and 227.0710 [M-R2OH-(OR1-H)+H]+ were observed as small peaks; however, they indicated the presence of the angeloate or senecioate group at C-3′. Therefore, peaks 15 and 18 were tentatively identified as 3′-O-angeloyl-4′-O-isobutyrylkhellactone [45,48] and 3′-O-senecioyl-4′-O-isobutyrylkhellactone [49], respectively, but they are interchangeable. Both structures have not been found in P. japonicum; however, they were reported in P. prearuptorum [45,48,49]. Peaks 16 (tR 15.442 min) and 17 (tR 15.667 min) produced the same molecular ion at m/z 437.1577 [M + Na]+ (calculated for C23H26O7Na+, 437.1576) as peaks 15 and 18, but exhibited different a fragment ion at m/z 315.1229 [M-R2OH+H]+, suggesting the existence of the angeloate or senecioate group at C-4′. The fragment ions at m/z 245.0813 [M-R2OH-(R1-H)+H]+ and 227.0702 [M-R2OH-(OR1-H)+H]+ indicated the presence of an isobutyrate group at C-3′. Thus, peaks 16 and 17 were tentatively identified as 3′-O- isobutyryl-4′-O-angeloylkhellactone and 3′-O-isobutyryl-4′-O-senecioylkhellactone, respectively, but they are interchangeable. Both structures have not been identified in P. japonicum; however, they have been reported in a study profiling pyranocoumarins in P. prearuptorum [45]. Peaks 19 (tR 16.016 min), 20 (tR 16.365 min), 21 (tR 16.747 min), and 22 (tR 16.993 min) produced identical molecular ions at m/z 449.1585 [M + Na]+ (calculated for C24H26O7Na+, 449.1576) and identical fragment ions at m/z 327.1234 [M-R2OH+H]+, indicating the presence of the angeloate or senecioate group at C-4′. The fragment ions at m/z 245.0814 [M-R2OH-(R1-H)+H]+ and m/z 227.0705 [M-R2OH-(OR1-H)+H]+ resulting from the fragmentation of the C-3′ substituent were observed as small peaks; however, this suggested the presence of the angeloate or senecioate group at C-3′. As a result, peaks 19–22 were tentatively identified as 3′,4′-O-diangeloylkhellactone, 3′-O-angeloyl-4′-O-senecioylkhellactone, 3′,4′-O-disenecioylkhellactone, and 3′-O-senecioyl-4′-O-angeloylkhellactone, respectively, but they are interchangeable. 3′-O-senecioyl-4′-O-angeloylkhellactone has not been found in P. japonicum; however, it has been reported in P. prearuptorum [48], whereas 3′,4′-O-diangeloylkhellactone [50], 3′-O-Angeloyl-4′-O-senecioylkhellactone [44], and 3′,4′-O-disenecioylkhellactone [51] have been isolated from P. japonicum. Peaks 23 (tR 17.273 min) and 24 (tR 17.542 min) yielded identical molecular ions at m/z 439.1729 [M + Na]+ (calculated for C23H28O7Na+, 439.1733). The fragment ions at m/z 329.1384 [M-R2OH+H]+ were observed by the loss of an isobutyrate group at C-4′. The fragment ions at m/z 245.0814 [M-R2OH-(R1-H)+H]+ and 227.0706 [M-R2OH-(OR1-H)+H]+ also indicated the presence of the 2-methyl butyrate or isovalerate group at C-3′. Therefore, peaks 23 and 24 were tentatively identified as 3′-O-(2-methyl butyryl)-4′-O-isobutyrylkhellactone and 3′-O-isovaleryl-4′-O-isobutyrylkhellactone, respectively, but they are interchangeable. 3′-O-(2-Methyl butyryl)-4′-O-isobutyrylkhellactone has been isolated from P. japonicum [6] and 3′-O-isovaleryl-4′-O-isobutyrylkhellactone has not; however, they have been reported in a profiling study of pyranocoumarins in P. prearuptorum [45]. Peak 25 (tR 18.371 min) showed a molecular ion at m/z 451.1734 [M + Na]+, corresponding to the molecular formula C24H28O7Na+. The fragment ions at m/z 329.1386 [M-R2OH+H]+ were observed by the loss of the angeloate or senecioate group at C-4′, and the fragment ions at m/z 245.0815 [M-R2OH-(R1-H)+H]+, and 227.0707 [M-R2OH-(OR1-H)+H]+ indicated the presence of the 2-methyl butyrate or isovalerate group at C-3′. Therefore, peak 25 was tentatively identified as a molecule among the four structures: 3′-O-(2-methyl butyryl)-4′-O-angeloylkhellactone, 3′-O-(2-methyl butyryl)-4′-O-senecioyl khellactone, 3′-O-isovaleryl-4′-O-angeloylkhellactone, and 3′-O-isovaleryl-4′-O-senecioylkhellactone. 3′-O-(2-Methyl butyryl)-4′-O-senecioyl khellactone [6], 3′-O-isovaleryl-4′-O-senecioylkhellactone [51], and 3′-O-isovaleryl-4′-O-angeloylkhellactone (paeruptorin C) [6] have been isolated from P. japonicum. 3′-O-(2-Methyl butyryl)-4′-O-angeloylkhellactone [52] has not been found in P. japonicum; however, it has been identified in P. prearuptorum [52]. Peak 26 (tR 18.799 min) exhibited the same molecular ion at m/z 451.1731 [M + Na]+ as peak 25, which corresponded to the molecular formula C24H28O7Na+; however, the fragment ions at m/z 327.1237 [M-R2OH+H]+ indicated that peaks 25 and 26 had substituents different from the substituent at C-4′ from the loss of the 2-methyl butyrate or isovalerate group at C-4′. The fragment ions at m/z 245.0819 [M-R2OH-(R1-H)+H]+ and 227.0710 [M-R2OH-(OR1-H)+H]+ suggested the presence of an angeloate or senecioate group at C-3′. Therefore, peak 26 was tentatively identified as a molecule among the four structures, 3′-O-angeloyl-4′-O-(2-methyl butyryl)khellactone, 3′-O-angeloyl-4′-O-isovaleryl khellactone, 3′-O-senecioyl-4′-O-(2-methyl butyryl)khellactone, or 3′-O-senecioyl-4′-O-isovaleryl khellactone. 3′-O-Angeloyl-4′-O-(2-methyl butyryl)khellactone [53] and 3′-O-angeloyl-4′-O-isovalerylkhellactone [53] have not been found in P. japonicum; however, they have been isolated from P. prearuptorum [53]. 3′-O-Senecioyl-4′-O-(2-methyl butyryl)khellactone [6] and 3′-O-senecioyl-4′-O-isovalerylkhellactone [3] have been isolated from P. japonicum. Peaks 27 (tR 20.919 min) and 28 (tR 21.268 min) produced the same molecular ion at m/z 453.1896 [M + Na]+ (calculated for C24H30O7Na+, 453.1889) and exhibited an identical fragment ion at m/z 329.1392 [M-R2OH+H]+, m/z 245.0817 [M-R2OH-(R1-H)+H]+, and m/z 227.0708 [M-R2OH-(OR1-H)+H]+, suggesting the presence of a 2-methyl butyrate or isovalerate group at C-4′ and a 2-methyl butyrate or isovalerate group at C-3′. Thus, peaks 27 and 28 were tentatively identified as two molecules among the four structures: 3′,4′-O-di(2-methyl butyryl)khellactone, 3′-O-(2-methyl butyryl)-4′-O- isovalerylkhellactone, 3′-O-isovaleryl-4′-O-(2-methyl butyryl)khellactone, or 3′,4′-O-diisovalerylkhellactone. 3′,4′-O-Di(2-methyl butyryl)khellactone [53], 3′-O-(2-methyl butyryl)-4′-O- isovalerylkhellactone [52], and 3′-O-isovaleryl-4′-O-(2-methyl butyryl)khellactone [53] have not been found in P. japonicum; however, they have been identified previously in P. prearuptorum [52,53], whereas 3′,4′-O-diisovalerylkhellactone has been isolated from P. japonicum [51].
2.2. Aldo–Keto Reductases Inhibitory Effects of the Different Parts for P. japonicum
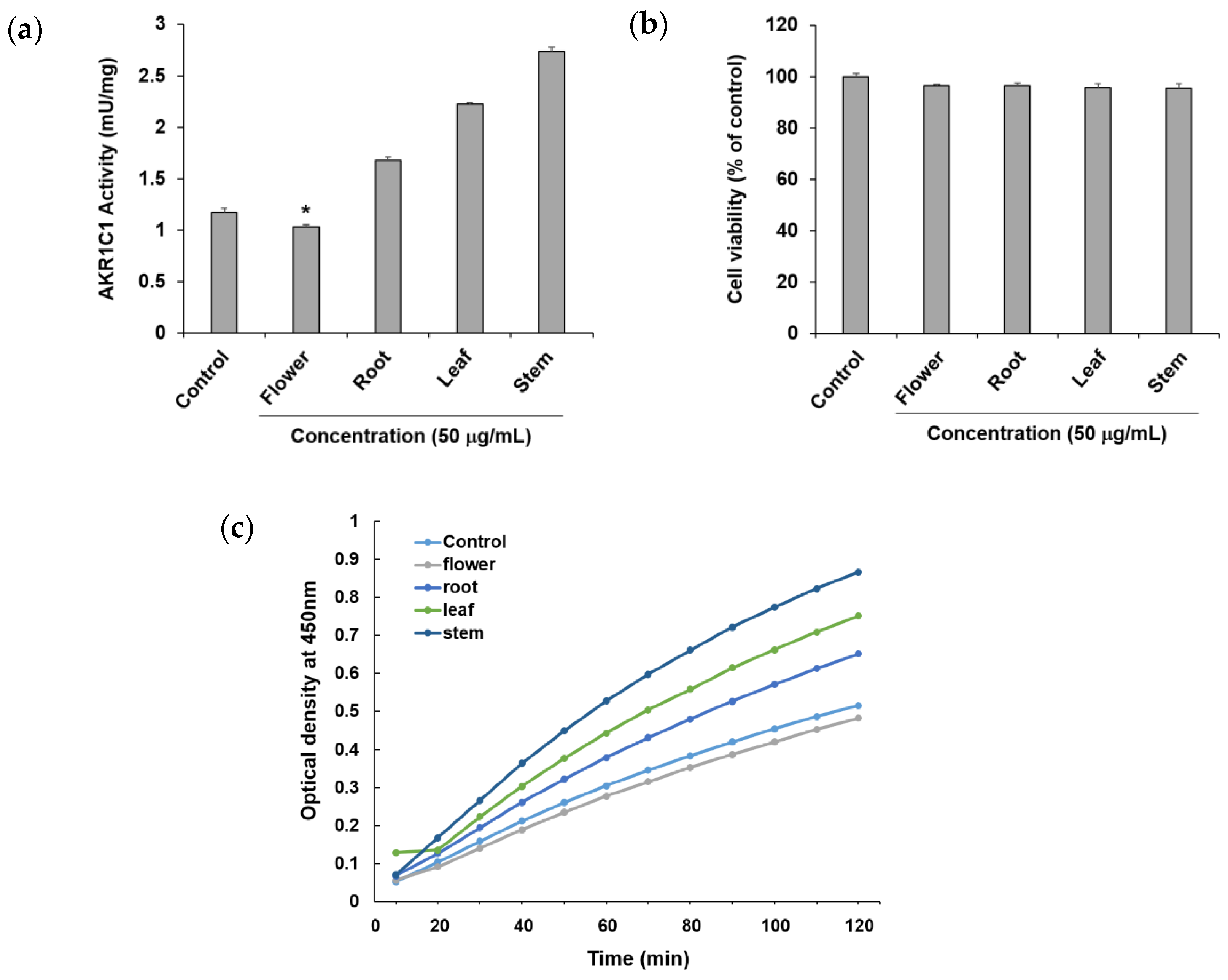
To confirm the ability of methanol extracts of four different parts of P. japonicum, flower (S1–S3), root (S13–S15), leaf (S7–S9), and stem (S19–S21), to circumvent anticancer drug resistance, they were tested using an in vitro AKR1C1 activity assay. Among the four extracts, the flower extract exhibited AKR1C1 activity inhibition of 11.74 ± 0.48% at a concentration of 50 μg/mL without causing cytotoxicity, whereas the other extracts increased AKR1C1 activity (Figure 3a). By observing optical density over time, the flower extract also showed a lower value compared with the control (Figure 3c). Thus, the flower extract was considered a potential natural source for compounds that prevent or overcome anticancer drug resistance in cancer. In the current reports of AKR1C1 inhibitors, non-competitive inhibitors include benzodiazepines, such as medazepam, phthalimide, pyrimidine, and anthranilic acid derivatives [54]. The representative competitive inhibitor of AKR1C1 is 3-bromo-5-phenylsalicylic acid [55]. Efforts for the development of AKR1C1 inhibitors are continuing through molecular docking and in vitro enzymatic studies using a number of synthetic compound derivatives [56,57,58]; however, there have been few reports of AKR1C1 inhibitors developed from natural products [59,60]. Therefore, the flower extract could be a promising source for the isolation of active lead compounds that inhibit AKR1C1 activity.
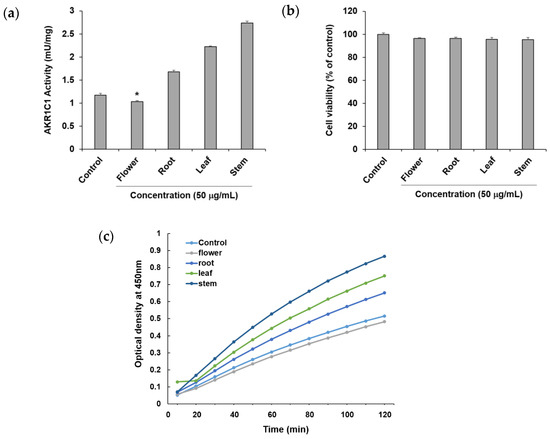
Figure 3.
Effects of the methanol extract of four different parts of Peucedanum japonicum (50 μg/mL) (a) on Aldo–keto reductase (AKR)1C1 activity, (b) cell viability, and (c) optical density (OD) over time, in A549 human non-small-cell lung cancer cells. Data are presented as means ± SD of three independent experiments. * p < 0.05 vs. control.
2.3. Multivariate Analysis
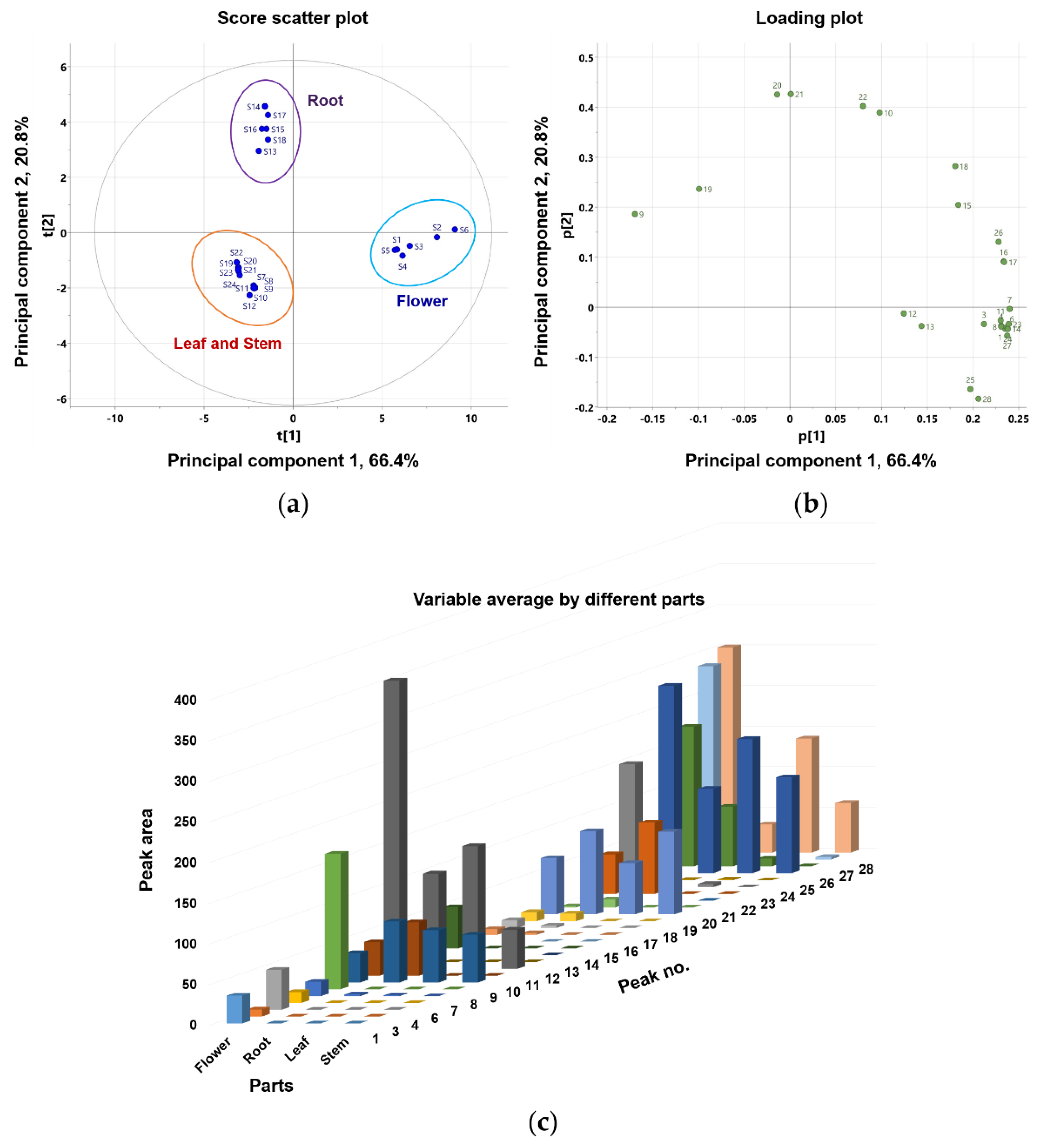
This study compared the differences in coumarin composition between the flowers, roots, leaves, and stems of P. japonicum. Using UPLC-QTof MS, the extracts from four different parts were analyzed. Twenty-four samples, six from each part containing 26 coumarins, were included in the multivariate analysis comparison study (peaks 2 and 5 were not included because they were not detected in methanol extracts). Principal component analysis (PCA) clearly distinguished flower (S1–S6), root (S13–S18), leaf (S7–S12), and stem (S19–S24) samples. As shown in Figure 4, leaf (S7–S12) and stem (S19–S24) samples clustered into one large group. PCA was conducted with three principal components (PC1–PC3) that described the variation, 0.913% R2X, and predictive capability, 0.826% Q2. Eigenvalues for PC1 and PC2 were 15.9 and 4.98, respectively, indicating that these first two principal components account for a substantial amount of the data variance. The relatively smaller eigenvalue of PC3 (0.996) led us to select only PC1 and PC2 for further examination. The first two principal components accounted for 87.2% of the variance (66.4% and 20.8% by PC1 and PC2, respectively). The corresponding PCA loading plot (Figure 4b) revealed that most of the peaks were coumarins, except for peaks 9, 10, and 19–21, which were chemical markers responsible for separating the flower sample from the root, leaf, and stem samples. Based on the PCA values, the coumarin profiles for the four parts of P. japonicum were distinct. Moreover, a variable average by different parts (Figure 4c) also clearly showed that most identified coumarins are abundant in the flower part compared with the root, leaf, and stem parts. Finally, leaves and stems share a chemical profile that is distinct from that of roots and flowers.
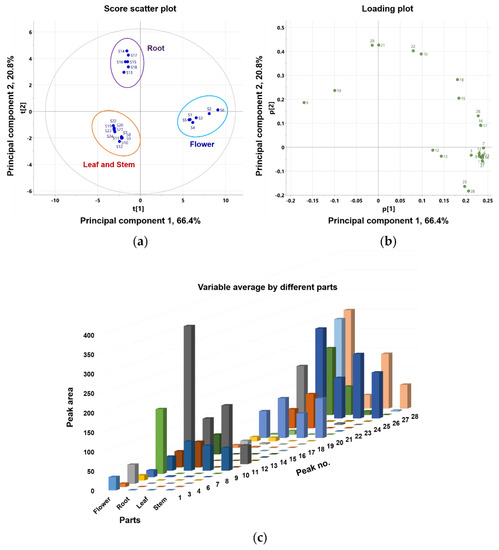
Figure 4.
Principal component analysis (PCA) (a) score plot and (b) loading plot of the metabolome analysis of flower, root, leaf, and stem of P. japonicum; (c) bar chart-showing the variable average for the different parts (flower, root, leaf, and stem) of P. japonicum.
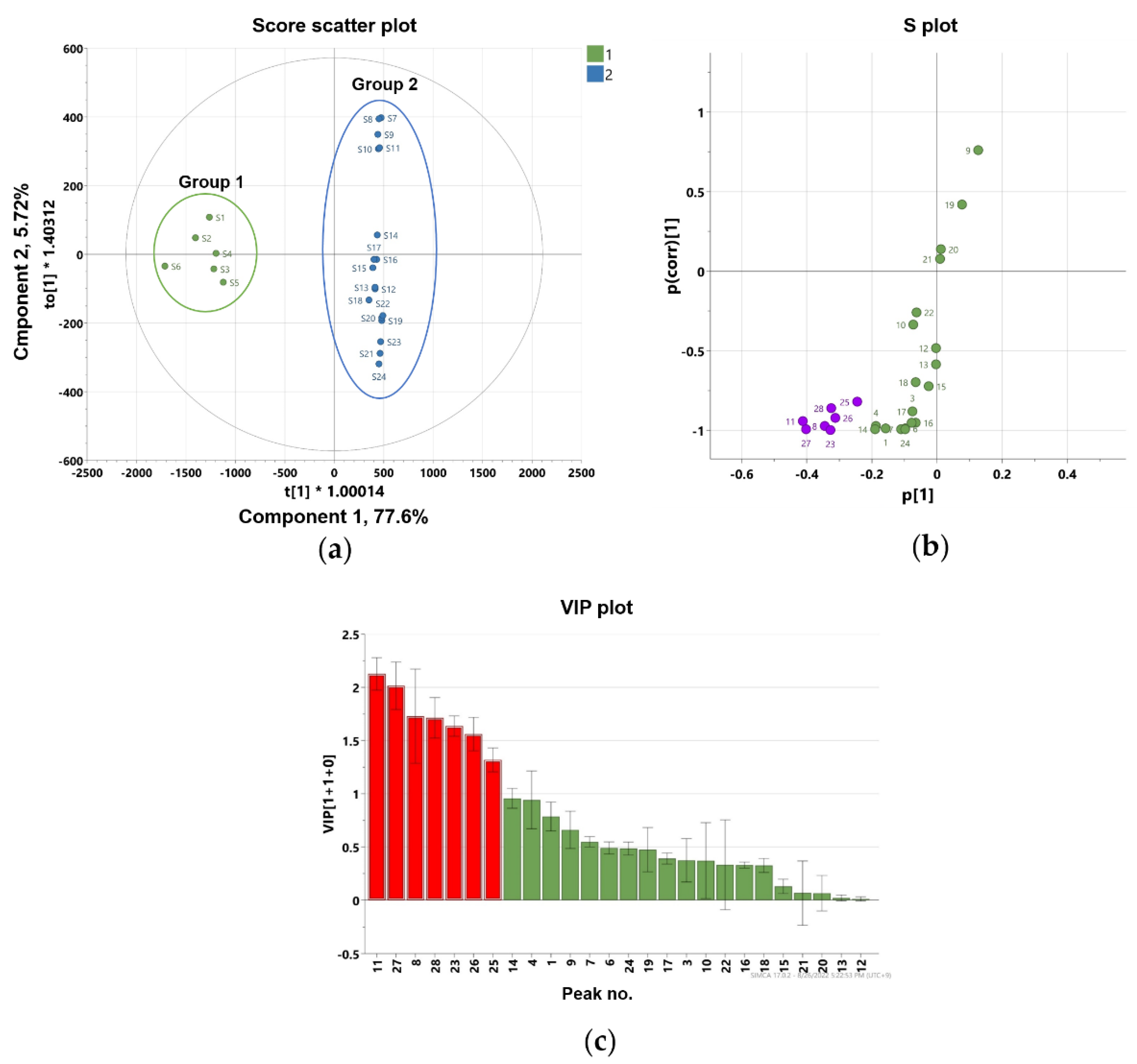
To further identify marker compounds responsible for the potency of flower samples, OPLS-DA with a VIP-plot and S-plot were used to compare the groups. Flower samples (S1–S6) were designated as Group 1, while root (S13–S18), leaf (S7–S12), and stem samples (S19–S24) were designated as Group 2. In the OPLS-DA model, flowers versus roots, leaves, and stems were clearly differentiated into two clusters (Figure 5a). The OPLS-DA model had a cumulative R2Y value of 1.00 and a cumulative Q2 value of 0.968. The internal validation of the OPLS-DA model was conducted by a permutation test (n = 200). The intercept values of R2 and Q2 in the permutation test were -0.0066 and -0.399, respectively. All permutations to the left of the R2 and Q2 values were less than the original points to the right (Figure S32). These values indicated that the OPLS-DA model for the analysis was robustly validated and not overfit. Several potential marker compounds capable of distinguishing the two groups were located apart from the center of the corresponding OPLS-DA S-plot (Figure 5b). Seven coumarins, peaks 8, 11, 23, 25–28, were shifted in the same direction as the Group 1 samples on the score scatter plot. The variable importance for the projection plot (VIP-plot, Figure 5c) indicates that these seven coumarins are the primarily responsible markers for discriminating between Group1 and Group 2 with significant VIP values (VIP ≥ 1). This indicates that the Group 1 samples contained higher concentrations of these coumarins compared with the Group 2 samples. Therefore, the composition and relative content of key markers that differentiate between flowers and other parts contribute to distinguishing active and inactive plant parts.
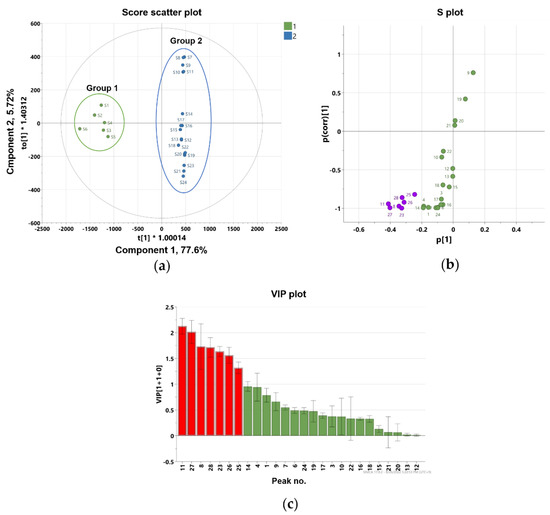
Figure 5.
Orthogonal partial least square-discriminant analysis (a) score plot and (b) S-plot exhibit markers for differentiating flower and root, stem, and leaf parts of P. japonicum (purple color shows the most deviated variables from the center), (c) the variable importance for the projection plot (VIP-plot) scores of the selected markers (red color shows the variables with VIP values ≥ 1).
3. Materials and Methods
3.1. Plant Materials
P. japonicum was grown under constant soil conditions in an experimental field located at 35.5114° N latitude by 126.8312° E longitude at the Advanced Radiation Technology Institute, Korea Atomic Energy Research Institute (Jeongeup-si, Jeollabuk-do 56212, Korea) for two years. This plant was bred and identified by Gileung Lee and Hong-Il Choi, co-authors of the study. The different parts of the P. japonicum plant (flowers, roots, leaves, and stems) were collected in October 2021, freeze-dried, and stored at –20 °C in polyethylene plastic bags until further use. The voucher specimens were deposited at the Advanced Radiation Technology Institute, Korea Atomic Energy Research Institute.
3.2. Sample Preparation
Freeze-dried parts of P. japonicum (flowers, roots, leaves, and stems) were ground into powder using a mixer. Six samples were prepared from each part and designated as follows: S1–S6 for flower, S13–S18 for root, S7–S12 for leaf, S19–S24 for stem parts. Then, 1 g of each sample was extracted with 20 mL of methanol in an ultrasonic bath for 1 h and then concentrated. The dried extract (10 mg) was dissolved in 1 mL of methanol and filtered using a 0.20 μm polyvinylidene fluoride filter. The samples were diluted with methanol to a concentration of 500 ppm for further LC–MS analysis. For the evaluation of bioactivity, each dried extract was initially dissolved in dimethyl sulfoxide (DMSO) at a concentration of 50 mg/mL. All extraction and chromatographic solvents used in this study were of analytical grade (J. T. Baker, Phillipsburg, NJ, USA).
3.3. UPLC-QTof MS Analysis
A Waters ACQUITY UPLC H-Class system combined with a SYNAPT XS QTof MS (Waters, Milford, MA, USA) was used. Each sample (1 μL) was injected into an ACQUITY UPLC BEH C18 column (100 × 2.1 mm i.d., 1.7 μm) at a flow rate of 0.4 mL/min. The temperature of the column oven was maintained at 40 °C. The mobile phase was composed of 0.1% formic acid in water (v/v; solvent A) and 0.1% formic acid in acetonitrile (v/v; solvent B). Gradient elution was carried out as follows: 0–2.0 min, 30% B; 2.0–6.0 min, 30–45% B; 6.0–11.0 min, 45–50% B; 11.0–14.0 min, 50–55% B; 14.0–18.0 min, 55% B; 18.0–21.0 min, 55–60% B; 21.0–25.0 min, 60–100% B; 25.0–27.0 min, 100% B; 27.0–27.1 min, 100–30% B; and 27.1–30.0 min, 30% B. The mass spectrometer was operated in positive ion mode using the following parameters: source temperature, 110 °C; desolvation temperature, 350 °C; capillary voltage, 2.5 kV; cone voltage, 50 V; cone gas flow: 50 L/h; flow rate of desolvation gas (N2), 800 L/h; mass scan range, 100–1200 Da; scan time, 0.5 s. Leucine-enkephalin was used for the lock mass ([M + H]+ m/z 556.2771). Full scan data, MS/MS spectra, accurate mass, and elemental composition were calculated using UNIFI software (version 3.03, Umetrics, Umeå, Sweden).
3.4. Aldo–Keto Reductases Activity Assay
The AKR1C1 activity of the extracts was measured using the EZDetectTM Aldo–keto Reductase Activity Assay Kit (Colorimetric) (BioVision Inc., Waltham, MA, USA), based on the manufacturer’s instructions. In this assay, AKR uses a general substrate to convert nicotinamide-adenine dinucleotide phosphate (NADP+) to dihydronicotinamide-adenine dinucleotide phosphate (NADPH). NADPH reacts with the AKR probe and generates a color which is proportional to the activity of AKR in the sample. To determine the effect of the extract on AKR enzyme activity in A549 non-small-cell lung cancer cells (American Type Culture Collection, Rockville, MD, USA), the cells were cultured in a 6-well plate at a density of 5 × 105 cells/mL for 24 h. They were subsequently treated with the extracts at a concentration of 50 μg/mL for an additional 24 h. After the incubation period, the cells were harvested according to the Kit manual for measuring AKR1C1 activity. The assay was performed using a single experiment and the samples were evaluated in triplets.
3.5. Statistical and Multivariate Analysis
The data set of the measured mass values and intensities (peak area) for each sample was exported to SIMCA 17.0.2 (Umetrics, Umeå, Sweden) and analyzed by multivariate statistical analysis including PCA and orthogonal partial least-squares discriminant analysis (OPLS-DA). An S-plot was used to identify the potential marker compounds that are responsible for the differentiation of the flower part from the root, leaf, and stem parts. Statistical analysis of the biological assay was performed to identify significant differences between the control and test groups (unpaired t-test) using GraphPad Prism9 software (GraphPad Prism version 9.0.0 (121) for Mac, San Diego, CA, USA).
4. Conclusions
A comparative study was done to identify coumarin composition and AKR1C1 inhibitory activities in different parts of P. japonicum. UPLC-QTof MS analysis was used to identify 28 compounds, including three coumarins which have not been reported in the genus Peucedanum. Among the methanol extracts of different parts of P. japonicum, the flower extract exhibited AKR1C1 activity inhibition in A549 cells at a concentration of 50 μg/mL without cytotoxicity. PCA results showed that the metabolites discriminate among the flower, leaf, stem, and root samples of P. japonicum and the OPLS-DA results revealed the responsible markers for discriminating between the active flower extract and the other three inactive parts. These markers were tentatively identified as isoimperatorin (peak 8), 3′-O-acetyl-4′-O-(2-methyl butanoate)khellactone (or 3′-O-acetyl-4′-O-isovalerylkhellactone; peak 11), 3′-O-(2-methyl butyryl)-4′-O-isobutyrylkhellactone (or 3′-O-isovaleryl-4′-O-isobutyrylkhellactone; peak 23), and compounds having a khellactone structure, in which four substituents, angeloate, senecioate, 2-methyl butyrate, and isovalerate groups, were frequently substituted at C-3 and C-4 (peaks 25–28). Therefore, these markers have the potential to have the ability to inhibit AKR1C1 activity, however further study involving the isolation of these compounds, the determination of their structures and stereochemistry, and the evaluation of their AKR1C1 activity inhibitory effects is required.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27217391/s1, Figure S1: LC–MS base peak ion chromatograms of the flowers of P. japonicum at positive ion mode (6 eV, ESI+), Figure S2: ESI-QTof-MS spectrum of oxypeucedanin hydrate (peak 1), Figure S3: ESI-QTof-MS spectrum of oxypeucedanin methanolate (peak 2), Figure S4: ESI-QTof-MS spectrum of pabulenol (peak 3), Figure S5: ESI-QTof-MS spectrum of 5-[(3-hydroxy-3-methyl-1-butenyl)oxy]psoralen (peak 4), Figure S6: ESI-QTof-MS spectrum of 3′-O-acetyl-4′-O-propanoylkhellactone (peak 5), Figure S7: ESI-QTof-MS spectrum of 3′- O-(2-methyl-butyryl)-4′-hydroxy khellactone (or 3′-O-(isovaleryl)-4′-hydroxy khellactone) (peak 6), Figure S8: ESI-QTof-MS spectrum of 3′-O-acetyl-4′-O-isobutyryl khellactone (hyuganin D) (peak 7), Figure S9: ESI-QTof-MS spectrum of isoimpertorin (peak 8), Figure S10: ESI-QTof-MS spectrum of 3′-O-acetyl-4′-O-angeloylkhellactone (pteryxin) (peak 9), Figure S11: ESI-QTof-MS spectrum of 3′-O-acetyl-4′-O-senecioylkhellactone (peak 10), Figure S12: ESI-QTof-MS spectrum of 3′-O-acetyl-4′-O-(2-methyl butanoate)khellactone (or 3′-O-acetyl-4′-O-isovalerylkhellactone) (peak 11), Figure S13: ESI-QTof-MS spectrum of 3′-O-propanoyl-4′-O-angeloyl khellactone (or 3′-O-propanoyl-4′-O-senecioylkhellactone) (peak 12), Figure S14: ESI-QTof-MS spectrum of 3′-O-angeloyl-4′-O-propanoylkhellactone (or 3′-O-senecioyl-4′-O-propanoylkhellactone) (peak 13), Figure S15: ESI-QTof-MS spectrum of 3′-O-isobutyryl-4′-O-isobutyrylkhellactone (peak 14), Figure S16: ESI-QTof-MS spectrum of 3′-O-angeloyl-4′-O-isobutyrylkhellactone (peak 15), Figure S17: ESI-QTof-MS spectrum of 3′-O- isobutyryl-4′-O-angeloylkhellactone (peak 16), Figure S18: ESI-QTof-MS spectrum of 3′-O-isobutyryl-4′-O-senecioylkhellactone (peak 17), Figure S19: ESI-QTof-MS spectrum of 3′-O-senecioyl-4′-O-isobutyrylkhellactone (peak 18), Figure S20: ESI-QTof-MS spectrum of 3′-O-angeloyl-4′-O-angeloylkhellactone (paeruptorin B) (peak 19), Figure S21: ESI-QTof-MS spectrum of 3′-O-angeloyl-4′-O-senecioylkhellactone (peak 20), Figure S22: ESI-QTof-MS spectrum of 3′-O-senecioyl-4′-O-senecioylkhellactone (peak 21), Figure S23: ESI-QTof-MS spectrum of 3′-O-senecioyl-4′-O-angeloylkhellactone (peak 22), Figure S24: ESI-QTof-MS spectrum of 3′-O-(2-methyl butyryl)-4′-O-isobutyrylkhellactone (peak23), Figure S25: ESI-QTof-MS spectrum of 3′-O-isovaleryl-4′-O-isobutyrylkhellactone (peak 24), Figure S26: ESI-QTof-MS spectrum of 3′-O-(2-methyl butyryl)-4′-O-angeloylkhellactone [3′-O-(2-methyl butyryl)-4′-O-senecioyl khellactone, 3′-O-isovaleryl-4′-O-angeloylkhellactone (paeruptorin C), or 3′-O-isovaleryl-4′-O-senecioylkhellactone] (peak 25), Figure S27: ESI-QTof-MS spectrum of 3′-O-angeloyl-4′-O-(2-methyl butyryl)khellactone [3′-O-angeloyl-4′-O-isovaleryl khellactone, 3′-O-senecioyl-4′-O-(2-methyl butyryl)khellactone, or 3′-O-senecioyl-4′-O-isovaleryl khellactone] (peak 26), Figure S28: ESI-QTof-MS spectrum of 3′-O-(2-methyl butyryl)-4′-O-(2-methyl butyryl)khellactone (or 3′-O-(2-methyl butyryl)-4′-O- isovalerylkhellactone) (peak 27), Figure S29: ESI-QTof-MS spectrum of 3′-O-isovaleryl-4′-O-(2-methyl butyryl)khellactone (or 3′-O-isovaleryl-4′-O-isovalerylkhellactone) (peak 28), Figure S30: Total scan PDA chromatograms of four extracts representing (a) flowers (S3), (b) roots (S15), (c) leaves (S9), and (d) stems (S21) of Peucedanum japonicum, Figure S31: UV-Vis spectra of all the peaks present in PDA chromatograms of the methanol extract of four parts of Peucedanum japonicum between 200–500 nm, Figure S32: Permutation plot for validation of OPLS-DA obtained from 200 permutation test.
Author Contributions
Conceptualization, A.-R.H. and C.-H.J.; methodology, A.-R.H., C.-H.J. and C.H.J.; software, J.P., S.B.P., G.-H.R. and B.-R.K.; validation, A.-R.H., C.-H.J., C.H.J., J.-W.N. and M.K.N.; formal analysis, J.P., S.B.P., G.-H.R. and B.-R.K.; investigation, J.P., S.B.P., C.H.J., G.L. and H.-I.C.; resources, G.L. and H.-I.C.; data curation, C.H.J., Y.-S.K. and C.-H.J.; writing—original draft preparation, J.P., S.B.P., C.H.J., and A.-R.H.; writing—review and editing, Y.-S.K., J.-W.N., C.-H.J., M.K.N. and A.-R.H.; visualization, J.P., S.B.P. and C.H.J.; supervision, A.-R.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the research program of Korea Atomic Energy Research Institute (Project No. 523310-22) and the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science and ICT) (No. 2022R1A2C2013345).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sarkhail, P. Traditional uses, phytochemistry and pharmacological properties of the genus Peucedanum: A review. J. Ethnopharmacol. 2014, 156, 235–270. [Google Scholar] [CrossRef] [PubMed]
- Morioka, T.; Suzui, M.; Nabandith, V.; Inamine, M.; Aniya, Y.; Nakayama, T.; Ichiba, T.; Mori, H.; Yoshimi, N. The modifying effect of Peucedanum japonicum, a herb in the Ryukyu Islands, on azoxymethane-induced colon preneoplastic lesions in male F344 rats. Cancer Lett. 2004, 205, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Ikeshiro, Y.; Mase, I.; Tomita, Y. Dihydropyranocoumarins from roots of Peucedanum japonicum. Phytochemistry 1992, 31, 4303–4306. [Google Scholar] [CrossRef]
- Lee, S.O.; Choi, S.Z.; Lee, J.H.; Chung, S.H.; Park, S.H.; Kang, H.C.; Yang, E.Y.; Cho, H.J.; Lee, K. Antidiabetic coumarin and cyclitol compounds from Peucedanum japonicum. Arch. Pharm. Res. 2004, 27, 1207–1210.21. [Google Scholar] [CrossRef]
- Yang, E.J.; Kim, S.S.; Oh, T.H.; Song, G.; Kim, K.N.; Kim, J.Y.; Lee, N.H.; Hyun, C.G. Peucedanum japonicum and Citrus unshiu essential oils inhibit the growth of antibiotic-resistant skin pathogens. Ann. Microbiol. 2009, 59, 623–628. [Google Scholar] [CrossRef]
- Hong, M.J.; Kim, J. Determination of the absolute configuration of khellactone esters from Peucedanum japonicum roots. J. Nat. Prod. 2017, 80, 1354–1360. [Google Scholar] [CrossRef]
- Won, H.J.; Lee, S.M.; Kim, D.-Y.; Kwon, O.-K.; Park, M.H.; Kim, J.-H.; Ryu, H.W.; Oh, S.-R. Rapid securing of reference substances from Peucedanum japonicum Thunberg by recycling preparative high-performance liquid chromatography. J. Chromatogr. B 2019, 1133, 121835. [Google Scholar] [CrossRef]
- Kim, J.-M.; Noh, E.-M.; Kim, H.-R.; Kim, M.-S.; Song, H.-K.; Lee, M.; Yang, S.-H.; Lee, G.-S.; Moon, H.-C.; Kwon, K.-B.; et al. Suppression of TPA-induced cancer cell invasion by Peucedanum japonicum Thunb. extract through the inhibition of PKCα/NF-κB-dependent MMP-9 expression in MCF-7 cells. Int. J. Mol. Med. 2016, 37, 108–114. [Google Scholar] [CrossRef]
- Choi, R.-Y.; Nam, S.-J.; Ham, J.-R.; Lee, H.-I.; Yee, S.-T.; Kang, K.-Y.; Seo, K.-I.; Lee, J.-H.; Kim, M.-J.; Lee, M.-K. Anti-adipogenic and anti-diabetic effects of cis-3′,4′-diisovalerylkhellactone isolated from Peucedanum japonicum Thunb leaves in vitro. Bioorg. Med. Chem. Lett. 2016, 26, 4655–4660. [Google Scholar] [CrossRef]
- Kim, J.-M.; Erkhembaatar, M.; Lee, G.S.; Lee, J.-H.; Noh, E.-M.; Lee, M.; Song, H.-K.; Lee, C.H.; Kwon, K.-B.; Kim, M.S.; et al. Peucedanum japonicum Thunb. ethanol extract suppresses RANKL-mediated osteoclastogenesis. Exp. Ther. Med. 2017, 14, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.M.; Lee, A.R.; Kim, H.S.; Lee, A.Y.; Gu, G.J.; Moon, B.C.; Kwon, B.-I. Peucedanum japonicum extract attenuates allergic airway inflammation by inhibiting Th2 cell activation and production of pro-inflammatory mediators. J. Ethnopharmacol. 2018, 211, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Jong, H.S.; Yoon, M.H.; Oh, S.H.; Jung, K.T. Antinociceptive effect of intrathecal sec-O-glucosylhamaudol on the formalin-induced pain in rats. Korean J. Pain 2017, 30, 98–103. [Google Scholar] [CrossRef]
- Suzuki, T.; Otsuka, A.; Ito, Y.; Yamada, S.; Miyake, H.; Ozono, S. Isosamidin, an extract of Peucedanum japonicum, inhibits phenylephrine-mediated contractions of the human prostate in vitro. Phytother. Res. 2018, 32, 949–952. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.M.; Lee, A.Y.; Kim, J.S.; Choi, G.; Kim, S.H. Protective Effects of Peucedanum japonicum extract against osteoarthritis in an animal model using a combined systems approach for compound-target prediction. Nutrients 2018, 10, 754. [Google Scholar] [CrossRef]
- Taira, J.; Ogi, T. Induction of antioxidant protein HO-1 through Nrf2-ARE signaling due to pteryxin in Peucedanum japonicum Thunb in RAW264.7 macrophage cells. Antioxidants 2019, 8, 621. [Google Scholar] [CrossRef] [PubMed]
- Hossin, A.Y.; Inafuku, M.; Oku, H. Dihydropyranocoumarins exerted anti-obesity activity in vivo and its activity was enhanced by nanoparticulation with polylactic-co-glycolic acid. Nutrients 2019, 11, 3053. [Google Scholar] [CrossRef] [PubMed]
- Do, M.H.; Lee, J.H.; Ahn, J.; Hong, M.J.; Kim, J.; Kim, S.Y. Isosamidin from Peucedanum japonicum roots prevents methylglyoxal-induced glucotoxicity in human umbilical vein endothelial cells via suppression of ROS-mediated Bax/Bcl-2. Antioxidants 2020, 9, 531. [Google Scholar] [CrossRef]
- Heo, J.H.; Eom, B.H.; Ryu, H.W.; Kang, M.-G.; Park, J.E.; Kim, D.-Y.; Kim, J.-H.; Park, D.; Oh, S.-R.; Kim, H. Acetylcholinesterase and butyrylcholinesterase inhibitory activities of khellactone coumarin derivatives isolated from Peucedanum japonicum Thurnberg. Sci. Rep. 2020, 10, 21695. [Google Scholar] [CrossRef]
- Kang, W.S.; Choi, H.; Lee, K.H.; Kim, E.; Kim, K.J.; Na, C.S.; Kim, S. Peucedanum japonicum Thunberg and its active components mitigate oxidative stress, inflammation and apoptosis after urban particulate matter-induced ocular surface damage. Antioxidants 2021, 10, 1717. [Google Scholar] [CrossRef]
- Gil, T.Y.; Jin, B.R.; An, H.J. Peucedanum japonicum Thunberg alleviates atopic dermatitis-like inflammation via STAT/MAPK signaling pathways in vivo and in vitro. Mol. Immunol. 2022, 144, 106–116. [Google Scholar] [CrossRef]
- Hwang, D.; Ryu, H.W.; Park, J.-W.; Kim, J.-H.; Kim, D.-Y.; Oh, J.-H.; Kwon, O.-K.; Han, S.-B.; Ahn, K.-S. Effects of 3′-isovaleryl-4′-senecioylkhellactone from Peucedanum japonicum Thunberg on PMA-stimulated inflammatory response in A549 human lung epithelial cells. J. Microbiol. Biotechnol. 2022, 32, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Gil, T.Y.; Jin, B.R.; Lee, J.H.; An, H.J. In vitro and in vivo experimental investigation of anti-inflammatory effects of Peucedanum japonicum aqueous extract by suppressing the LPS-induced NF-κB/MAPK JNK pathways. Am. J. Chin. Med. 2022, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, R.; Morimoto, R.; Horio, Y.; Sumitani, H.; Isegawa, Y. Inhibition of influenza virus replication by Apiaceae plants, with special reference to Peucedanum japonicum (Sacna) constituents. J. Ethnopharmacol. 2022, 292, 115243. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.M.; Chang, L.L.; Ying, M.D.; Cao, J.; He, Q.J.; Zhu, H.; Yang, B. Aldo-keto reductase AKR1C1-AKR1C4: Functions, regulation, and intervention for anti-cancer therapy. Front. Pharmacol. 2017, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Penning, T.M.; Jonnalagadda, S.; Trippier, P.C.; Rižner, T.L. Aldo-keto reductases and cancer drug resistance. Pharmacol. Rev. 2021, 73, 1150–1171. [Google Scholar] [CrossRef] [PubMed]
- Barski, O.A.; Tipparaju, S.M.; Bhatnagar, A. The aldo-keto reductase superfamily and its role in drug metabolism and detoxification. Drug. Metab. Rev. 2008, 40, 553–624. [Google Scholar] [CrossRef] [PubMed]
- Minotti, G.; Menna, P.; Salvatorelli, E.; Cairo, G.; Gianni, L. Anthracyclines: Molecular advances and pharmacologie developments in antitumor activity and cardiotoxicity. Pharmacol. Rev. 2004, 56, 185–229. [Google Scholar] [CrossRef]
- Hofman, J.; Malcekova, B.; Skarka, A.; Novotna, E.; Wsol, V. Anthracycline resistance mediated by reductive metabolism in cancer cells: The role of aldo-keto reductase 1C3. Toxicol. Appl. Pharmacol. 2014, 278, 238–248. [Google Scholar] [CrossRef]
- Wsol, V.; Szotakova, B.; Martin, H.J.; Maser, E. Aldo-keto reductases (AKR) from the AKR1C subfamily catalyze the carbonyl reduction of the novel anticancer drug oracin in man. Toxicology 2007, 238, 111–118. [Google Scholar] [CrossRef]
- Matsunaga, T.; Hojo, A.; Yamane, Y.; Endo, S.; El-Kabbani, O.; Hara, A. Pathophysiological roles of aldo-keto reductases (AKR1C1 and AKR1C3) in development of cisplatin resistance in human colon cancers. Chem. Biol. Interact. 2013, 202, 234–242. [Google Scholar] [CrossRef]
- Chang, W.M.; Chang, Y.C.; Yang, Y.C.; Lin, S.K.; Chang, P.M.; Hsiao, M. AKR1C1 controls cisplatin-resistance in head and neck squamous cell carcinoma through cross-talk with the STAT1/3 signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 245. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Shen, G.; Yang, F.; Duan, J.; Wu, Z.; Yang, M.; Liu, Y.; Du, X.; Zhang, X.; Xiao, S. Loss of AKR1C1 is a good prognostic factor in advanced NPC cases and increases chemosensitivity to cisplatin in NPC cells. J. Cell Mol. Med. 2020, 24, 6438–6447. [Google Scholar] [CrossRef] [PubMed]
- Bortolozzi, R.; Bresolin, S.; Rampazzo, E.; Paganin, M.; Maule, F.; Mariotto, E.; Boso, D.; Minuzzo, S.; Agnusdei, V.; Viola, G.; et al. AKR1C enzymes sustain therapy resistance in paediatric T-ALL. Br. J. Cancer. 2018, 118, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Yonezawa, A.; Takasawa, H.; Nagao, Y.; Iguchi, K.; Endo, S.; Ikari, A.; Matsunaga, T. Development of cisplatin resistance in breast cancer MCF7 cells by up-regulating aldo-keto reductase 1C3 expression, glutathione synthesis and proteasomal proteolysis. J. Biochem. 2022, 171, 97–108. [Google Scholar] [CrossRef]
- Zhou, C.; Wang, Z.; Li, J.; Wu, X.; Fan, N.; Li, D.; Liu, F.; Plum, P.S.; Hoppe, S.; Hillmer, A.M.; et al. Aldo-keto reductase 1C3 mediates chemotherapy resistance in esophageal adenocarcinoma via ROS detoxification. Cancers 2021, 13, 2403. [Google Scholar] [CrossRef]
- Ji, Q.; Aoyama, C.; Nien, Y.D.; Liu, P.I.; Chen, P.K.; Chang, L.; Stanczyk, F.Z.; Stolz, A. Selective loss of AKR1C1 and AKR1C2 in breast cancer and their potential effect on progesterone signaling. Cancer Res. 2004, 64, 7610–7617. [Google Scholar] [CrossRef]
- Shi, H.; Chang, Y.Q.; Feng, X.; Yang, G.Y.; Zheng, Y.G.; Zheng, Q.; Zhang, L.L.; Zhang, D.; Guo, L. Chemical comparison and discrimination of two plant sources of Angelicae dahuricae Radix, Angelica dahurica and Angelica dahurica var. formosana, by HPLC-Q/TOF-MS and quantitative analysis of multiple components by a single marker. Phytochem. Anal. 2022, 33, 776–791. [Google Scholar] [CrossRef]
- Chen, I.S.; Chang, C.T.; Sheen, W.S.; Teng, C.M.; Tsai, I.L.; Duh, C.Y.; Ko, F.N. Coumarins and antiplatelet aggregation constituents from Formosan Peucedanum japonicum. Phytochemistry 1996, 41, 525–530. [Google Scholar] [CrossRef]
- Reisch, J.; Khaled, S.A.; Szendrei, K.; Novak, I. Natural product chemistry. 51. 5-Alkoxy-furanocoumarins from Peucedanum ostruthium. Phytochemistry 1975, 14, 1889–1890. [Google Scholar] [CrossRef]
- Varga, E.; Simokovics, J.; Szendrei, K.; Reisch, J. Furanocoumarins and chromones from the fruits of Peucedanum ostrutbium L. (Koch) (Umbelliferae). Fitoterapia 1979, 50, 259–264. [Google Scholar]
- Lee, J.; Lee, Y.J.; Kim, J.; Bang, O.-S. Pyranocoumarins from root extracts of Peucedanum praeruptorum dunn with multidrug resistance reversal and anti-inflammatory activities. Molecules 2015, 20, 20967–20978. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Lin, Z.; Niu, F.; Ding, J. Studies on Chinese herbs in Umbelliferae. II. New coumarins in Peucedanum turgeniifolium Wolff. Yunnan Zhiwu Yanjiu 1981, 3, 173–180. [Google Scholar]
- Song, Y.-L.; Jing, W.-H.; Tu, P.-F.; Wang, Y.-T. Enantiomeric separation of angular-type pyranocoumarins from Peucedani Radix using AD-RH chiral column. Nat. Prod. Res. 2014, 28, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Taira, N.; Nugara, R.N.; Inafuku, M.; Takara, K.; Ogi, T.; Ichiba, T.; Iwasaki, H.; Okabe, T.; Oku, H. In vivo and in vitro anti-obesity activities of dihydropyranocoumarins derivatives from Peucedanum japonicum Thunb. J. Funct. Foods. 2017, 29, 19–28. [Google Scholar] [CrossRef]
- Yang, W.; Li, Y.; Kang, C.; Zhao, H.; Xiang, L.; Li, C.; Wang, Q. Sodiation-based in-source collision for profiling of pyranocoumarins in Radix Peucedani (Qianhu): Utility of sodium adducts’ stability with in-source collision. J. Mass Spectrom. 2017, 52, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Lee, C.; Jin, Q.; Yeon, E.T.; Lee, D.; Kim, S.-Y.; Han, S.B.; Hong, J.T.; Lee, M.K.; Hwang, B.Y. Pyranocoumarins from Glehnia littoralis inhibit the LPS-induced NO production in macrophage RAW 264.7 cells. Bioorg. Med. Chem. Lett. 2014, 24, 2717–2719. [Google Scholar] [CrossRef]
- Olennikov, D.N.; Fedorov, I.A.; Kashchenko, N.I.; Chirikova, N.K.; Vennos, C. Khellactone derivatives and other phenolics of Phlojodicarpus sibiricus (Apiaceae): HPLC-DAD-ESI-QQQ-MS/MS and HPLC-UV profile, and antiobesity potential of dihydrosamidin. Molecules 2019, 24, 2286. [Google Scholar] [CrossRef]
- Wang, X.-Y.; Li, J.-F.; Jian, Y.-M.; Wu, Z.; Fang, M.-J.; Qiu, Y.-K. On-line comprehensive two-dimensional normal-phase liquid chromatography × reversed-phase liquid chromatography for preparative isolation of Peucedanum praeruptorum. J. Chromatogr. A 2015, 1387, 60–68. [Google Scholar] [CrossRef]
- Li, X.-M.; Jiang, X.-J.; Yang, K.; Wang, L.-X.; Wen, S.-Z.; Wang, F. Prenylated Coumarins from Heracleum stenopterum, Peucedanum praeruptorum, Clausena lansium, and Murraya paniculata. Nat. Prod. Bioprospect. 2016, 6, 233–237. [Google Scholar] [CrossRef]
- Hata, K.; Kozawa, M.; Ikeshiro, Y.; Yen, K.-Y. New coumarins isolated from the root of Peucedanum formosanum and Peucedanum japonicum. Yakugaku Zasshi 1968, 88, 513–520. [Google Scholar] [CrossRef]
- Jong, T.T.; Hwang, H.C.; Jean, M.Y.; Wu, T.S.; Teng, C.M. An antiplatelet aggregation principle and X-ray structural analysis of cis-khellactone diester from Peucedanum japonicum. J. Nat. Prod. 1992, 5, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Song, Q.; Liu, Y.; Li, J.; Wan, J.-B.; Wang, Y.; Jiang, Y.; Tu, P. Integrated work-flow for quantitative metabolome profiling of plants, Peucedani Radix as a case. Anal. Chim. Acta 2017, 953, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Luo, J.; Wang, X.; Kong, L. Application of UPLC-Quadrupole-TOF-MS coupled with recycling preparative HPLC in isolation and preparation of coumarin isomers with similar polarity from Peucedanum praeruptorum. Chromatographia 2013, 76, 141–148. [Google Scholar] [CrossRef]
- Brozic, P.; Cesar, J.; Kovac, A.; Davies, M.; Johnson, A.P.; Fishwick, C.W.G.; Lanisnik Rizner, T.; Gobec, S. Derivatives of pyrimidine, phthalimide and anthranilic acid as inhibitors of human hydroxysteroid dehydrogenase AKR1C1. Chem. Biol. Interact 2009, 178, 158–164. [Google Scholar] [CrossRef]
- El-Kabbani, O.; Scammells, P.J.; Gosling, J.; Dhagat, U.; Endo, S.; Matsunaga, T.; Soda, M.; Hara, A. Structure-guided design, synthesis, and evaluation of salicylic acid-based inhibitors targeting a selectivity pocket in the active site of human 20alpha-hydroxysteroid dehydrogenase (AKR1C1). J. Med. Chem. 2009, 52, 3259–3264. [Google Scholar] [CrossRef]
- Verma, P.; Hassan, M.I.; Singh, A.; Singh, I.K. Design and development of novel inhibitors of aldo-ketoreductase 1C1 as potential lead molecules in treatment of breast cancer. Mol. Cell Biochem. 2021, 476, 2975–2987. [Google Scholar] [CrossRef]
- He, S.; Liu, Y.; Chu, X.; Li, Q.; Lyu, W.; Liu, Y.; Xing, S.; Feng, F.; Liu, W.; Guo, Q.; et al. Discovery of novel aldo-keto reductase 1C3 inhibitors as chemotherapeutic potentiators for cancer drug resistance. ACS Med. Chem. Lett. 2022, 13, 1286–1294. [Google Scholar] [CrossRef]
- Kljun, J.; Pavlič, R.; Hafner, E.; Lipec, T.; Moreno-Da Silva, S.; Tič, P.; Turel, I.; Büdefeld, T.; Stojan, J.; Rižner, T.L. Ruthenium complexes show potent inhibition of AKR1C1, AKR1C2, and AKR1C3 enzymes and anti-proliferative action against chemoresistant ovarian cancer cell line. Front Pharmacol. 2022, 13, 920379. [Google Scholar] [CrossRef]
- Zeng, C.; Zhu, D.; You, J.; Dong, X.; Yang, B.; Zhu, H.; He, Q. Liquiritin, as a natural inhibitor of AKR1C1, could interfere with the progesterone metabolism. Front. Physiol. 2019, 10, 833. [Google Scholar] [CrossRef]
- Fu, Z.; Li, S.; Liu, J.; Zhang, C.; Jian, C.; Wang, L.; Zhang, Y.; Shi, C. Natural product alantolactone targeting AKR1C1 suppresses cell proliferation and metastasis in non-small-cell lung cancer. Front. Pharmacol. 2022, 13, 847906. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).