Identification of Small Molecule Inhibitors against Mycobacteria in Activated Macrophages
Abstract
1. Introduction
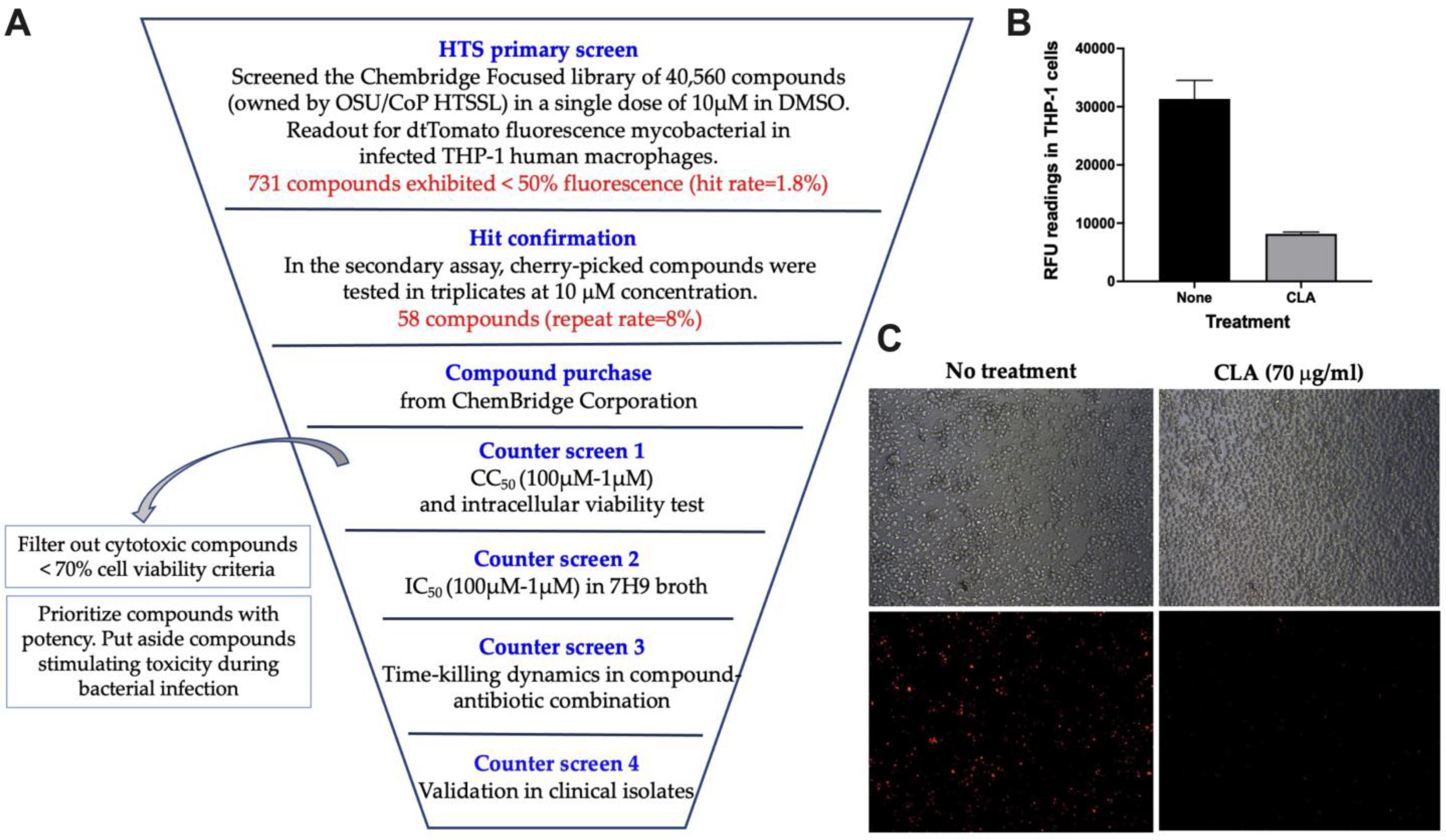
2. Materials and Methods
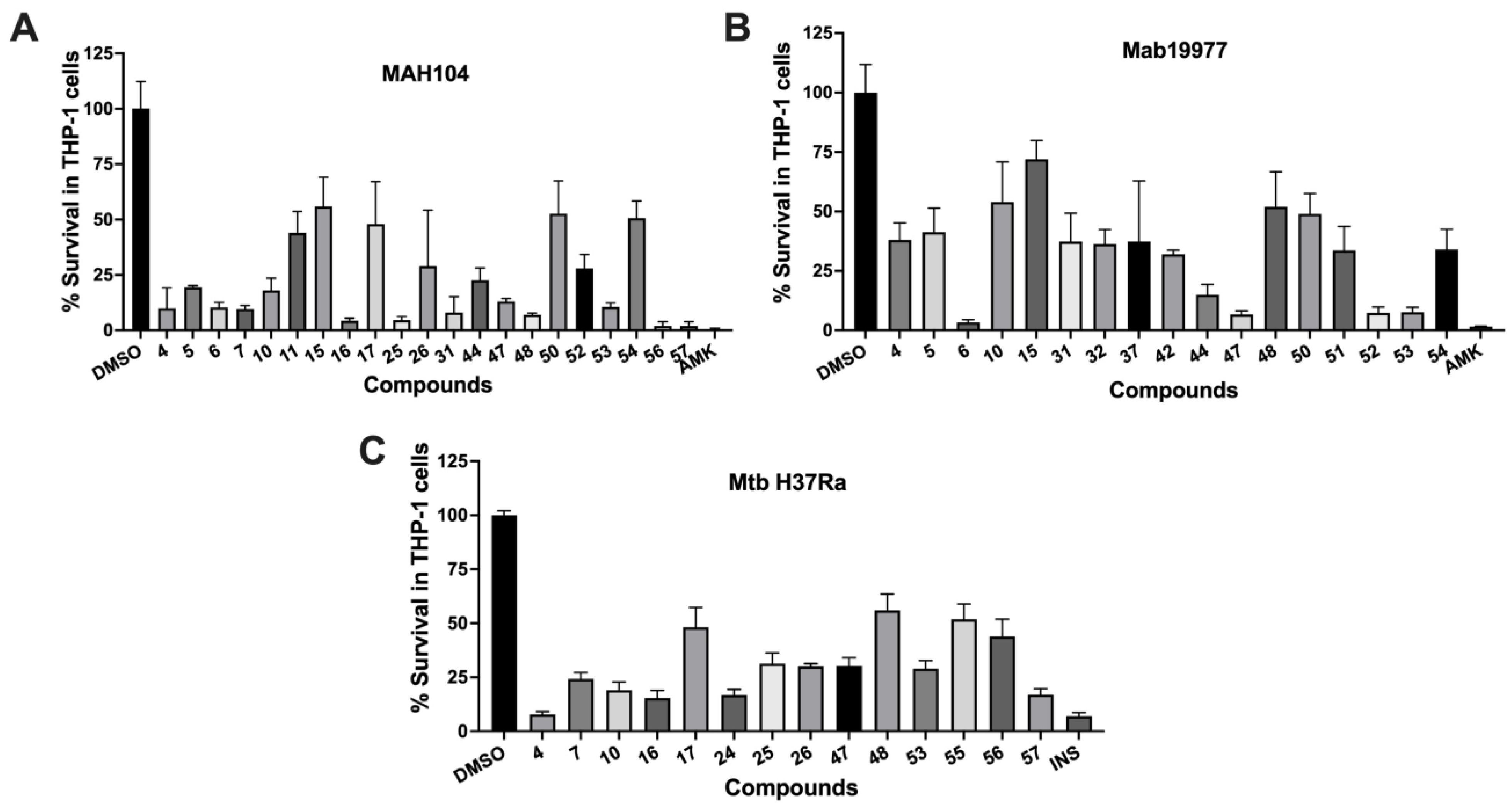
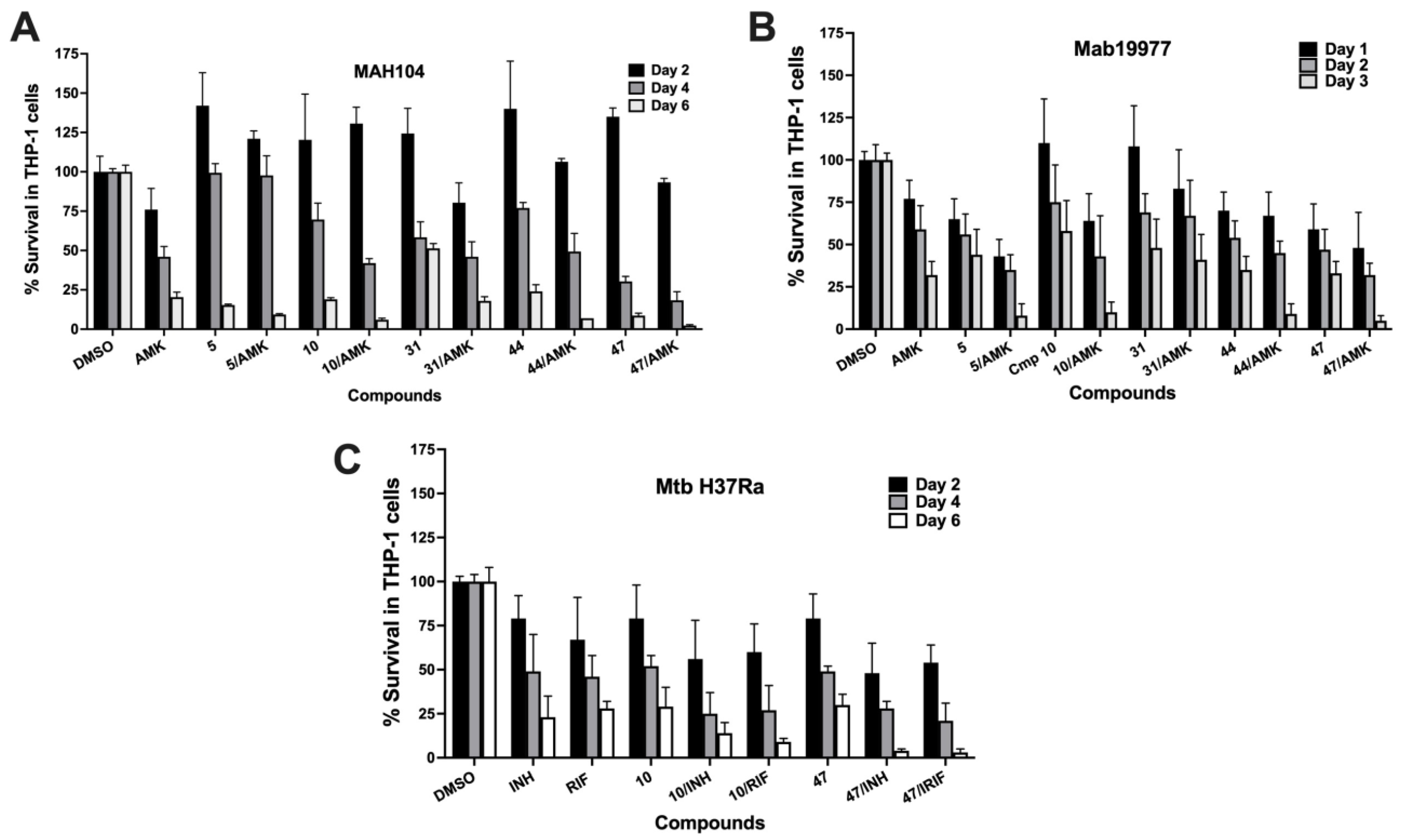
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gopalaswamy, R.; Shanmugam, S.; Mondal, R.; Subbian, S. Of tuberculosis and non-tuberculous mycobacterial infections—A comparative analysis of epidemiology, diagnosis and treatment. J. Biomed. Sci. 2020, 27, 74. [Google Scholar] [CrossRef]
- Johansen, M.D.; Herrmann, J.L.; Kremer, L. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat. Rev. Microbiol. 2020, 18, 392–407. [Google Scholar] [CrossRef]
- Ratnatunga, C.N.; Lutzky, V.P.; Kupz, A.; Doolan, D.L.; Reid, D.W.; Field, M.; Bell, S.C.; Thomson, R.M.; Miles, J.J. The Rise of Non-Tuberculosis Mycobacterial Lung Disease. Front. Immunol. 2020, 11, 303. [Google Scholar] [CrossRef]
- Maiz Carro, L.; Barbero Herranz, E.; Nieto Royo, R. Respiratory infections due to nontuberculous mycobacterias. Med. Clin. 2018, 150, 191–197. [Google Scholar] [CrossRef]
- van Ingen, J.; Obradovic, M.; Hassan, M.; Lesher, B.; Hart, E.; Chatterjee, A.; Daley, C.L. Nontuberculous mycobacterial lung disease caused by Mycobacterium avium complex-disease burden, unmet needs, and advances in treatment developments. Expert Rev. Respir. Med. 2021, 15, 1387–1401. [Google Scholar] [CrossRef]
- Gorzynski, M.; Week, T.; Jaramillo, T.; Dzalamidze, E.; Danelishvili, L. Mycobacterium abscessus Genetic Determinants Associated with the Intrinsic Resistance to Antibiotics. Microorganisms 2021, 9, 2527. [Google Scholar] [CrossRef]
- Luthra, S.; Rominski, A.; Sander, P. The Role of Antibiotic-Target-Modifying and Antibiotic-Modifying Enzymes in Mycobacterium abscessus Drug Resistance. Front. Microbiol. 2018, 9, 2179. [Google Scholar] [CrossRef]
- Vianna, J.S.; Machado, D.; Ramis, I.B.; Silva, F.P.; Bierhals, D.V.; Abril, M.A.; von Groll, A.; Ramos, D.F.; Lourenco, M.C.S.; Viveiros, M.; et al. The Contribution of Efflux Pumps in Mycobacterium abscessus Complex Resistance to Clarithromycin. Antibiotics 2019, 8, 153. [Google Scholar] [CrossRef]
- Batt, S.M.; Burke, C.E.; Moorey, A.R.; Besra, G.S. Antibiotics and resistance: The two-sided coin of the mycobacterial cell wall. Cell Surf. 2020, 6, 100044. [Google Scholar] [CrossRef]
- Rojony, R.; Danelishvili, L.; Campeau, A.; Wozniak, J.M.; Gonzalez, D.J.; Bermudez, L.E. Exposure of Mycobacterium abscessus to Environmental Stress and Clinically Used Antibiotics Reveals Common Proteome Response among Pathogenic Mycobacteria. Microorganisms 2020, 8, 698. [Google Scholar] [CrossRef]
- Silva, C.; Rojony, R.; Bermudez, L.E.; Danelishvili, L. Short-Chain Fatty Acids Promote Mycobacterium avium subsp. hominissuis Growth in Nutrient-Limited Environments and Influence Susceptibility to Antibiotics. Pathogens 2020, 9, 700. [Google Scholar] [CrossRef]
- Rojony, R.; Martin, M.; Campeau, A.; Wozniak, J.M.; Gonzalez, D.J.; Jaiswal, P.; Danelishvili, L.; Bermudez, L.E. Quantitative analysis of Mycobacterium avium subsp. hominissuis proteome in response to antibiotics and during exposure to different environmental conditions. Clin. Proteom. 2019, 16, 39. [Google Scholar] [CrossRef] [PubMed]
- Parker, H.; Lorenc, R.; Ruelas Castillo, J.; Karakousis, P.C. Mechanisms of Antibiotic Tolerance in Mycobacterium avium Complex: Lessons From Related Mycobacteria. Front. Microbiol. 2020, 11, 573983. [Google Scholar] [CrossRef]
- Quang, N.T.; Jang, J. Current Molecular Therapeutic Agents and Drug Candidates for Mycobacterium abscessus. Front. Pharmacol. 2021, 12, 724725. [Google Scholar] [CrossRef] [PubMed]
- Daley, C.L.; Iaccarino, J.M.; Lange, C.; Cambau, E.; Wallace, R.J.; Andrejak, C.; Bottger, E.C.; Brozek, J.; Griffith, D.E.; Guglielmetti, L.; et al. Treatment of Nontuberculous Mycobacterial Pulmonary Disease: An Official ATS/ERS/ESCMID/IDSA Clinical Practice Guideline. Clin. Infect. Dis. 2020, 71, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.R.; Sheng, W.H.; Hung, C.C.; Yu, C.J.; Lee, L.N.; Hsueh, P.R. Mycobacterium abscessus Complex Infections in Humans. Emerg. Infect. Dis. 2015, 21, 1638–1646. [Google Scholar] [CrossRef]
- Diel, R.; Lipman, M.; Hoefsloot, W. High mortality in patients with Mycobacterium avium complex lung disease: A systematic review. BMC Infect. Dis. 2018, 18, 206. [Google Scholar] [CrossRef]
- Gupta, R.; Netherton, M.; Byrd, T.F.; Rohde, K.H. Reporter-Based Assays for High-Throughput Drug Screening against Mycobacterium abscessus. Front. Microbiol. 2017, 8, 2204. [Google Scholar] [CrossRef]
- Christophe, T.; Jackson, M.; Jeon, H.K.; Fenistein, D.; Contreras-Dominguez, M.; Kim, J.; Genovesio, A.; Carralot, J.P.; Ewann, F.; Kim, E.H.; et al. High content screening identifies decaprenyl-phosphoribose 2’ epimerase as a target for intracellular antimycobacterial inhibitors. PLoS Pathog. 2009, 5, e1000645. [Google Scholar] [CrossRef]
- Ananthan, S.; Faaleolea, E.R.; Goldman, R.C.; Hobrath, J.V.; Kwong, C.D.; Laughon, B.E.; Maddry, J.A.; Mehta, A.; Rasmussen, L.; Reynolds, R.C.; et al. High-throughput screening for inhibitors of Mycobacterium tuberculosis H37Rv. Tuberculosis 2009, 89, 334–353. [Google Scholar] [CrossRef]
- Chengalroyen, M.D.; Jordaan, A.; Seldon, R.; Ioerger, T.; Franzblau, S.G.; Nasr, M.; Warner, D.F.; Mizrahi, V. Biological Profiling Enables Rapid Mechanistic Classification of Phenotypic Screening Hits and Identification of KatG Activation-Dependent Pyridine Carboxamide Prodrugs With Activity Against Mycobacterium tuberculosis. Front. Cell. Infect. Microbiol. 2020, 10, 582416. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.; Shapira, T.; Av-Gay, Y. THP-1 and Dictyostelium Infection Models for Screening and Characterization of Anti-Mycobacterium abscessus Hit Compounds. Antimicrob. Agents Chemother. 2019, 64, e01601-19. [Google Scholar] [CrossRef] [PubMed]
- Early, J.; Bermudez, L.E. Mimicry of the pathogenic mycobacterium vacuole in vitro elicits the bacterial intracellular phenotype, including early-onset macrophage death. Infect. Immun. 2011, 79, 2412–2422. [Google Scholar] [CrossRef]
- Chinison, J.J.; Danelishvili, L.; Gupta, R.; Rose, S.J.; Babrak, L.M.; Bermudez, L.E. Identification of Mycobacterium avium subsp. hominissuis secreted proteins using an in vitro system mimicking the phagosomal environment. BMC Microbiol. 2016, 16, 270. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.; Maser, J.; Lai, B.; Cai, Z.; Barry, C.E., 3rd; Honer Zu Bentrup, K.; Russell, D.G.; Bermudez, L.E. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell’s endosomal system. J. Immunol. 2005, 174, 1491–1500. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, L.E.; Petrofsky, M.; Kolonoski, P.; Young, L.S. An animal model of Mycobacterium avium complex disseminated infection after colonization of the intestinal tract. J. Infect. Dis. 1992, 165, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Danelishvili, L.; Stang, B.; Bermudez, L.E. Identification of Mycobacterium avium genes expressed during in vivo infection and the role of the oligopeptide transporter OppA in virulence. Microb. Pathog. 2014, 76, 67–76. [Google Scholar] [CrossRef][Green Version]
- Martin, A.; Camacho, M.; Portaels, F.; Palomino, J.C. Resazurin microtiter assay plate testing of Mycobacterium tuberculosis susceptibilities to second-line drugs: Rapid, simple, and inexpensive method. Antimicrob. Agents Chemother. 2003, 47, 3616–3619. [Google Scholar] [CrossRef]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef]
- Danelishvili, L.; Armstrong, E.; Miyasako, E.; Jeffrey, B.; Bermudez, L.E. Exposure of Mycobacterium avium subsp. homonissuis to Metal Concentrations of the Phagosome Environment Enhances the Selection of Persistent Subpopulation to Antibiotic Treatment. Antibiotics 2020, 9, 927. [Google Scholar] [CrossRef]
- Bussi, C.; Gutierrez, M.G. Mycobacterium tuberculosis infection of host cells in space and time. FEMS Microbiol. Rev. 2019, 43, 341–361. [Google Scholar] [CrossRef]
- Ganbat, D.; Seehase, S.; Richter, E.; Vollmer, E.; Reiling, N.; Fellenberg, K.; Gaede, K.I.; Kugler, C.; Goldmann, T. Mycobacteria infect different cell types in the human lung and cause species dependent cellular changes in infected cells. BMC Pulm. Med. 2016, 16, 19. [Google Scholar] [CrossRef]
- Maertzdorf, J.; Tonnies, M.; Lozza, L.; Schommer-Leitner, S.; Mollenkopf, H.; Bauer, T.T.; Kaufmann, S.H.E. Mycobacterium tuberculosis Invasion of the Human Lung: First Contact. Front. Immunol. 2018, 9, 1346. [Google Scholar] [CrossRef]
- Cohen, S.B.; Gern, B.H.; Delahaye, J.L.; Adams, K.N.; Plumlee, C.R.; Winkler, J.K.; Sherman, D.R.; Gerner, M.Y.; Urdahl, K.B. Alveolar Macrophages Provide an Early Mycobacterium tuberculosis Niche and Initiate Dissemination. Cell Host Microbe 2018, 24, 439–446.e434. [Google Scholar] [CrossRef]
- Cambier, C.J.; Takaki, K.K.; Larson, R.P.; Hernandez, R.E.; Tobin, D.M.; Urdahl, K.B.; Cosma, C.L.; Ramakrishnan, L. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 2014, 505, 218–222. [Google Scholar] [CrossRef]
- Danelishvili, L.; Shulzhenko, N.; Chinison, J.J.J.; Babrak, L.; Hu, J.; Morgun, A.; Burrows, G.; Bermudez, L.E. Mycobacterium tuberculosis Proteome Response to Antituberculosis Compounds Reveals Metabolic “Escape” Pathways That Prolong Bacterial Survival. Antimicrob. Agents Chemother. 2017, 61, e00430-17. [Google Scholar] [CrossRef]
- Kalsum, S.; Otrocka, M.; Andersson, B.; Welin, A.; Schon, T.; Jenmalm-Jensen, A.; Lundback, T.; Lerm, M. A high content screening assay for discovery of antimycobacterial compounds based on primary human macrophages infected with virulent Mycobacterium tuberculosis. Tuberculosis 2022, 135, 102222. [Google Scholar] [CrossRef]
- Subhash, N.; Sundaramurthy, V. Advances in host-based screening for compounds with intracellular anti-mycobacterial activity. Cell. Microbiol. 2021, 23, e13337. [Google Scholar] [CrossRef]
- Lele, A.C.; Raju, A.; Khambete, M.P.; Ray, M.K.; Rajan, M.G.; Arkile, M.A.; Jadhav, N.J.; Sarkar, D.; Degani, M.S. Design and Synthesis of a Focused Library of Diamino Triazines as Potential Mycobacterium tuberculosis DHFR Inhibitors. ACS Med. Chem. Lett. 2015, 6, 1140–1144. [Google Scholar] [CrossRef]
- Wang, X.; Inoyama, D.; Russo, R.; Li, S.G.; Jadhav, R.; Stratton, T.P.; Mittal, N.; Bilotta, J.A.; Singleton, E.; Kim, T.; et al. Antitubercular Triazines: Optimization and Intrabacterial Metabolism. Cell Chem. Biol. 2020, 27, 172–185.e11. [Google Scholar] [CrossRef]
- Selwood, T.; Larsen, B.J.; Mo, C.Y.; Culyba, M.J.; Hostetler, Z.M.; Kohli, R.M.; Reitz, A.B.; Baugh, S.D.P. Advancement of the 5-Amino-1-(Carbamoylmethyl)-1H-1,2,3-Triazole-4-Carboxamide Scaffold to Disarm the Bacterial SOS Response. Front. Microbiol. 2018, 9, 2961. [Google Scholar] [CrossRef] [PubMed]
- Scherman, M.S.; North, E.J.; Jones, V.; Hess, T.N.; Grzegorzewicz, A.E.; Kasagami, T.; Kim, I.H.; Merzlikin, O.; Lenaerts, A.J.; Lee, R.E.; et al. Screening a library of 1600 adamantyl ureas for anti-Mycobacterium tuberculosis activity in vitro and for better physical chemical properties for bioavailability. Bioorg. Med. Chem. 2012, 20, 3255–3262. [Google Scholar] [CrossRef] [PubMed]
- Dal Molin, M.; Selchow, P.; Schafle, D.; Tschumi, A.; Ryckmans, T.; Laage-Witt, S.; Sander, P. Identification of novel scaffolds targeting Mycobacterium tuberculosis. J. Mol. Med. 2019, 97, 1601–1613. [Google Scholar] [CrossRef] [PubMed]
- Kamat, V.; Santosh, R.; Poojary, B.; Nayak, S.P.; Kumar, B.K.; Sankaranarayanan, M.; Faheem; Khanapure, S.; Barretto, D.A.; Vootla, S.K. Pyridine- and Thiazole-Based Hydrazides with Promising Anti-inflammatory and Antimicrobial Activities along with Their In Silico Studies. ACS Omega 2020, 5, 25228–25239. [Google Scholar] [CrossRef]
- Hampannavar, G.A.; Karpoormath, R.; Palkar, M.B.; Shaikh, M.S.; Chandrasekaran, B. Dehydrozingerone Inspired Styryl Hydrazine Thiazole Hybrids as Promising Class of Antimycobacterial Agents. ACS Med. Chem. Lett. 2016, 7, 686–691. [Google Scholar] [CrossRef]
- Makam, P.; Kannan, T. 2-Aminothiazole derivatives as antimycobacterial agents: Synthesis, characterization, in vitro and in silico studies. Eur. J. Med. Chem. 2014, 87, 643–656. [Google Scholar] [CrossRef]
- Mori, G.; Chiarelli, L.R.; Esposito, M.; Makarov, V.; Bellinzoni, M.; Hartkoorn, R.C.; Degiacomi, G.; Boldrin, F.; Ekins, S.; de Jesus Lopes Ribeiro, A.L.; et al. Thiophenecarboxamide Derivatives Activated by EthA Kill Mycobacterium tuberculosis by Inhibiting the CTP Synthetase PyrG. Chem. Biol. 2015, 22, 917–927. [Google Scholar] [CrossRef]
- Strzelecka, M.; Swiatek, P. 1,2,4-Triazoles as Important Antibacterial Agents. Pharmaceuticals 2021, 14, 224. [Google Scholar] [CrossRef]
- Naz, S.; Farooq, U.; Ali, S.; Sarwar, R.; Khan, S.; Abagyan, R. Identification of new benzamide inhibitor against alpha-subunit of tryptophan synthase from Mycobacterium tuberculosis through structure-based virtual screening, anti-tuberculosis activity and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2019, 37, 1043–1053. [Google Scholar] [CrossRef]
- Hopfner, S.M.; Lee, B.S.; Kalia, N.P.; Miller, M.J.; Pethe, K.; Moraski, G.C. Structure guided generation of thieno[3,2-d]pyrimidin-4-amine Mycobacterium tuberculosis bd oxidase inhibitors. RSC Med. Chem. 2021, 12, 73–77. [Google Scholar] [CrossRef]
- Friedrich, T.; Wohlwend, D.; Borisov, V.B. Recent Advances in Structural Studies of Cytochrome bd and Its Potential Application as a Drug Target. Int. J. Mol. Sci. 2022, 23, 3166. [Google Scholar] [CrossRef] [PubMed]
- Ayers, B.; Long, H.; Sim, E.; Smellie, I.A.; Wilkinson, B.L.; Fairbanks, A.J. Stereoselective synthesis of beta-arabino glycosyl sulfones as potential inhibitors of mycobacterial cell wall biosynthesis. Carbohydr. Res. 2009, 344, 739–746. [Google Scholar] [CrossRef] [PubMed]

| Cluster | Compound | Structure | Name |
|---|---|---|---|
| Thiazole- hydrazine | 24 | 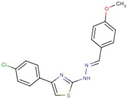 | 4-(4-chlorophenyl)-N-[(Z)-(4methoxyphenyl)methylideneamino]-1,3-thiazol-2-amine |
| 25 | 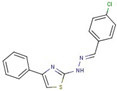 | N-[(Z)-(4-chlorophenyl)methylideneamino]-4-phenyl-1,3-thiazol-2-amine | |
| 26 |  | N-[(E)-benzylideneamino]-4-(4-methylphenyl)-1,3-thiazol-2-amine | |
| 47 |  | N-[(Z)-(4-methylphenyl)methylideneamino]-4-phenyl-1,3-thiazol-2-amine | |
| Pyridine- hydrazide | 55 | 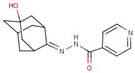 | N-[(5-hydroxy-2-adamantylidene)amino]pyridine-4-carboxamide |
| 56 | 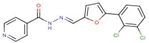 | N-[(Z)-[5-(2,3-dichlorophenyl)furan-2-yl]methylideneamino]pyridine-4-carboxamide | |
| 57 | 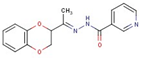 | N-[(Z)-1-(2,3-dihydro-1,4-benzodioxin-3-yl)ethylideneamino]pyridine-3-carboxamide | |
| 58 | 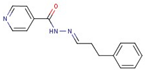 | N-[(Z)-3-phenylpropylideneamino]pyridine-4-carboxamide | |
| Triazole- carboxamides | 30 |  | 1-(4-ethylphenyl)-N-(3-fluorophenyl)-5-methyltriazole-4-carboxamide |
| 31 | 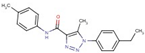 | 1-(4-ethylphenyl)-5-methyl-N-(4-methylphenyl)triazole-4-carboxamide | |
| 35 | 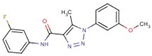 | N-(3-fluorophenyl)-1-(3-methoxyphenyl)-5-methyltriazole-4-carboxamide | |
| 37 |  | 1-(3-methoxyphenyl)-5-methyl-N-(4-propoxyphenyl)triazole-4-carboxamide | |
| Pyrimidine | 19 | 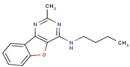 | N-butyl-2-methyl-[1]benzofuro[3,2-d]pyrimidin-4-amine |
| 36 | 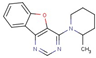 | 4-(2-methylpiperidin-1-yl)-[1]benzofuro[3,2-d]pyrimidine | |
| 50 |  | N-cyclopentylquinazolin-4-amine | |
| Phenyl- urea | 44 | 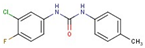 | 1-(3-chloro-4-fluorophenyl)-3-(4-methylphenyl)urea |
| 46 | 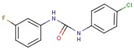 | 1-(4-chlorophenyl)-3-(3-fluorophenyl)urea | |
| 54 | 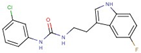 | 1-(3-chlorophenyl)-3-[2-(5-fluoro-1H-indol-3-yl)ethyl]urea | |
| Triazine diamine | 5 |  | 4-N-ethyl-2-N-propan-2-yl-6-pyrrolidin-1-yl-1,3,5-triazine-2,4-diamine |
| 10 | 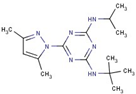 | 2-N-tert-butyl-6-(3,5-dimethylpyrazol- 1-yl)-4-N-propan-2-yl-1,3,5-triazine-2,4-diamine | |
| Benzamide | 16 | 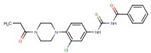 | N-[[3-chloro-4-(4-propanoylpiperazin-1-yl)phenyl]carbamothioyl]benzamide |
| 18 | 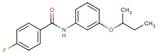 | N-(3-butan-2-yloxyphenyl)-4-fluorobenzamide | |
| Thiophene- carboxamide | 11 |  | N-(3-fluorophenyl)-5-methylthiophene-3-carboxamide |
| 51 |  | 5-ethyl-N-pyridin-4-yl-4,5,6,7-tetrahydro-1-benzothiophene-2-carboxamide | |
| Alcohol with piperazine | 17 | 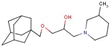 | 1-(1-adamantylmethoxy)-3-(3-methylpiperidin-1-yl)propan-2-ol |
| 42 | 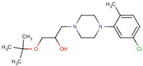 | 1-tert-butoxy-3-[4-(5-chloro-2-methylphenyl)-1-piperazinyl]-2-propanol hydrochloride | |
| Thiourea | 48 | 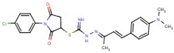 | [1-(4-chlorophenyl)-2,5-dioxopyrrolidin-3-yl] N’-[(Z)-[(E)-4[4(dimethylamino)phenyl]but-3-en-2-ylidene]amino] carbamimidothioate |
| 53 | 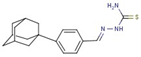 | [(Z)-[4-(1-adamantyl)phenyl]methylideneamino]thiourea | |
| Sulfone | 15 | 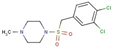 | 1-[(3,4-dichlorophenyl)methylsulfonyl]-4-methylpiperazine |
| 49 | 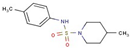 | 4-methyl-N-(4-methylphenyl)piperidine-1-sulfonamide | |
| Pyridine- carboxamides | 4 |  | N’-(4-biphenylylmethylene)isonicotinohydrazide |
| Carboxylic acid | 6 | 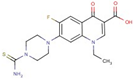 | 7-(4carbamothioylpiperazin-1-yl)-1-ethyl-6-fluoro-4-oxoquinoline-3-carboxylic acid |
| Aminothiazole | 7 | 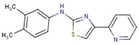 | N-(3,4-dimethylphenyl)-4-pyridin-2-yl-1,3-thiazol-2-amine |
| Acetamide | 32 |  | N-[4-(4-acetylpiperazin-1-yl)-3-chlorophenyl]-2-(4-chlorophenoxy) acetamide |
| Imidazole | 40 | 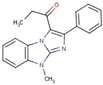 | 1-(4-methyl-2-phenylimidazo[1,2-a]benzimidazol-1-yl)propan-1-one |
| Pyridine- thioether | 52 |  | 4-[2-(2-phenylethylsulfanyl)ethyl]pyridine |
| Compound | THP-1 Cytotoxicity [μM] | Intracellular Killing in THP-1 Cells | MIC50 [μM] | ||||
|---|---|---|---|---|---|---|---|
| MAH104 | Mab19977 | MtbH37Ra | MAH104 | Mab19977 | MtbH37Ra | ||
| 4 | 32 | Yes | Yes | Yes | 10 | - | 3 |
| 5 | 32 | Yes | Yes | - | - | - | - |
| 6 | - | Yes | Yes | - | 3 | 32 | 10 |
| 7 | 10 | Yes | - | Yes | 10 | 10 | 10 |
| 10 | 32 | Yes | Yes | Yes | - | - | - |
| 11 | 32 | Yes | - | - | - | 32 | - |
| 15 | - | Yes | Yes | - | - | - | - |
| 16 | 10 | Yes | - | Yes | 32 | - | - |
| 17 | 32 | Yes | - | Yes | 100 | - | 10 |
| 18 | 32 | * | - | - | 100 | - | - |
| 19 | - | - | - | - | 100 | 100 | 10 |
| 24 | - | - | - | Yes | - | - | - |
| 25 | - | Yes | - | Yes | 32 | - | 10 |
| 26 | 32 | Yes | - | Yes | - | - | 10 |
| 30 | - | - | - | - | - | - | 10 |
| 31 | - | Yes | Yes | - | - | - | - |
| 32 | 32 | - | Yes | - | - | - | - |
| 35 | 32 | * | - | - | - | 100 | - |
| 36 | 32 | * | * | - | 100 | 100 | - |
| 37 | 32 | - | Yes | - | 100 | - | - |
| 40 | 32 | - | - | - | - | - | 10 |
| 42 | - | - | Yes | - | - | - | 10 |
| 44 | - | Yes | Yes | - | - | - | - |
| 46 | 32 | * | * | - | 10 | 32 | - |
| 47 | - | Yes | Yes | Yes | - | - | - |
| 48 | 32 | Yes | Yes | Yes | 10 | - | 10 |
| 49 | 32 | - | - | - | - | - | 10 |
| 50 | - | Yes | Yes | - | - | - | - |
| 51 | 32 | - | Yes | - | - | - | 10 |
| 52 | - | Yes | Yes | - | 10 | - | - |
| 53 | 32 | Yes | Yes | Yes | - | - | 10 |
| 54 | 10 | Yes | Yes | - | 32 | - | 10 |
| 55 | - | * | - | Yes | 32 | - | 10 |
| 56 | 32 | Yes | - | Yes | 100 | 10 | - |
| 57 | 32 | Yes | - | Yes | 32 | - | - |
| 58 | - | * | - | - | 100 | - | - |
| Compound | MIC50 [μM] | |||||||
|---|---|---|---|---|---|---|---|---|
| MAH104 | NJH 0133 | MAH B | MAH C | Mab19977 | DNA 01627 | NR49093 Strain DJO44274 | NR44273 Strain 4529 | |
| 4 | 10 | 10 | 100 | 10 | - | - | - | - |
| 6 | 3 | 3 | - | 3 | 32 | 32 | 32 | 32 |
| 7 | 10 | 10 | - | - | 10 | 10 | 10 | 10 |
| 11 | - | - | - | - | 32 | 32 | 32 | 32 |
| 16 | 32 | 32 | 100 | 100 | - | - | - | 100 |
| 17 | 100 | 100 | 100 | - | - | - | - | - |
| 18 | 100 | 100 | 100 | 100 | - | - | - | 100 |
| 19 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| 25 | 32 | - | - | - | - | - | - | - |
| 35 | - | - | - | - | 100 | 100 | 32 | 32 |
| 36 | 100 | 100 | - | 100 | 100 | 100 | 32 | 32 |
| 37 | 100 | 100 | - | 100 | - | - | - | - |
| 46 | 10 | 10 | - | 10 | 32 | 32 | 10 | 32 |
| 48 | 10 | 10 | 32 | 32 | - | - | - | - |
| 52 | 10 | - | - | - | - | - | - | - |
| 54 | 32 | 100 | - | - | - | - | - | - |
| 55 | 32 | 10 | 10 | 10 | - | - | - | - |
| 56 | 100 | 10 | 10 | 10 | 10 | 10 | 32 | 32 |
| 57 | 32 | 10 | 10 | 100 | - | - | - | - |
| 58 | 100 | 10 | 10 | 100 | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vande Voorde, R.; Dzalamidze, E.; Nelson, D.; Danelishvili, L. Identification of Small Molecule Inhibitors against Mycobacteria in Activated Macrophages. Molecules 2022, 27, 5824. https://doi.org/10.3390/molecules27185824
Vande Voorde R, Dzalamidze E, Nelson D, Danelishvili L. Identification of Small Molecule Inhibitors against Mycobacteria in Activated Macrophages. Molecules. 2022; 27(18):5824. https://doi.org/10.3390/molecules27185824
Chicago/Turabian StyleVande Voorde, Rebecca, Elizaveta Dzalamidze, Dylan Nelson, and Lia Danelishvili. 2022. "Identification of Small Molecule Inhibitors against Mycobacteria in Activated Macrophages" Molecules 27, no. 18: 5824. https://doi.org/10.3390/molecules27185824
APA StyleVande Voorde, R., Dzalamidze, E., Nelson, D., & Danelishvili, L. (2022). Identification of Small Molecule Inhibitors against Mycobacteria in Activated Macrophages. Molecules, 27(18), 5824. https://doi.org/10.3390/molecules27185824






