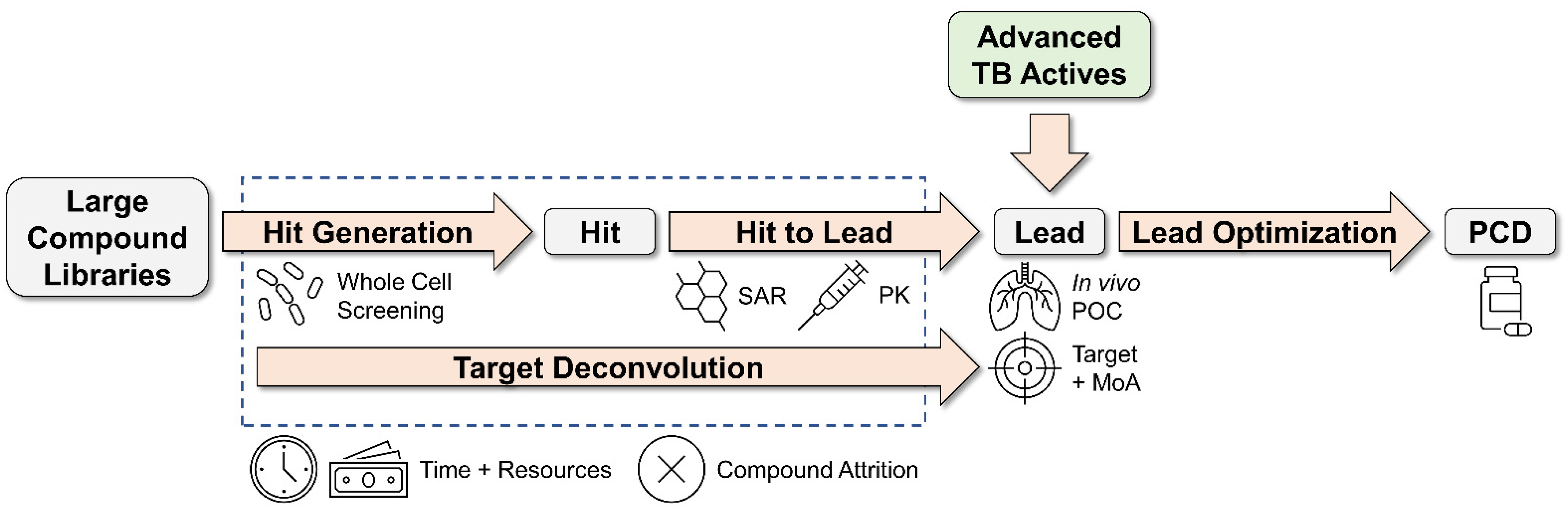
Why Matter Matters: Fast-Tracking Mycobacterium abscessus Drug Discovery
Abstract
1. Introduction
2. EC/11770 (Benzoxaboroles)
3. IC25 (Indole 2-Carboxamides)
4. Cyclohexyl-Griselimycin (Cyclic Depsipeptides)
5. OZ439 (Synthetic Trioxolanes)
6. EC/11716 (Novel Bacterial Topoisomerase Inhibitors)
7. TPP8 (Tricyclic Pyrrolopyrimidines)
8. CRS400226 (Benzothiazole Amides)
9. TBAJ-876 (Diarylquinolines)
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Daley, C.L.; Iaccarino, J.M.; Lange, C.; Cambau, E.; Wallace, R.J.; Andrejak, C.; Bottger, E.C.; Brozek, J.; Griffith, D.E.; Guglielmetti, L.; et al. Treatment of Nontuberculous Mycobacterial Pulmonary Disease: An Official ATS/ERS/ESCMID/IDSA Clinical Practice Guideline: Executive Summary. Clin. Infect. Dis. 2020, 71, e1–e36. [Google Scholar] [CrossRef] [PubMed]
- Strnad, L.; Winthrop, K.L. Treatment of Mycobacterium abscessus Complex. Semin. Respir. Crit. Care Med. 2018, 39, 362–376. [Google Scholar] [CrossRef] [PubMed]
- Jarand, J.; Levin, A.; Zhang, L.; Huitt, G.; Mitchell, J.D.; Daley, C.L. Clinical and microbiologic outcomes in patients receiving treatment for Mycobacterium abscessus pulmonary disease. Clin. Infect. Dis. 2011, 52, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Maurer, F.P.; Bruderer, V.L.; Ritter, C.; Castelberg, C.; Bloemberg, G.V.; Bottger, E.C. Lack of antimicrobial bactericidal activity in Mycobacterium abscessus. Antimicrob. Agents Chemother. 2014, 58, 3828–3836. [Google Scholar] [CrossRef] [PubMed]
- Luthra, S.; Rominski, A.; Sander, P. The Role of Antibiotic-Target-Modifying and Antibiotic-Modifying Enzymes in Mycobacterium abscessus Drug Resistance. Front. Microbiol. 2018, 9, 2179. [Google Scholar] [CrossRef]
- Falkinham, J.O., 3rd. Challenges of NTM Drug Development. Front. Microbiol. 2018, 9, 1613. [Google Scholar] [CrossRef]
- Philley, J.V.; Griffith, D.E. Disease Caused by Mycobacterium abscessus and Other Rapidly Growing Mycobacteria (RGM). In Nontuberculous Mycobacterial Disease: A Comprehensive Approach to Diagnosis and Management; Griffith, D.E., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 369–399. [Google Scholar]
- Machado, D.; Cannalire, R.; Santos Costa, S.; Manfroni, G.; Tabarrini, O.; Cecchetti, V.; Couto, I.; Viveiros, M.; Sabatini, S. Boosting Effect of 2-Phenylquinoline Efflux Inhibitors in Combination with Macrolides against Mycobacterium smegmatis and Mycobacterium avium. ACS Infect. Dis. 2015, 1, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Dal Molin, M.; Gut, M.; Rominski, A.; Haldimann, K.; Becker, K.; Sander, P. Molecular Mechanisms of Intrinsic Streptomycin Resistance in Mycobacterium abscessus. Antimicrob. Agents Chemother. 2018, 62, e01427-17. [Google Scholar] [CrossRef]
- Rominski, A.; Roditscheff, A.; Selchow, P.; Bottger, E.C.; Sander, P. Intrinsic rifamycin resistance of Mycobacterium abscessus is mediated by ADP-ribosyltransferase MAB_0591. J. Antimicrob. Chemother. 2017, 72, 376–384. [Google Scholar] [CrossRef]
- Rominski, A.; Schulthess, B.; Muller, D.M.; Keller, P.M.; Sander, P. Effect of beta-lactamase production and beta-lactam instability on MIC testing results for Mycobacterium abscessus. J. Antimicrob. Chemother. 2017, 72, 3070–3078. [Google Scholar] [CrossRef] [PubMed]
- Rominski, A.; Selchow, P.; Becker, K.; Brulle, J.K.; Dal Molin, M.; Sander, P. Elucidation of Mycobacterium abscessus aminoglycoside and capreomycin resistance by targeted deletion of three putative resistance genes. J. Antimicrob. Chemother. 2017, 72, 2191–2200. [Google Scholar] [CrossRef] [PubMed]
- Rudra, P.; Hurst-Hess, K.; Lappierre, P.; Ghosh, P. High Levels of Intrinsic Tetracycline Resistance in Mycobacterium abscessus Are Conferred by a Tetracycline-Modifying Monooxygenase. Antimicrob. Agents Chemother. 2018, 62, e00119-18. [Google Scholar] [CrossRef] [PubMed]
- Griffith, D.E.; Daley, C.L. Treatment of Mycobacterium abscessus Pulmonary Disease. Chest 2022, 161, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Nash, K.A.; Brown-Elliott, B.A.; Wallace, R.J., Jr. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob. Agents Chemother. 2009, 53, 1367–1376. [Google Scholar] [CrossRef]
- Daniel-Wayman, S.; Abate, G.; Barber, D.L.; Bermudez, L.E.; Coler, R.N.; Cynamon, M.H.; Daley, C.L.; Davidson, R.M.; Dick, T.; Floto, R.A.; et al. Advancing Translational Science for Pulmonary Nontuberculous Mycobacterial Infections. A Road Map for Research. Am. J. Respir. Crit. Care Med. 2019, 199, 947–951. [Google Scholar] [CrossRef]
- Daley, C.L.; Olivier, K.N. ALIS (Amikacin Liposome Inhalation Suspension): The Beginning of a Wonderland? Am. J. Respir. Crit. Care Med. 2018, 198, 1473–1475. [Google Scholar] [CrossRef]
- Henriette Zweijpfenning, S.M.; Chiron, R.; Essink, S.; Schildkraut, J.; Akkerman, O.W.; Aliberti, S.; Altenburg, J.; Arets, B.; van Braeckel, E.; Delaere, B.; et al. Safety and Outcomes of Amikacin Liposome Inhalation Suspension for Mycobacterium abscessus Pulmonary Disease: A NTM-NET study. Chest 2022, 162, 76–81. [Google Scholar] [CrossRef]
- Yang, B.; Jhun, B.W.; Moon, S.M.; Lee, H.; Park, H.Y.; Jeon, K.; Kim, D.H.; Kim, S.Y.; Shin, S.J.; Daley, C.L.; et al. Clofazimine-Containing Regimen for the Treatment of Mycobacterium abscessus Lung Disease. Antimicrob. Agents Chemother. 2017, 61, e02052-16. [Google Scholar] [CrossRef]
- Dick, T. Rifabutin: A Repurposing Candidate for Mycobacterium abscessus Lung Disease. Front. Microbiol. 2020, 11, 371. [Google Scholar] [CrossRef]
- Cheng, A.; Sun, H.-Y.; Wu, U.-i.; Sheng, W.-H.; Chen, Y.-C.; Chang, S.-C. 1360. Disseminated Mycobacterium abscessus Infections in Patients with Neutralizing Anti-interferon-gamma Autoantibodies Treated with Rifabutin-based Combination Regimens. Open Forum Infect. Dis. 2019, 6, S492–S493. [Google Scholar] [CrossRef][Green Version]
- Ganapathy, U.S.; Dartois, V.; Dick, T. Repositioning rifamycins for Mycobacterium abscessus lung disease. Expert Opin. Drug Discov. 2019, 14, 867–878. [Google Scholar] [CrossRef]
- Ganapathy, U.S.; Lan, T.; Krastel, P.; Lindman, M.; Zimmerman, M.D.; Ho, H.; Sarathy, J.P.; Evans, J.C.; Dartois, V.; Aldrich, C.C.; et al. Blocking Bacterial Naphthohydroquinone Oxidation and ADP-Ribosylation Improves Activity of Rifamycins against Mycobacterium abscessus. Antimicrob. Agents Chemother. 2021, 65, e0097821. [Google Scholar] [CrossRef]
- Wu, M.L.; Aziz, D.B.; Dartois, V.; Dick, T. NTM drug discovery: Status, gaps and the way forward. Drug Discov. Today 2018, 23, 1502–1519. [Google Scholar] [CrossRef]
- Egorova, A.; Jackson, M.; Gavrilyuk, V.; Makarov, V. Pipeline of anti-Mycobacterium abscessus small molecules: Repurposable drugs and promising novel chemical entities. Med. Res. Rev. 2021, 41, 2350–2387. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov (accessed on 1 August 2022).
- Malin, J.J.; Winter, S.; van Gumpel, E.; Plum, G.; Rybniker, J. Extremely Low Hit Rate in a Diverse Chemical Drug Screen Targeting Mycobacterium abscessus. Antimicrob. Agents Chemother. 2019, 63, e01008-19. [Google Scholar] [CrossRef]
- Tortoli, E. The Taxonomy of the Genus Mycobacterium. In Nontuberculous Mycobacteria (NTM); Velayati, A.A., Farnia, P., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 1–10. [Google Scholar]
- Low, J.L.; Wu, M.L.; Aziz, D.B.; Laleu, B.; Dick, T. Screening of TB Actives for Activity against Nontuberculous Mycobacteria Delivers High Hit Rates. Front. Microbiol. 2017, 8, 1539. [Google Scholar] [CrossRef]
- Raynaud, C.; Daher, W.; Johansen, M.D.; Roquet-Baneres, F.; Blaise, M.; Onajole, O.K.; Kozikowski, A.P.; Herrmann, J.L.; Dziadek, J.; Gobis, K.; et al. Active Benzimidazole Derivatives Targeting the MmpL3 Transporter in Mycobacterium abscessus. ACS Infect. Dis. 2020, 6, 324–337. [Google Scholar] [CrossRef]
- Ganapathy, U.S.; Del Rio, R.G.; Cacho-Izquierdo, M.; Ortega, F.; Lelievre, J.; Barros-Aguirre, D.; Lindman, M.; Dartois, V.; Gengenbacher, M.; Dick, T. A Leucyl-tRNA Synthetase Inhibitor with Broad-Spectrum Anti-Mycobacterial Activity. Antimicrob. Agents Chemother. 2021, 65, e02420-20. [Google Scholar] [CrossRef]
- Pandya, A.N.; Prathipati, P.K.; Hegde, P.; Li, W.; Graham, K.F.; Mandal, S.; Drescher, K.M.; Destache, C.J.; Ordway, D.; Jackson, M.; et al. Indole-2-Carboxamides Are Active against Mycobacterium abscessus in a Mouse Model of Acute Infect.ion. Antimicrob. Agents Chemother. 2019, 63, e02245-18. [Google Scholar] [CrossRef]
- Aragaw, W.W.; Roubert, C.; Fontaine, E.; Lagrange, S.; Zimmerman, M.D.; Dartois, V.; Gengenbacher, M.; Dick, T. Cyclohexyl-griselimycin Is Active against Mycobacterium abscessus in Mice. Antimicrob. Agents Chemother. 2022, 66, e0140021. [Google Scholar] [CrossRef] [PubMed]
- Belardinelli, J.M.; Verma, D.; Li, W.; Avanzi, C.; Wiersma, C.J.; Williams, J.T.; Johnson, B.K.; Zimmerman, M.; Whittel, N.; Angala, B.; et al. Therapeutic efficacy of antimalarial drugs targeting DosRS signaling in Mycobacterium abscessus. Sci. Transl. Med. 2022, 14, eabj3860. [Google Scholar] [CrossRef]
- Ganapathy, U.S.; Del Rio, R.G.; Cacho-Izquierdo, M.; Ortega, F.; Lelievre, J.; Barros-Aguirre, D.; Aragaw, W.W.; Zimmerman, M.D.; Lindman, M.; Dartois, V.; et al. A Mycobacterium tuberculosis NBTI DNA Gyrase Inhibitor Is Active against Mycobacterium abscessus. Antimicrob. Agents Chemother. 2021, 65, e0151421. [Google Scholar] [CrossRef] [PubMed]
- Madani, A.; Negatu, D.A.; El Marrouni, A.; Miller, R.R.; Boyce, C.W.; Murgolo, N.; Bungard, C.J.; Zimmerman, M.D.; Dartois, V.; Gengenbacher, M.; et al. Activity of Tricyclic Pyrrolopyrimidine Gyrase B Inhibitor against Mycobacterium abscessus. Antimicrob. Agents Chemother. 2022, 66, e0066922. [Google Scholar] [CrossRef]
- De Groote, M.A.; Jarvis, T.C.; Wong, C.; Graham, J.; Hoang, T.; Young, C.L.; Ribble, W.; Day, J.; Li, W.; Jackson, M.; et al. Optimization and Lead Selection of Benzothiazole Amide Analogs Toward a Novel Antimycobacterial Agent. Front. Microbiol. 2018, 9, 2231. [Google Scholar] [CrossRef]
- Sarathy, J.P.; Ganapathy, U.S.; Zimmerman, M.D.; Dartois, V.; Gengenbacher, M.; Dick, T. TBAJ-876, a 3,5-Dialkoxypyridine Analogue of Bedaquiline, Is Active against Mycobacterium abscessus. Antimicrob. Agents Chemother. 2020, 64, e02404-19. [Google Scholar] [CrossRef]
- Baker, S.J.; Zhang, Y.K.; Akama, T.; Lau, A.; Zhou, H.; Hernandez, V.; Mao, W.; Alley, M.R.; Sanders, V.; Plattner, J.J. Discovery of a new boron-containing antifungal agent, 5-fluoro-1,3-dihydro-1-hydroxy-2,1- benzoxaborole (AN2690), for the potential treatment of onychomycosis. J. Med. Chem. 2006, 49, 4447–4450. [Google Scholar] [CrossRef]
- Rock, F.L.; Mao, W.; Yaremchuk, A.; Tukalo, M.; Crepin, T.; Zhou, H.; Zhang, Y.K.; Hernandez, V.; Akama, T.; Baker, S.J.; et al. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 2007, 316, 1759–1761. [Google Scholar] [CrossRef]
- Hernandez, V.; Crepin, T.; Palencia, A.; Cusack, S.; Akama, T.; Baker, S.J.; Bu, W.; Feng, L.; Freund, Y.R.; Liu, L.; et al. Discovery of a novel class of boron-based antibacterials with activity against gram-negative bacteria. Antimicrob. Agents Chemother. 2013, 57, 1394–1403. [Google Scholar] [CrossRef]
- Mendes, R.E.; Alley, M.R.; Sader, H.S.; Biedenbach, D.J.; Jones, R.N. Potency and spectrum of activity of AN3365, a novel boron-containing protein synthesis inhibitor, tested against clinical isolates of Enterobacteriaceae and nonfermentative Gram-negative bacilli. Antimicrob. Agents Chemother. 2013, 57, 2849–2857. [Google Scholar] [CrossRef]
- Palencia, A.; Li, X.; Bu, W.; Choi, W.; Ding, C.Z.; Easom, E.E.; Feng, L.; Hernandez, V.; Houston, P.; Liu, L.; et al. Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium tuberculosis That Target Leucyl-tRNA Synthetase. Antimicrob. Agents Chemother. 2016, 60, 6271–6280. [Google Scholar] [CrossRef]
- Li, X.; Hernandez, V.; Rock, F.L.; Choi, W.; Mak, Y.S.L.; Mohan, M.; Mao, W.; Zhou, Y.; Easom, E.E.; Plattner, J.J.; et al. Discovery of a Potent and Specific M. tuberculosis Leucyl-tRNA Synthetase Inhibitor: (S)-3-(Aminomethyl)-4-chloro-7-(2-hydroxyethoxy)benzo[c][1,2]oxaborol-1(3H)-ol (GSK656). J. Med. Chem. 2017, 60, 8011–8026. [Google Scholar] [CrossRef]
- Ganapathy, U.S.; Gengenbacher, M.; Dick, T. Epetraborole Is Active against Mycobacterium abscessus. Antimicrob. Agents Chemother. 2021, 65, e0115621. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.R.; Lupien, A.; Kalthoff, E.; Hamela, C.; Taylor, L.; Munro, K.A.; Schmeing, T.M.; Kremer, L.; Behr, M.A. Efficacy of epetraborole against Mycobacterium abscessus is increased with norvaline. PLoS Pathog. 2021, 17, e1009965. [Google Scholar] [CrossRef]
- Wu, W.; He, S.; Li, A.; Guo, Q.; Tan, Z.; Liu, S.; Wang, X.; Zhang, Z.; Li, B.; Chu, H. A Novel Leucyl-tRNA Synthetase Inhibitor, MRX-6038, Expresses Anti-Mycobacterium abscessus Activity In Vitro and In Vivo. Antimicrob. Agents Chemother. 2022, 66, e0060122. [Google Scholar] [CrossRef]
- Collins, L.; Franzblau, S.G. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob. Agents Chemother. 1997, 41, 1004–1009. [Google Scholar] [CrossRef]
- Ballell, L.; Bates, R.H.; Young, R.J.; Alvarez-Gomez, D.; Alvarez-Ruiz, E.; Barroso, V.; Blanco, D.; Crespo, B.; Escribano, J.; Gonzalez, R.; et al. Fueling open-source drug discovery: 177 small-molecule leads against tuberculosis. Chem. Med. Chem. 2013, 8, 313–321. [Google Scholar] [CrossRef]
- Kondreddi, R.R.; Jiricek, J.; Rao, S.P.; Lakshminarayana, S.B.; Camacho, L.R.; Rao, R.; Herve, M.; Bifani, P.; Ma, N.L.; Kuhen, K.; et al. Design, synthesis, and biological evaluation of indole-2-carboxamides: A promising class of antituberculosis agents. J. Med. Chem. 2013, 56, 8849–8859. [Google Scholar] [CrossRef]
- Lun, S.; Guo, H.; Onajole, O.K.; Pieroni, M.; Gunosewoyo, H.; Chen, G.; Tipparaju, S.K.; Ammerman, N.C.; Kozikowski, A.P.; Bishai, W.R. Indoleamides are active against drug-resistant Mycobacterium tuberculosis. Nat. Commun. 2013, 4, 2907. [Google Scholar] [CrossRef]
- Rao, S.P.; Lakshminarayana, S.B.; Kondreddi, R.R.; Herve, M.; Camacho, L.R.; Bifani, P.; Kalapala, S.K.; Jiricek, J.; Ma, N.L.; Tan, B.H.; et al. Indolcarboxamide is a preclinical candidate for treating multidrug-resistant tuberculosis. Sci. Transl. Med. 2013, 5, 214ra168. [Google Scholar] [CrossRef]
- Li, W.; Upadhyay, A.; Fontes, F.L.; North, E.J.; Wang, Y.; Crans, D.C.; Grzegorzewicz, A.E.; Jones, V.; Franzblau, S.G.; Lee, R.E.; et al. Novel insights into the mechanism of inhibition of MmpL3, a target of multiple pharmacophores in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2014, 58, 6413–6423. [Google Scholar] [CrossRef]
- Stec, J.; Onajole, O.K.; Lun, S.; Guo, H.; Merenbloom, B.; Vistoli, G.; Bishai, W.R.; Kozikowski, A.P. Indole-2-carboxamide-based MmpL3 Inhibitors Show Exceptional Antitubercular Activity in an Animal Model of Tuberculosis Infection. J. Med. Chem. 2016, 59, 6232–6247. [Google Scholar] [CrossRef] [PubMed]
- Onajole, O.K.; Pieroni, M.; Tipparaju, S.K.; Lun, S.; Stec, J.; Chen, G.; Gunosewoyo, H.; Guo, H.; Ammerman, N.C.; Bishai, W.R.; et al. Preliminary structure-activity relationships and biological evaluation of novel antitubercular indolecarboxamide derivatives against drug-susceptible and drug-resistant Mycobacterium tuberculosis strains. J. Med. Chem. 2013, 56, 4093–4103. [Google Scholar] [CrossRef] [PubMed]
- Kozikowski, A.P.; Onajole, O.K.; Stec, J.; Dupont, C.; Viljoen, A.; Richard, M.; Chaira, T.; Lun, S.; Bishai, W.; Raj, V.S.; et al. Targeting Mycolic Acid Transport by Indole-2-carboxamides for the Treatment of Mycobacterium abscessus Infections. J. Med. Chem. 2017, 60, 5876–5888. [Google Scholar] [CrossRef] [PubMed]
- Franz, N.D.; Belardinelli, J.M.; Kaminski, M.A.; Dunn, L.C.; Calado Nogueira de Moura, V.; Blaha, M.A.; Truong, D.D.; Li, W.; Jackson, M.; North, E.J. Design, synthesis and evaluation of indole-2-carboxamides with pan anti-mycobacterial activity. Bioorg. Med. Chem. 2017, 25, 3746–3755. [Google Scholar] [CrossRef] [PubMed]
- Terlain, B.; Thomas, J.P. Structure of griselimycin, polypeptide antibiotic extracted Streptomyces cultures. I. Identification of the products liberated by hydrolysis. Bull. Soc. Chim. Fr. 1971, 6, 2349–2356. [Google Scholar]
- Terlain, B.; Thomas, J.P. Structure of griselimycin, polypeptide antibiotic extracted from streptomyces cultures. II. Structure of griselimycin. Bull. Soc. Chim. Fr. 1971, 6, 2357–2362. [Google Scholar]
- Terlain, B.; Thomas, J.P. Structure of griselimycin, polypeptide antibiotic extracted from streptomyces cultures. 3. Products related to griselimycin. Bull. Soc. Chim. Fr. 1971, 6, 2363–2365. [Google Scholar]
- Noufflard-Guy-Loe, H.; Berteaux, S. Experimental antituberculous action of a new antibiotic: RP 11,072. Rev. Tuberc. Pneumol. 1965, 29, 301–326. [Google Scholar]
- Toyohara, M. Aspects of the antituberculous activity of 27753-RP, a new semisynthetic derivative of griselimycine. Ann. Inst. Pasteur. Microbiol. 1987, 138, 737–744. [Google Scholar] [CrossRef]
- Kling, A.; Lukat, P.; Almeida, D.V.; Bauer, A.; Fontaine, E.; Sordello, S.; Zaburannyi, N.; Herrmann, J.; Wenzel, S.C.; Konig, C.; et al. Antibiotics. Targeting DnaN for tuberculosis therapy using novel griselimycins. Science 2015, 348, 1106–1112. [Google Scholar] [CrossRef]
- Bunting, K.A.; Roe, S.M.; Pearl, L.H. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. EMBO J. 2003, 22, 5883–5892. [Google Scholar] [CrossRef] [PubMed]
- Burnouf, D.Y.; Olieric, V.; Wagner, J.; Fujii, S.; Reinbolt, J.; Fuchs, R.P.; Dumas, P. Structural and biochemical analysis of sliding clamp/ligand interactions suggest a competition between replicative and translesion DNA polymerases. J. Mol. Biol. 2004, 335, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Wayne, L.G.; Hayes, L.G. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 1996, 64, 2062–2069. [Google Scholar] [CrossRef]
- Boon, C.; Dick, T. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J. Bacteriol. 2002, 184, 6760–6767. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Colvin, C.J.; Johnson, B.K.; Kirchhoff, P.D.; Wilson, M.; Jorgensen-Muga, K.; Larsen, S.D.; Abramovitch, R.B. Inhibitors of Mycobacterium tuberculosis DosRST signaling and persistence. Nat. Chem. Biol. 2017, 13, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Vennerstrom, J.L.; Arbe-Barnes, S.; Brun, R.; Charman, S.A.; Chiu, F.C.; Chollet, J.; Dong, Y.; Dorn, A.; Hunziker, D.; Matile, H.; et al. Identification of an antimalarial synthetic trioxolane drug development candidate. Nature 2004, 430, 900–904. [Google Scholar] [CrossRef] [PubMed]
- Falzon, D.; Schunemann, H.J.; Harausz, E.; Gonzalez-Angulo, L.; Lienhardt, C.; Jaramillo, E.; Weyer, K. World Health Organization treatment guidelines for drug-resistant tuberculosis, 2016 update. Eur. Respir. J. 2017, 49, 1602308. [Google Scholar] [CrossRef]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of quinolone action and resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef]
- Cowman, S.; Burns, K.; Benson, S.; Wilson, R.; Loebinger, M.R. The antimicrobial susceptibility of non-tuberculous mycobacteria. J. Infect. 2016, 72, 324–331. [Google Scholar] [CrossRef]
- Lee, S.H.; Yoo, H.K.; Kim, S.H.; Koh, W.J.; Kim, C.K.; Park, Y.K.; Kim, H.J. The drug resistance profile of Mycobacterium abscessus group strains from Korea. Ann. Lab. Med. 2014, 34, 31–37. [Google Scholar] [CrossRef]
- Chew, K.L.; Octavia, S.; Go, J.; Ng, S.; Tang, Y.E.; Soh, P.; Yong, J.; Jureen, R.; Lin, R.T.P.; Yeoh, S.F.; et al. In vitro susceptibility of Mycobacterium abscessus complex and feasibility of standardizing treatment regimens. J. Antimicrob. Chemother. 2021, 76, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Negatu, D.A.; Beuchel, A.; Madani, A.; Alvarez, N.; Chen, C.; Aragaw, W.W.; Zimmerman, M.D.; Laleu, B.; Gengenbacher, M.; Dartois, V.; et al. Piperidine-4-carboxamides target DNA gyrase in Mycobacterium abscessus. Antimicrob. Agents Chemother. 2021, 65, e0067621. [Google Scholar] [CrossRef] [PubMed]
- Talley, A.K.; Thurston, A.; Moore, G.; Gupta, V.K.; Satterfield, M.; Manyak, E.; Stokes, S.; Dane, A.; Melnick, D. First-in-Human Evaluation of the Safety, Tolerability, and Pharmacokinetics of SPR720, a Novel Oral Bacterial DNA Gyrase (GyrB) Inhibitor for Mycobacterial Infections. Antimicrob. Agents Chemother. 2021, 65, e0120821. [Google Scholar] [CrossRef]
- Coates, W.; Gwynn, M.; Hatton, I.; Masters, P.; Pearson, N.; Rahman, S.; Slocombe, B.; Warrack, J. Preparation of Piperidinylalkylquinolines as Antibacterials. Patent WO1999037635, 29 July 1999. [Google Scholar]
- Gomez, L.; Hack, M.D.; Wu, J.; Wiener, J.J.; Venkatesan, H.; Santillan, A., Jr.; Pippel, D.J.; Mani, N.; Morrow, B.J.; Motley, S.T.; et al. Novel pyrazole derivatives as potent inhibitors of type II topoisomerases. Part 1: Synthesis and preliminary SAR analysis. Bioorg. Med. Chem. Lett. 2007, 17, 2723–2727. [Google Scholar] [CrossRef] [PubMed]
- Wiener, J.J.; Gomez, L.; Venkatesan, H.; Santillan, A., Jr.; Allison, B.D.; Schwarz, K.L.; Shinde, S.; Tang, L.; Hack, M.D.; Morrow, B.J.; et al. Tetrahydroindazole inhibitors of bacterial type II topoisomerases. Part 2: SAR development and potency against multidrug-resistant strains. Bioorg. Med. Chem. Lett. 2007, 17, 2718–2722. [Google Scholar] [CrossRef] [PubMed]
- Black, M.T.; Stachyra, T.; Platel, D.; Girard, A.M.; Claudon, M.; Bruneau, J.M.; Miossec, C. Mechanism of action of the antibiotic NXL101, a novel nonfluoroquinolone inhibitor of bacterial type II topoisomerases. Antimicrob. Agents Chemother. 2008, 52, 3339–3349. [Google Scholar] [CrossRef] [PubMed]
- Bax, B.D.; Chan, P.F.; Eggleston, D.S.; Fosberry, A.; Gentry, D.R.; Gorrec, F.; Giordano, I.; Hann, M.M.; Hennessy, A.; Hibbs, M.; et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature 2010, 466, 935–940. [Google Scholar] [CrossRef]
- Miles, T.J.; Axten, J.M.; Barfoot, C.; Brooks, G.; Brown, P.; Chen, D.; Dabbs, S.; Davies, D.T.; Downie, D.L.; Eyrisch, S.; et al. Novel amino-piperidines as potent antibacterials targeting bacterial type IIA topoisomerases. Bioorg. Med. Chem. Lett. 2011, 21, 7489–7495. [Google Scholar] [CrossRef]
- Miles, T.J.; Barfoot, C.; Brooks, G.; Brown, P.; Chen, D.; Dabbs, S.; Davies, D.T.; Downie, D.L.; Eyrisch, S.; Giordano, I.; et al. Novel cyclohexyl-amides as potent antibacterials targeting bacterial type IIA topoisomerases. Bioorg. Med. Chem. Lett. 2011, 21, 7483–7488. [Google Scholar] [CrossRef]
- Dougherty, T.J.; Nayar, A.; Newman, J.V.; Hopkins, S.; Stone, G.G.; Johnstone, M.; Shapiro, A.B.; Cronin, M.; Reck, F.; Ehmann, D.E. NBTI 5463 is a novel bacterial type II topoisomerase inhibitor with activity against gram-negative bacteria and in vivo efficacy. Antimicrob. Agents Chemother. 2014, 58, 2657–2664. [Google Scholar] [CrossRef][Green Version]
- Gibson, E.G.; Bax, B.; Chan, P.F.; Osheroff, N. Mechanistic and Structural Basis for the Actions of the Antibacterial Gepotidacin against Staphylococcus aureus Gyrase. ACS Infect. Dis. 2019, 5, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Gibson, E.G.; Blower, T.R.; Cacho, M.; Bax, B.; Berger, J.M.; Osheroff, N. Mechanism of Action of Mycobacterium tuberculosis Gyrase Inhibitors: A Novel Class of Gyrase Poisons. ACS Infect. Dis. 2018, 4, 1211–1222. [Google Scholar] [CrossRef] [PubMed]
- Blanco, D.; Perez-Herran, E.; Cacho, M.; Ballell, L.; Castro, J.; Gonzalez Del Rio, R.; Lavandera, J.L.; Remuinan, M.J.; Richards, C.; Rullas, J.; et al. Mycobacterium tuberculosis gyrase inhibitors as a new class of antitubercular drugs. Antimicrob. Agents Chemother. 2015, 59, 1868–1875. [Google Scholar] [CrossRef] [PubMed]
- Tari, L.W.; Li, X.; Trzoss, M.; Bensen, D.C.; Chen, Z.; Lam, T.; Zhang, J.; Lee, S.J.; Hough, G.; Phillipson, D.; et al. Tricyclic GyrB/ParE (TriBE) inhibitors: A new class of broad-spectrum dual-targeting antibacterial agents. PLoS ONE 2013, 8, e84409. [Google Scholar] [CrossRef]
- McGarry, D.H.; Cooper, I.R.; Walker, R.; Warrilow, C.E.; Pichowicz, M.; Ratcliffe, A.J.; Salisbury, A.M.; Savage, V.J.; Moyo, E.; Maclean, J.; et al. Design, synthesis and antibacterial properties of pyrimido[4,5-b]indol-8-amine inhibitors of DNA gyrase. Bioorg. Med. Chem. Lett. 2018, 28, 2998–3003. [Google Scholar] [CrossRef] [PubMed]
- Henderson, S.R.; Stevenson, C.E.M.; Malone, B.; Zholnerovych, Y.; Mitchenall, L.A.; Pichowicz, M.; McGarry, D.H.; Cooper, I.R.; Charrier, C.; Salisbury, A.M.; et al. Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase. J. Antimicrob. Chemother. 2020, 75, 2835–2842. [Google Scholar] [CrossRef]
- Franzblau, S.G.; DeGroote, M.A.; Cho, S.H.; Andries, K.; Nuermberger, E.; Orme, I.M.; Mdluli, K.; Angulo-Barturen, I.; Dick, T.; Dartois, V.; et al. Comprehensive analysis of methods used for the evaluation of compounds against Mycobacterium tuberculosis. Tuberculosis 2012, 92, 453–488. [Google Scholar] [CrossRef]
- Graham, J.; Wong, C.E.; Day, J.; McFaddin, E.; Ochsner, U.; Hoang, T.; Young, C.L.; Ribble, W.; DeGroote, M.A.; Jarvis, T.; et al. Discovery of benzothiazole amides as potent antimycobacterial agents. Bioorg. Med. Chem. Lett. 2018, 28, 3177–3181. [Google Scholar] [CrossRef]
- Andries, K.; Verhasselt, P.; Guillemont, J.; Gohlmann, H.W.; Neefs, J.M.; Winkler, H.; Van Gestel, J.; Timmerman, P.; Zhu, M.; Lee, E.; et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 2005, 307, 223–227. [Google Scholar] [CrossRef]
- Koul, A.; Dendouga, N.; Vergauwen, K.; Molenberghs, B.; Vranckx, L.; Willebrords, R.; Ristic, Z.; Lill, H.; Dorange, I.; Guillemont, J.; et al. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat. Chem. Biol. 2007, 3, 323–324. [Google Scholar] [CrossRef]
- Mahajan, R. Bedaquiline: First FDA-approved tuberculosis drug in 40 years. Int. J. Appl. Basic Med. Res. 2013, 3, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Diacon, A.H.; Pym, A.; Grobusch, M.; Patientia, R.; Rustomjee, R.; Page-Shipp, L.; Pistorius, C.; Krause, R.; Bogoshi, M.; Churchyard, G.; et al. The diarylquinoline TMC207 for multidrug-resistant tuberculosis. N. Engl. J. Med. 2009, 360, 2397–2405. [Google Scholar] [CrossRef]
- Philley, J.V.; Wallace, R.J., Jr.; Benwill, J.L.; Taskar, V.; Brown-Elliott, B.A.; Thakkar, F.; Aksamit, T.R.; Griffith, D.E. Preliminary Results of Bedaquiline as Salvage Therapy for Patients With Nontuberculous Mycobacterial Lung Disease. Chest 2015, 148, 499–506. [Google Scholar] [CrossRef]
- Janssen Pharmaceutical, C. Briefing Document, TMC207 (Bedaquiline), Treatment of Patients with MDR-TB, NDA 204–384; Janssen Pharmaceutical Companies: Titusville, FL, USA, 2012. [Google Scholar]
- Diacon, A.H.; Donald, P.R.; Pym, A.; Grobusch, M.; Patientia, R.F.; Mahanyele, R.; Bantubani, N.; Narasimooloo, R.; De Marez, T.; van Heeswijk, R.; et al. Randomized pilot trial of eight weeks of bedaquiline (TMC207) treatment for multidrug-resistant tuberculosis: Long-term outcome, tolerability, and effect on emergence of drug resistance. Antimicrob. Agents Chemother. 2012, 56, 3271–3276. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, H.S.; Tong, A.S.T.; Choi, P.J.; Blaser, A.; Conole, D.; Franzblau, S.G.; Lotlikar, M.U.; Cooper, C.B.; Upton, A.M.; Denny, W.A.; et al. 3,5-Dialkoxypyridine analogues of bedaquiline are potent antituberculosis agents with minimal inhibition of the hERG channel. Bioorg. Med. Chem. 2019, 27, 1292–1307. [Google Scholar] [CrossRef]
- Sarathy, J.P.; Ragunathan, P.; Shin, J.; Cooper, C.B.; Upton, A.M.; Gruber, G.; Dick, T. TBAJ-876 Retains Bedaquiline’s Activity against Subunits c and epsilon of Mycobacterium tuberculosis F-ATP Synthase. Antimicrob. Agents Chemother. 2019, 63, e01191-19. [Google Scholar] [CrossRef]
- Sarathy, J.P.; Ragunathan, P.; Cooper, C.B.; Upton, A.M.; Gruber, G.; Dick, T. TBAJ-876 Displays Bedaquiline-Like Mycobactericidal Potency without Retaining the Parental Drug’s Uncoupler Activity. Antimicrob. Agents Chemother. 2020, 64, e01540-19. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.; Viljoen, A.; Thomas, S.; Roquet-Baneres, F.; Herrmann, J.L.; Pethe, K.; Kremer, L. Bedaquiline Inhibits the ATP Synthase in Mycobacterium abscessus and Is Effective in Infect.ed Zebrafish. Antimicrob. Agents Chemother. 2017, 61, e01225-17. [Google Scholar] [CrossRef]
| Compound | Structure | Class | Target | Pathway | Reference a |
|---|---|---|---|---|---|
| EC/11770 | 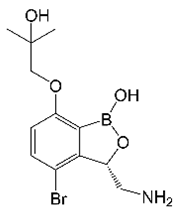 | Benzoxaborole | Leucyl-tRNA synthetase | Protein Biosynthesis | [31] |
| IC25 | 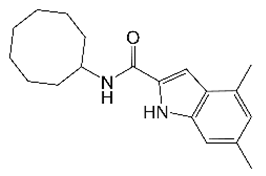 | Indole 2-carboxamide | Membrane Transporter MmpL3 | Cell Wall Biosynthesis | [32] |
| Cyclohexyl-griselimycin | 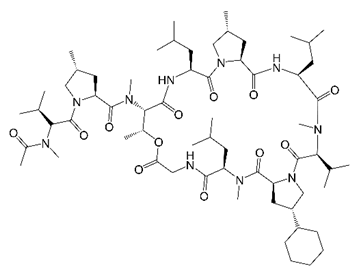 | Cyclic depsipeptide | DNA Sliding Clamp DnaN | DNA Replication | [33] |
| OZ439 |  | Synthetic trioxolane | Sensor Histidine Kinase DosS | Dormancy Response | [34] |
| EC/11716 | 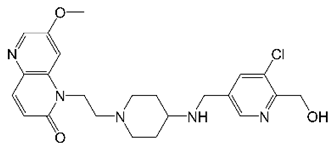 | NBTI | DNA Gyrase | DNA Topology | [35] |
| TPP8 | 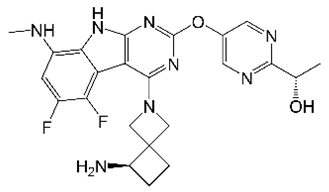 | Tricyclic pyrrolo- pyrimidine | DNA Gyrase | DNA Topology | [36] |
| CRS400226 | 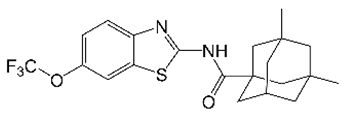 | Benzothiazole amide | Membrane Transporter MmpL3 | Cell Wall Biosynthesis | [37] |
| TBAJ-876 |  | Diarylquinoline | ATP Synthase | ATP Synthesis | [38] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganapathy, U.S.; Dick, T. Why Matter Matters: Fast-Tracking Mycobacterium abscessus Drug Discovery. Molecules 2022, 27, 6948. https://doi.org/10.3390/molecules27206948
Ganapathy US, Dick T. Why Matter Matters: Fast-Tracking Mycobacterium abscessus Drug Discovery. Molecules. 2022; 27(20):6948. https://doi.org/10.3390/molecules27206948
Chicago/Turabian StyleGanapathy, Uday S., and Thomas Dick. 2022. "Why Matter Matters: Fast-Tracking Mycobacterium abscessus Drug Discovery" Molecules 27, no. 20: 6948. https://doi.org/10.3390/molecules27206948
APA StyleGanapathy, U. S., & Dick, T. (2022). Why Matter Matters: Fast-Tracking Mycobacterium abscessus Drug Discovery. Molecules, 27(20), 6948. https://doi.org/10.3390/molecules27206948





