Abstract
In order to discover more promising antifungal and antibacterial agents, a series of new derivatives were designed and synthesized by structure modification based on the naturally occurring antimicrobial compound lophanic acid. The structures of all the target compounds were well characterized by spectroscopic data. The stereochemistry of these compounds was further determined through the X-ray diffraction analysis of 6a. The synthetic compounds were evaluated for their antimicrobial activities against filamentous fungi (T. rubrum, T. mentagrophytes), yeasts (C. neoformans, C. albicans) and Gram-positive and Gram-negative bacteria (MRSA, S. mutans, S. sobrinus, and E. coli). Among them, 3d and 3i are found as the most promising leads that showed potent inhibitory effects against all the tested fungal and bacterial strains except for E. coli. The presence of the C-20 carboxylic ester groups and the free hydroxy group at C-13 was found to be essential for the antifungal and antibacterial activities of the lophanic acid derivatives.
1. Introduction
Tinea pedis is a chronic or recurring disease characterized by dermatophytic infection of the feet and toes, which can involve the interdigital web spaces or the sides of the feet, and it is often caused by anthropophiles, including Trichophyton rubrum sensu stricto, T. interdigitale and Epidermophyton floccosum [1]. Currently, the effective treatments for tinea pedis are topical or oral antifungals or a combination of both, such as terbinafine and clotrimazole [2,3]. Although synthetic chemical drugs have been used to treat tinea pedis, their overuse over the years has led to considerable concern for fungus resistance and other adverse effects on human health [4,5]. Thus, there is an urgent need to discover new promising alternatives from sustainable natural bioresources to effectively and selectively treat tinea pedis.
For decades, medicinal plants have been considered a rich source of lead compounds for drug discovery and development [6,7]. Abietane-type diterpenoid is one of the most prevalent classes of diterpenes widely found in the plants of the Isodon genus [8,9]. They have shown a variety of pharmacological activities, including anticancer, anti-inflammatory, and antivirus activities [10,11]. Meanwhile, abietane-type diterpenoids were also found to show exceeding antimicrobial activities. For example, our previous studies described that rubesanolide D (Figure 1I) had antibacterial activity against biofilm formation of the dental bacterium Streptococcus mutans [12], kunminolide A (Figure 1II) and fladin A (Figure 1III) displayed inhibitory effects against the dental pathogens S. mutans, Porphyromonas gingivalis and Candida albicans and the athlete’s foot fungus T. rubrum [13,14]. Abietane-type diterpenoids appear to be attractive molecules for further structure modification to discover novel antifungal and antibacterial agents.
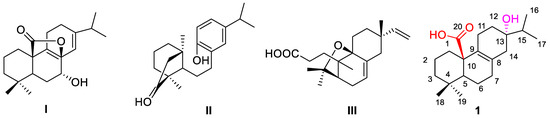
Figure 1.
Chemical structures of rubesanolide D (I), kunminolide A (II), fladin A (III), and lophanic acid (1).
Lophanic acid (1, Figure 1), a naturally occurring abietane-type diterpenoid, was found abundant in the medicinal plants I. flavidus [13] and I. lophanthoides [14]. Our study has demonstrated that the compound possesses potent biological activities against a wide range of fungi and bacteria, including T. rubrum, P. gingivalis, S. mutans, and S. albus [15]. However, little attention has been paid to the structural modification to synthesize the derivatives of 1 as antifungal and antibacterial agents. In our previous research on the bioactive compounds from Isodon plants, we obtained a rich amount of lophanic acid (1), which allowed us to carry out a synthetic study by using lophanic acid as a scaffold. Herein we report the design and synthesis of a series of new lophanic acid derivatives, and the antifungal and antibacterial activity evaluation of the synthetic derivatives against two filamentous fungi (T. rubrum, T. mentagrophytes), two yeasts (C. neoformans, C. albicans), and four Gram-positive and Gram-negative bacteria (MRSA, S. mutans, S. sobrinus, and E. coli).
2. Results and Discussion
2.1. Chemistry
As shown in Scheme 1, fourteen C-20 ester derivatives (3a–3n) were first synthesized by reaction of lophanic acid (1) with the corresponding alkyl halides in the presence of K2CO3 at room temperature. The carboxyl group of 3a was further reduced to an alcohol group by LiAlH4 to obtain compound 4, which was substituted with different alkoxy groups at C-20 to produce 5a–5h. In addition, as outlined in Scheme 2, the hydroxyl groups of compounds 3a and 3d underwent an esterification reaction with acetyl chloride in the presence of pyridine to afford 6a and 6b. Interestingly, the acetyl group at C-13 of 6a was further acetylated to form acetylacetic ester (6c) through a C–C bond. The structure determination of 6c was evidenced by the compassion among the 1H NMR spectra of 3a, 6a, and 6c as depicted in Figure 2. The stereochemistry of 6a was further confirmed by the X-ray crystallographic analysis (Figure 3).
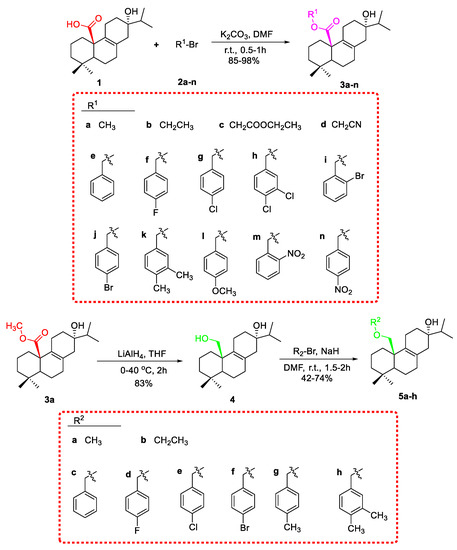
Scheme 1.
The synthetic route of compounds 3a–n and 5a–h.
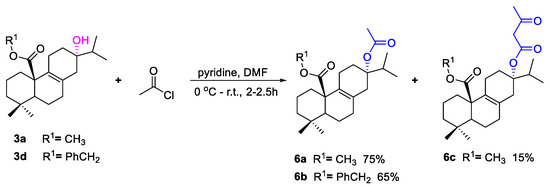
Scheme 2.
The synthetic route of compounds 6a–c.
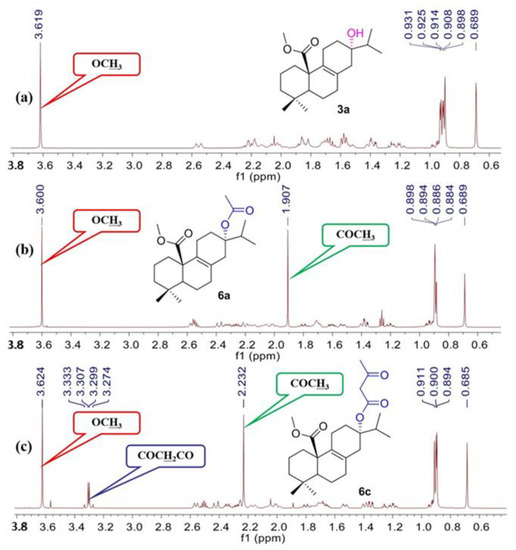
Figure 2.
The 1H NMR spectra of 3a (a), 6a (b), and 6c (c).
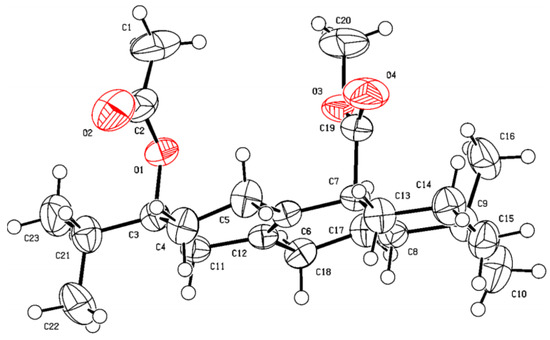
Figure 3.
X-ray crystallographic structure of 6a.
2.2. In Vitro Antifungal Activity Evaluation against Filamentous Fungi and Yeasts
The antifungal activities of lophanic acid derivatives (3a–6c) were investigated against two filamentous fungi (T. rubrum, T. mentagrophytes) and two yeasts (C. neoformans, C. albicans) in vitro at 100 μg/mL with miconazole served as a positive control agent.
As shown in Table 1, compounds 3d and 3i exhibited a broad spectrum of antifungal activities against T. rubrum, T. mentagrophytes, and C. neoformans with inhibitory rates over 60%. On the other hand, some compounds were found to exhibit selective activities against filamentous fungi. For example, compound 3a inhibited the growth of T. rubrum by 81.61%, but it showed only a 49.1% inhibitory rate against C. neoformans. Compounds 3b, 3e, 5c, and 5h displayed mild antifungal effects against T. rubrum and C. neoformans. By analyzing the structure-activity relationship (SAR) of the lophanic acid derivatives, we observed that the C-13 hydroxy is essential for retaining their antifungal activities. For example, the antimicrobial inhibitory rates of compounds 3a and 3b were measured over 60%, whereas their corresponding derivatives 6a–6c showed almost no antimicrobial activities. Based on the antimicrobial activities of the lophanic acid derivatives (e.g., 3a, 3d, 5a, and 5b), the structural modification at C-20 could be performed to improve the bioactivity. On the other hand, when a phenyl ring was introduced with an electron-donating group, the corresponding compounds exhibited better activity potency than those with an electron-withdrawing group. For example, 3i showed growth inhibitory rates against T. rubrum, T. mentagrophytes, and C. neoformans at 74.91, 63.00, and 88.54%, respectively, while 3n displayed no antimicrobial activities against these fungi. Interestingly, no lophanic acid derivatives were found to show antifungal activities against C. albicans at the concentration of 100 µg/mL.

Table 1.
Antifungal activities of compounds 3a–6c at 100 μg/mL.
2.3. In Vitro Antibacterial Activity Evaluation against Gram-Positive and Gram-Negative Bacteria
The antibacterial activities of compounds 3a–6c were further tested against Gram-positive bacteria (MRSA, S. mutans, and S. sobrinus) and Gram-negative bacterium (E. coli) in vitro at the concentration of 100 μg/mL with tetracycline as the positive control agent (Table 2). Compounds 3a, 3b, 3d, 3e, 3i, 5c, and 5h displayed more potent antibacterial activities against MRSA than they are against the other bacteria. Especially, 3b, 3d, 5c, and 5h were found to possess antibacterial activities with inhibitory rates greater than 90%. However, no antimicrobial activities were observed for the synthetic derivatives against S. mutans, S. sobrinus, and E. coli. Preliminary SAR analysis showed that the C-13 hydroxy plays an indispensable role in the antibacterial activities of the lophanic acid derivatives. For instance, compounds 3a and 3b showed strong activities with inhibitory rates of 84.43% and 95.89%, respectively. On the contrary, the introduction of an acyl group on the C-13 hydroxy group of 3a and 3b resulted in the loss of the antibacterial activities (e.g., 6a-6c). Furthermore, a proper ester or alkyloxy group substituted at C-20 was found important for retaining the antibacterial activity potency of a lophanic acid derivative (e.g., 3a, 3b, 3d, 5c, and 5h).

Table 2.
Antibacterial activities of compounds 3a–6c at 100 μg/mL.
3. Materials and Methods
3.1. Chemistry
All chemical reagents were purchased and utilized without further purification. Solvents were used directly or treated with standard methods before use. Melting points (m.p.) were determined on an X-6a digital melting point apparatus (Gongyi Tech Instrument Co., Ltd., Gongyi, China) and were uncorrected. Infrared spectra (IR) were recorded on a Bruker TENSOR 27 spectrometer. Proton nuclear magnetic resonance spectra (1H NMR) and carbon nuclear magnetic resonance spectra (13C NMR) were recorded in CDCl3 on a Bruker Avance 400, 500, or 600 MHz instruments using tetramethylsilane (TMS) as the internal standard. High-resolution mass spectra (HRMS) were carried out with IonSpec 4.7 Tesla FTMS instrument. The purities of all the title compounds were determined on an UltiMate 3000 (Dionex, Sunnyvale, CA, USA) HPLC system and were of > 95% purity.
3.1.1. General Procedure for the Synthesis of Compound 3a–n
Lophanic acid (1, 100 mg, 0.31 mmol) and potassium carbonate (86 mg, 0.62 mmol) were dissolved in N, N-Dimethylformamide (DMF, 5 mL), and the solution was stirred at room temperature. Then a solution of substituent haloalkane (0.62 mmol) in DMF (2 mL) was added dropwise for 10 min. When the reaction was complete, checked by thin-layer chromatography (TLC) analysis, pure water (30 mL) was added to the reaction, which was extracted with ethyl acetate (3 × 30 mL). The combined organic phase was dried over anhydrous Na2SO4, filtered, concentrated under reduced pressure, and purified by silica gel column chromatography eluting with petroleum ether/ethyl acetate to afford compound 3a-n in 85–98%.
Data for 3a: White solid, yield: 98%, m.p. 99–101 °C; IR cm−1: 3527, 1709, 1213, 1141; 1H NMR (400 MHz, CDCl3): δ 3.61 (s, 3H, OCH3), 2.57 (d, J = 12.6 Hz, 1H), 2.27–2.13 (m, 3H), 2.07–1.98 (m, 2H), 1.89–1.81 (m, 3H), 1.72–1.65 (m, 3H), 1.59–1.51 (m, 3H), 1.42–1.36 (m, 2H), 1.26–1.20 (m, 1H), 0.98–0.94 (m, 1H), 0.92–0.91 (m, 6H, H-16, 17), 0.89 (s, 3H, H-19), 0.68 (s, 3H, H-18); 13C NMR (100 MHz, CDCl3): δ 176.2 (C-20), 130.1 (C-9), 129.6 (C-8), 72.0 (C-13), 52.4 (OCH3), 51.4 (C-5), 48.7 (C-10), 41.8 (C-3), 41.2 (C-14), 34.5 (C-1), 34.4 (C-4), 33.7 (C-15), 32.2 (C-12), 32.0 (C-7), 31.6 (C-18), 21.5 (C-11), 20.1 (C-2), 19.8 (C-19), 18.1 (C-6), 16.7 (C-16), 16.7 (C-17); HRMS m/z calcd for C21H34O3Na ([M + Na]+) 357.2400, found 357.2401 (Figures S1–S3).
Data for 3b: White solid, yield: 95%, m.p. 52–54 °C; IR cm−1: 3540, 1709, 1214, 1145; 1H NMR (600 MHz, CDCl3): δ 4.13–4.03 (m, 2H, CH2CH3), 2.56 (d, J = 12.0 Hz, 1H), 2.29–2.05 (m, 4H), 2.02–1.97 (m, 1H), 1.92–1.81 (m, 3H), 1.71–1.64 (m, 3H), 1.58–1.51 (m, 3H), 1.41–1.36 (m, 2H), 1.25–1.18 (m, 4H), 0.96–0.93 (m, 1H), 0.92–0.89 (m, 6H, H-16, 17), 0.90 (s, 3H, H-19), 0.72 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 175.6 (C-20), 130.3 (C-9), 129.5 (C-8), 72.0 (C-13), 60.2 (CH2CH3), 52.4 (C-5), 48.7 (C-10), 41.9 (C-3), 41.2 (C-14), 34.5 (C-1), 34.5 (C-4), 33.8 (C-15), 32.2 (C-12), 32.0 (C-7), 31.6 (C-18), 21.5 (C-11), 20.1 (C-2), 20.0 (C-19), 18.2 (C-6), 16.8 (C-16), 16.8 (C-17), 14.2 (CH2CH3); HRMS m/z calcd for C22H36O3Na ([M + Na]+) 371.2557, found 371.2558 (Figures S4–S6).
Data for 3c: colorless oil, yield: 89%; IR cm−1: 3660, 1735, 1726, 1381, 1130; 1H NMR (600 MHz, CDCl3): δ 4.74 (d, J = 15.6 Hz, 1H, CH2COOCH2CH3), 4.32 (d, J = 15.6 Hz, 1H, CH2COOCH2CH3), 4.25–4.16 (m, 2H, CH2COOCH2CH3), 2.63 (d, J = 12.1 Hz, 1H), 2.26–2.12 (m, 4H), 2.06–2.00 (m, 2H), 1.85–1.78 (m, 2H), 1.76–1.64 (m, 3H), 1.56–1.51 (m, 3H), 1.47–1.39 (m, 2H), 1.27–1.20 (m, 4H), 1.04–0.99 (m, 1H), 0.96 (d, J = 6.6 Hz, 3H, H-16), 0.93 (d, J = 6.6 Hz, 3H, H-17), 0.90 (s, 3H, H-19), 0.71 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 175.1 (C-20), 168.2 (CH2COOCH2CH3), 130.6 (C-9), 129.2 (C-8), 71.4 (C-13), 61.5 (CH2COOCH2CH3), 60.2 (CH2COOCH2CH3), 51.9 (C-5), 48.5 (C-10), 41.9 (C-3), 40.3 (C-14), 36.1 (C-1), 34.7 (C-4), 33.9 (C-15), 32.0 (C-12), 31.9 (C-7), 31.4 (C-18), 20.5 (C-11), 20.0 (C-2), 19.9 (C-19), 18.2 (C-6), 17.1 (C-16), 16.9 (C-17), 14.1 (CH2COOCH2CH3); HRMS m/z calcd for C24H38O5Na ([M + Na]+) 429.2611, found 429.2613 (Figures S7–S9).
Data for 3d: colorless oil, yield: 92%; IR cm−1: 1732, 1457, 1115; 1H NMR (600 MHz, CDCl3): δ 4.68 (s, 2H, CH2CN), 2.59 (d, J = 13.8 Hz, 1H), 2.21–2.13 (m, 3H), 2.09–2.01 (m, 2H), 1.87–1.74 (m, 4H), 1.67–1.56 (m, 5H), 1.45–1.42 (m, 2H), 1.25–1.19 (m, 1H), 1.03–0.98 (m, 1H), 0.89–0.91 (m, 9H, H-16, 17, 19), 0.71 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 174.3 (C-20), 131.3 (C-9), 128.9 (C-8), 114.5 (CH2CN), 71.9 (C-13), 52.4 (C-5), 49.1 (CH2CN), 48.2 (C-10), 41.7 (C-3), 41.3 (C-14), 34.6 (C-1), 34.5 (C-4), 33.9 (C-15), 31.9 (C-12), 31.9 (C-7), 29.8 (C-18), 21.6 (C-11), 20.2 (C-2), 20.1 (C-19), 18.2 (C-6), 16.8 (C-16), 16.8 (C-17); HRMS m/z calcd for C22H33O3NNa ([M + Na]+) 382.2352, found 382.2352 (Figures S10–S12).
Data for 3e: colorless oil, yield: 97%; IR cm−1: 1716, 1456, 1205, 1129; 1H NMR (600 MHz, CDCl3): δ 7.26–7.35 (m, 5H, H-Ph), 5.16 (d, J = 12.6 Hz, 1H, PhCH2), 5.00 (d, J = 12.6 Hz, 1H, PhCH2), 2.58 (d, J = 13.2 Hz, 1H), 2.30–2.17 (m, 2H), 2.13–1.97 (m, 3H), 1.90–1.78 (m, 3H), 1.70–1.63 (m, 2H), 1.59–1.45 (m, 3H), 1.40–1.37 (m, 3H), 1.22–1.17 (m, 1H), 0.97–0.92 (m, 1H), 0.90 (s, 3H, H-16), 0.89 (s, 3H, H-17), 0.88 (s, 3H, H-19), 0.67 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 175.5 (C-20), 136.3 (C-Ph), 130.1 (C-9), 130.0 (C-Ph), 128.6 (C-8), 128.1 (C-Ph), 128.0 (C-Ph), 71.9 (C-13), 66.2 (PhCH2), 52.5 (C-5), 48.9 (C-10), 42.0 (C-3), 41.5 (C-14), 34.6 (C-1), 34.0 (C-4), 33.9 (C-15), 32.1 (C-12), 32.0 (C-7), 32.0 (C-18), 21.8 (C-11), 20.2 (C-2), 20.2 (C-19), 18.3 (C-6), 16.8 (C-16), 16.8 (C-17); HRMS m/z calcd for C27H38O3Na ([M + Na]+) 433.2713, found 433.2713 (Figures S13–S15).
Data for 3f: colorless oil, yield: 85%; IR cm−1: 1715, 1513, 1224; 1H NMR (600 MHz, CDCl3): δ 7.30–7.26 (m, 2H, H-Ph), 7.03–7.01 (m, 2H, H-Ph), 5.11 (d, J = 12.0 Hz, 1H, PhCH2), 4.97 (d, J = 12.0 Hz, 1H, PhCH2), 2.56 (d, J = 13.8 Hz, 1H), 2.26–2.17 (m, 2H), 2.13–1.95 (m, 3H), 1.87–1.76 (m, 3H), 1.70–1.63 (m, 2H), 1.59–1.46 (m, 4H), 1.39–1.37 (m, 2H), 1.22–1.17 (m, 1H), 0.97–0.92 (m, 1H), 0.91 (s, 3H, H-16), 0.90 (s, 3H, H-17), 0.87 (s, 3H, H-19), 0.63 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 175.3 (C-20), 163.3 (C-Ph), 132.0 (C-Ph), 131.9 (C-9), 130.1 (C-Ph), 130.0 (C-8), 115.3 (C-Ph), 71.9 (C-13), 65.4 (PhCH2), 52.3 (C-5), 48.8 (C-10), 41.8 (C-3), 41.3 (C-14), 34.4 (C-1), 34.0 (C-4), 33.8 (C-15), 31.9 (C-12), 31.9 (C-7), 31.8 (C-18), 21.6 (C-11), 20.1 (C-2), 20.0 (C-19), 18.1 (C-6), 16.7 (C-16), 16.7 (C-17); HRMS m/z calcd for C27H37O3FNa ([M + Na]+) 451.2619, found 451.2617 (Figures S16–S18).
Data for 3g: White solid, yield: 98%, m.p. 76–78 °C; IR cm−1: 1720, 1456, 1201, 1146, 1128, 1089, 981, 806; 1H NMR (600 MHz, CDCl3): δ 7.31 (d, J = 8.4 Hz, 2H, H-Ph), 7.25 (d, J = 8.4 Hz, 2H, H-Ph), 5.11 (d, J = 12.6 Hz, 1H, PhCH2), 4.95 (d, J = 12.6 Hz, 1H, PhCH2), 2.56 (d, J = 12.6 Hz, 1H), 2.26–2.17 (m, 2H), 2.13–1.96 (m, 3H), 1.85–1.76 (m, 3H), 1.70–1.46 (m, 6H), 1.40–1.37 (m, 2H), 1.22–1.17 (m, 1H), 0.97–0.92 (m, 1H), 0.91 (s, 3H, H-16), 0.90 (s, 3H, H-17), 0.88 (s, 3H, H-19), 0.64 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 175.4 (C-20), 134.8 (C-Ph), 133.9 (C-Ph), 130.1 (C-9), 129.9 (C-Ph), 129.5 (C-8), 128.8 (C-Ph), 71.9 (C-13), 65.4 (PhCH2), 52.4 (C-5), 48.9 (C-10), 41.9 (C-3), 41.4 (C-14), 34.5 (C-1), 34.1 (C-4), 33.9 (C-15), 32.0 (C-12), 32.0 (C-7), 32.0 (C-18), 21.8 (C-11), 20.2 (C-2), 20.1 (C-19), 18.3 (C-6), 16.8 (C-16), 16.8 (C-17); HRMS m/z calcd for C27H37O3ClNa ([M + Na]+) 467.2323, found 467.2326 (Figures S19–S21).
Data for 3h: colorless oil, yield: 88%; IR cm−1: 1716, 1472, 1205, 1128; 1H NMR (600 MHz, CDCl3): δ 7.41–7.40 (m, 2H, H-Ph), 7.14–7.13 (m, 1H, H-Ph), 5.14 (d, J = 12.6 Hz, 1H, PhCH2), 4.89 (d, J = 12.6 Hz, 1H, PhCH2), 2.57 (d, J = 13.2 Hz, 1H), 2.22–2.16 (m, 2H), 2.14–1.98 (m, 3H), 1.88–1.77 (m, 3H), 1.73–1.48 (m, 6H), 1.42–1.39 (m, 2H), 1.23–1.18 (m, 1H), 0.99–0.94 (m, 1H), 0.92 (s, 3H, H-16), 0.90 (s, 3H, H-17), 0.89 (s, 3H, H-19), 0.66 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 175.4 (C-20), 136.7 (C-Ph), 132.7 (C-Ph), 132.1 (C-Ph), 130.6 (C-9), 130.2 (C-Ph), 129.9 (C-8), 129.8 (C-Ph), 127.2 (C-Ph), 72.0 (C-13), 64.6 (PhCH2), 52.4 (C-5), 49.0 (C-10), 41.9 (C-3), 41.5 (C-14), 34.6 (C-1), 34.3 (C-4), 33.9 (C-15), 32.0 (C-12), 32.0 (C-7), 29.7 (C-18), 21.8 (C-11), 20.2 (C-2), 20.1 (C-19), 18.4 (C-6), 16.8 (C-16), 16.8 (C-17); HRMS m/z calcd for C27H36O3Cl2Na ([M + Na]+) 501.1934, found 501.1935 (Figures S22–S24).
Data for 3i: White solid, yield: 96%, m.p. 65–67 °C; IR cm−1: 1720, 1199, 1145, 1129, 1112, 746; 1H NMR (600 MHz, CDCl3): δ 7.55–7.52 (m, 1H, H-Ph), 7.37–7.28 (m, 2H, H-Ph), 7.17–7.15 (m, 1H, H-Ph), 5.28 (d, J = 13.2 Hz, 1H, PhCH2), 5.03 (d, J = 13.2 Hz, 1H, PhCH2), 2.59 (d, J = 13.8 Hz, 1H), 2.30–2.17 (m, 2H), 2.11–1.98 (m, 3H), 1.88–1.76 (m, 3H), 1.71–1.57 (m, 4H), 1.47–1.38 (m, 4H), 1.23–1.18 (m, 1H), 0.97–0.92 (m, 1H), 0.90 (s, 3H, H-16), 0.89 (s, 3H, H-17), 0.88 (s, 3H, H-19), 0.67 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 175.3 (C-20), 135.6 (C-Ph), 132.8 (C-Ph), 132.6 (C-Ph), 130.2 (C-9), 129.8 (C-Ph), 129.7 (C-8), 127.7 (C-Ph), 123.7 (C-Ph), 71.9 (C-13), 65.7 (PhCH2), 52.4 (C-5), 49.0 (C-10), 42.0 (C-3), 41.5 (C-14), 34.6 (C-1), 33.9 (C-4), 33.8 (C-15), 32.1 (C-12), 32.0 (C-7), 32.0 (C-18), 21.8 (C-11), 20.2 (C-2), 20.1 (C-19), 18.3 (C-6), 16.8 (C-16), 16.8 (C-17); HRMS m/z calcd for C27H37O3BrNa ([M + Na]+): 511.1818, found 511.1822 (Figures S25–S27).
Data for 3j: White solid, yield: 93%, m.p. 63–65 °C; IR cm−1: 1719, 1455, 1200, 1146, 1127; 1H NMR (600 MHz, CDCl3): δ 7.47 (d, J = 8.4 Hz, 2H, H-Ph), 7.19 (d, J = 8.4 Hz, 2H, H-Ph), 5.10 (d, J = 12.6 Hz, 1H, PhCH2), 4.93 (d, J = 12.6 Hz, 1H, PhCH2), 2.57 (d, J = 13.2 Hz, 1H), 2.25–2.17 (m, 2H), 2.13–1.96 (m, 3H), 1.87–1.77 (m, 3H), 1.71–1.47 (m, 6H), 1.40–1.37 (m, 2H), 1.22–1.17 (m, 1H), 0.98–0.92 (m, 1H), 0.91 (s, 3H, H-16), 0.90 (s, 3H, H-17), 0.88 (s, 3H, H-19), 0.65 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 175.4 (C-20), 135.3 (C-Ph), 131.7 (C-Ph), 130.1 (C-9), 130.0 (C-Ph), 129.9 (C-8), 122.1 (C-Ph), 72.0 (C-13), 65.5 (PhCH2), 52.5 (C-5), 49.0 (C-10), 41.9 (C-3), 41.5 (C-14), 34.6 (C-1), 34.2 (C-4), 33.9 (C-15), 32.1 (C-12), 32.0 (C-7), 32.0 (C-18), 21.8 (C-11), 20.2 (C-2), 20.2 (C-19), 18.3 (C-6), 16.8 (C-16), 16.8 (C-17); HRMS m/z calcd for C27H37O3BrNa ([M + Na]+) 511.1818, found 511.1822 (Figures S28–S30).
Data for 3k: colorless oil, yield: 95%; IR cm−1: 1714, 1455, 1202, 1126; 1H NMR (600 MHz, CDCl3): δ 7.09 (d, J = 7.8 Hz, 1H, H-Ph), 7.06 (s, 1H, H-Ph), 7.03 (d, J = 7.2 Hz, 1H, H-Ph), 5.09 (d, J = 12.6 Hz, 1H, PhCH2), 4.92 (d, J = 12.0 Hz, 1H, PhCH2), 2.57 (d, J = 13.2 Hz, 1H), 2.30–2.17 (m, 8H), 2.13–1.96 (m, 3H), 1.91–1.78 (m, 3H), 1.69–1.63 (m, 2H), 1.59–1.48 (m, 3H), 1.39–1.38 (m, 3H), 1.22–1.17 (m, 1H), 0.96–0.91 (m, 1H), 0.90 (s, 3H, H-16), 0.89 (s, 3H, H-17), 0.88 (s, 3H, H-19), 0.68 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 175.5 (C-20), 136.7 (C-Ph), 136.4 (C-Ph), 133.7 (C-Ph), 130.2 (C-9), 129.8 (C-Ph), 129.7 (C-8), 129.4 (C-Ph), 125.5 (C-Ph), 71.9 (C-13), 66.2 (PhCH2), 52.4 (C-5), 48.9 (C-10), 42.0(C-3), 41.5 (C-14), 34.5 (C-1), 34.0 (C-4), 33.9 (C-15), 32.1 (C-12), 32.1 (C-7), 32.0 (C-18), 21.8 (C-11), 20.2 (C-2), 20.2 (C-19), 19.8 (CH3Ph), 19.6 (CH3Ph), 18.3 (C-6), 16.8 (C-16), 16.8 (C-17); HRMS m/z calcd for C29H42O3Na([M + Na]+) 461.3026, found 461.3025 (Figures S31–S33).
Data for 3l: colorless oil, yield: 94%; IR cm−1: 1713, 1515, 1249; 1H NMR (600 MHz, CDCl3): δ 7.25 (d, J = 8.0 Hz, 2H, H-Ph), 6.86 (d, J = 8.0 Hz, 2H, H-Ph), 5.06 (d, J = 12.0 Hz, 1H, PhCH2), 4.95 (d, J = 12.0 Hz, 1H, PhCH2), 3.79 (s, 3H, OCH3), 2.55 (d, J = 13.2 Hz, 1H), 2.28–2.16 (m, 2H), 2.12–1.96 (m, 3H), 1.88–1.76 (m, 3H), 1.68–1.63 (m, 2H), 1.58–1.45 (m, 4H), 1.39–1.35 (m, 2H), 1.21–1.16 (m, 1H), 0.95–0.92 (m, 1H), 0.90 (s, 3H, H-16), 0.89 (s, 3H, H-17), 0.87 (s, 3H, H-19), 0.65 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 175.5 (C-20), 159.5 (C-Ph), 130.1 (C-9), 129.9 (C-Ph), 129.8 (C-8), 128.3 (C-Ph), 113.9 (C-Ph), 71.9 (C-13), 66.0 (PhCH2), 55.3 (OCH3), 52.4 (C-5), 48.9 (C-10), 41.9 (C-3), 41.4 (C-14), 34.5 (C-1), 34.1 (C-4), 33.9 (C-15), 32.1 (C-12), 32.0 (C-7), 31.9 (C-18), 21.7 (C-11), 20.2 (C-2), 20.1 (C-19), 18.2 (C-6), 16.8 (C-16), 16.8 (C-17); HRMS m/z calcd for C28H40O4Na ([M + Na]+) 463.2819, found 463.2820 (Figures S34–S36).
Data for 3m: White solid, yield: 91%, m.p. 163–165 °C; IR cm−1: 1715, 1525, 1358, 1199, 736; 1H NMR (600 MHz, CDCl3):δ 8.06 (d, J = 7.8 Hz, 1H, H-Ph), 7.60 (td, J = 7.8, 1.2 Hz, 1H, H-Ph), 7.52 (d, J = 9.0 Hz, 1H, H-Ph), 7.48 (t, J = 7.2 Hz, 1H, H-Ph), 5.72 (d, J = 14.4 Hz, 1H, PhCH2), 5.21 (d, J = 14.4 Hz, 1H, PhCH2), 2.60 (d, J = 13.2 Hz, 1H), 2.23–1.98 (m, 5H), 1.91–1.78 (m, 3H), 1.72–1.64 (m, 2H), 1.60–1.51 (m, 4H), 1.42–1.40 (m, 2H), 1.24–1.18 (m, 1H), 0.99–0.94 (m, 1H), 0.93 (s, 3H, H-16), 0.92 (s, 3H, H-17), 0.88 (s, 3H, H-19), 0.69 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3):δ 175.1 (C-20), 147.8 (C-Ph), 133.9 (C-Ph), 132.5 (C-Ph), 130.6 (C-9) 129.7 (C-Ph), 129.5 (C-8), 128.9 (C-Ph), 125.1 (C-Ph), 71.9 (C-13), 63.0 (PhCH2), 52.3 (C-5), 49.0 (C-10), 42.0 (C-3), 41.5 (C-14), 34.8 (C-1), 34.5 (C-4), 34.0 (C-15), 32.2 (C-12), 32.1 (C-7), 31.9 (C-18), 21.5 (C-11), 20.1 (C-2), 20.1 (C-19), 18.4 (C-6), 16.9 (C-16), 16.8 (C-17); HRMS m/z calcd for C27H37O5NNa([M + Na]+) 478.2564, found 478.2568 (Figures S37–S39).
Data for 3n: White solid, yield: 95%, m.p. 99–101 °C; IR cm−1: 1710, 1518, 1345, 1162, 1122; 1H NMR (600 MHz, CDCl3): δ 8.21 (d, J = 8.4 Hz, 2H, H-Ph), 7.48 (d, J = 8.4 Hz, 2H, H-Ph), 5.29 (d, J = 13.2 Hz, 1H, PhCH2), 5.05 (d, J = 13.5 Hz, 1H, PhCH2), 2.60 (d, J = 13.2 Hz, 1H), 2.26–2.19 (m, 2H), 2.13–1.99 (m, 3H), 1.87–1.77 (m, 3H), 1.75–1.45 (m, 6H), 1.44–1.39 (m, 2H), 1.25–1.19 (m, 1H), 1.01–0.96 (m, 1H), 0.92 (s, 3H, H-16), 0.91 (s, 3H, H-17), 0.89 (s, 3H, H-19), 0.66 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 175.3 (C-20), 147.7 (C-Ph), 143.7 (C-Ph), 130.4 (C-9), 129.7 (C-8), 128.3 (C-Ph), 123.9 (C-Ph), 72.0 (C-13), 64.8 (PhCH2), 52.4 (C-5), 49.0 (C-10), 41.8 (C-3), 41.4 (C-14), 34.6 (C-1), 34.6 (C-4), 33.9 (C-15), 32.0 (C-12), 32.0 (C-7), 32.0 (C-18), 21.7 (C-11), 20.2 (C-2), 20.1 (C-19), 18.3 (C-6), 16.8 (C-16), 16.8 (C-17); HRMS m/z calcd for C27H37O5NNa ([M + Na]+) 478.2564, found 478.2568 (Figures S40–S42).
3.1.2. Synthesis of Compound 4
To a suspension of compound 3a (100 mg, 0.30 mmol) in dry tetrahydrofuran (THF, 10 mL) at 0 °C under N2 was added lithium aluminum hydride (56 mg, 1.5 mmol) in dry THF (2 mL) dropwise over 10 min. The resulting mixture was allowed to heat to 40 °C for 1 h. When the reaction was complete, pure water (30 mL) was added to the reaction, The solvent was removed and the residue was diluted by ethyl acetate (30 mL), washed with saturated brine (30 mL), dried over anhydrous Na2SO4, concentrated in vacuo, and purified by silica gel column chromatography to afford 4 in 83%.
Data for 4: White solid, yield: 83%, m.p. 168–170 °C; IR cm−1: 3294, 2943, 1362, 1046, 935; 1H NMR (600 MHz, CDCl3): δ 3.97 (d, J = 11.4 Hz, 1H, H-20), 3.53 (d, J = 11.4 Hz, 1H, H-20), 2.15–2.01 (m, 6H), 1.97–1.85 (m, 3H), 1.81–1.78 (m, 1H), 1.67–1.60 (m, 3H), 1.52–1.43 (m, 4H), 1.31 (dd, J = 13.2, 3.0 Hz, 1H), 1.20–1.11 (m, 2H), 0.94–0.92 (m, 9H, H-16, 17, 19), 0.90 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 132.4 (C-9), 129.1 (C-8), 72.5 (C-13), 66.0 (C-20), 51.0 (C-5), 42.8 (C-10), 41.9 (C-3), 40.3 (C-14), 36.8 (C-1), 34.9 (C-4), 33.6 (C-15), 32.5 (C-12), 32.5 (C-7), 31.4 (C-18), 21.9 (C-11), 21.9 (C-2), 19.2 (C-19), 18.8 (C-6), 17.0 (C-16), 16.9 (C-17); HRMS m/z calcd for C20H34O2Na p([M + Na]+) 329.2451, found 329.2451 (Figures S43–S45).
3.1.3. General Procedure for the Synthesis of Compound 5a–i
To a solution of 4 (100 mg, 0.33 mmol) and Sodium hydride (26 mg, 1.0 mmol) in DMF (10 mL) was added substituent haloalkane (0.66 mmol) in DMF (2 mL) was added dropwise for 10 min. The resulting mixture was stirred at room temperature for 1.5–2 h until the starting materials were completely transformed. After termination by pure water (30 mL), the reaction was extracted with ethyl acetate (3 × 30 mL). The combined organic phase was dried over anhydrous Na2SO4, filtered, concentrated under reduced pressure, and purified by silica gel column chromatography eluting with petroleum ether/ethyl acetate to afford compound 5a-i in 42–74%.
Data for 5a: colorless oil, yield: 43%; IR cm−1: 2925, 2358, 988, 957; 1H NMR (600 MHz, CDCl3): δ 3.51–3.48 (m, 2H, H-20), 3.25 (s, 3H, OCH3), 2.24–2.05 (m, 6H), 1.94–1.90 (m, 1H), 1.74–1.58 (m, 5H), 1.52–1.42 (m, 4H), 1.30–1.27 (m, 1H), 1.20–1.15 (m, 1H), 1.11–1.08 (m, 1H), 0.95–0.92 (m, 6H, H-16, 17), 0.89 (s, 3H, H-19), 0.86 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 134.9 (C-9), 126.2 (C-8), 76.8 (C-20), 72.3 (C-13), 59.3 (OCH3), 51.1 (C-5), 41.9 (C-10), 41.6 (C-3), 40.3 (C-14), 36.1 (C-1), 34.2 (C-4), 33.5 (C-15), 33.4 (C-12), 32.6 (C-7), 31.1 (C-18), 22.9 (C-11), 22.1 (C-2), 19.2 (C-19), 18.6 (C-6), 17.2 (C-16), 17.2 (C-17); HRMS m/z calcd for C21H36O2Na ([M + Na]+): 343.2608, found 343.2607 (Figures S46–S48).
Data for 5b: colorless oil, yield: 73%; IR cm−1: 2925, 2358, 988, 957; 1H NMR (600 MHz, CDCl3): δ 3.53 (s, 2H, H-20), 3.37 (q, J = 7.2 Hz, 2H, CH2CH3), 2.30–2.06 (m, 6H), 1.94–1.90 (m, 1H), 1.74–1.65 (m, 4H), 1.60–1.56 (m, 1H), 1.53–1.41 (m, 4H), 1.30–1.28 (m, 1H), 1.20–1.15 (m, 1H), 1.14 (t, J = 7.2 Hz, 3H, CH2CH3), 1.10–1.06 (m, 1H), 0.95–0.92 (m, 6H, H-16, 17), 0.89 (s, 3H, H-19), 0.85 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 134.9 (C-9), 126.1 (C-8), 74.4 (C-20), 72.3 (C-13), 66.7 (CH2CH3), 51.0 (C-5), 42.0 (C-10), 41.5 (C-3), 40.2 (C-14), 36.2 (C-1), 34.2 (C-4), 33.5 (C-15), 33.4 (C-12), 32.5 (C-7), 31.1 (C-18), 22.9 (C-11), 22.1 (C-2), 19.2 (C-19), 18.6 (C-6), 17.2 (C-16), 17.2 (C-17), 15.4 (CH2CH3); HRMS m/z calcd for C22H38O2Na ([M + Na]+): 357.2764, found 357.2764 (Figures S49–S51).
Data for 5c: colorless oil, yield: 43%; IR cm−1: 2941, 1455, 1386, 1090; 1H NMR (600 MHz, CDCl3): δ 7.33–7.23(m, 5H, H-Ph), 4.47 (d, J = 12.6 Hz, 1H, PhCH2), 4.41 (d, J = 12.6 Hz, 1H, PhCH2), 3.59–3.55 (m, 2H, H-20), 2.34–2.02 (m, 6H), 1.95–1.91 (m, 1H), 1.76–1.58 (m, 5H), 1.51–1.37 (m, 4H), 1.28–1.25 (m, 1H), 1.17–1.12 (m, 1H), 1.07–1.02 (m, 1H), 0.94–0.92 (m, 6H, H-16, 17), 0.86 (s, 3H, H-19), 0.74 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3):δ 138.9 (C-Ph), 135.1 (C-Ph), 128.4 (C-9), 127.6 (C-8), 127.4 (C-Ph), 126.1 (C-Ph), 74.0 (C-20), 73.5 (PhCH2), 72.3 (C-13), 51.2 (C-5), 41.9 (C-10), 41.5 (C-3), 40.4 (C-14), 35.8 (C-1), 34.0 (C-4), 33.5 (C-15), 33.5 (C-12), 32.4 (C-7), 31.4 (C-18), 22.8 (C-11), 22.0 (C-2), 19.1 (C-19), 18.6 (C-6), 17.2 (C-16), 17.1 (C-17); HRMS m/z calcd for C27H40O2Na ([M + Na]+) 419.2921, found 419.2913 (Figures S52–S54).
Data for 5d: colorless oil, yield: 46%; IR cm−1: 2941, 1455, 1386, 1090; 1H NMR (600 MHz, CDCl3): δ 7.26–7.23 (m, 2H, H-Ph), 7.02–6.99 (m, 2H, H-Ph), 4.43 (d, J = 12.0 Hz, 1H, PhCH2), 4.36 (d, J = 12.0 Hz, 1H, PhCH2), 3.57–3.53 (m, 2H, H-20), 2.31–2.04 (m, 6H), 1.94–1.91 (m, 1H), 1.75–1.57 (m, 5H), 1.51–1.38 (m, 4H), 1.28–1.25 (m, 1H), 1.18–1.13 (m, 1H), 1.08–1.03 (m, 1H), 0.94–0.92 (m, 6H, H-16, 17), 0.87 (s, 3H, H-19), 0.75 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 163.1 (C-Ph), 135.0 (C-Ph), 129.4 (C-9), 129.3 (C-8), 126.2 (C-Ph), 115.3 (C-Ph), 74.05 (C-20), 72.8 (PhCH2), 72.3 (C-13), 51.2 (C-5), 41.9 (C-10), 41.5 (C-3), 40.4 (C-14), 35.9 (C-1), 34.0 (C-4), 33.5 (C-15), 33.5 (C-12), 32.4 (C-7), 31.4 (C-18), 22.8 (C-11), 22.0 (C-2), 19.1 (C-19), 18.6 (C-6), 17.2 (C-16), 17.1 (C-17); HRMS m/z calcd for C27H39O2FNa ([M + Na]+) 437.2828, found 437.2823 (Figures S55–S57).
Data for 5e: colorless oil, yield: 53%; IR cm−1: 2921, 1490, 1455, 1089, 1015, 806; 1H NMR (600 MHz, CDCl3): δ 7.29 (d, J = 7.8 Hz, 2H, H-Ph), 7.22 (d, J = 8.4 Hz, 2H, H-Ph), 4.43 (d, J = 12.0 Hz, 1H, PhCH2), 4.36 (d, J = 12.0 Hz, 1H, PhCH2), 3.59–3.54 (m, 2H, H-20), 2.31–2.04 (m, 6H), 1.95–1.91 (m, 1H), 1.76–1.57 (m, 5H), 1.51–1.39 (m, 4H), 1.29–1.26 (m, 1H), 1.18–1.13 (m, 1H), 1.09–1.03 (m, 1H), 0.94–0.92 (m, 6H, H-16, 17), 0.87 (s, 3H, H-19), 0.76 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 137.4 (C-Ph), 135.0 (C-Ph), 133.2 (C-Ph), 128.9 (C-9), 128.6 (C-8), 126.3 (C-Ph), 74.3 (C-20), 72.8 (PhCH2), 72.3 (C-13), 51.2 (C-5), 41.8 (C-10), 41.5 (C-3), 40.4 (C-14), 35.9 (C-1), 34.0 (C-4), 33.5 (C-15), 32.4 (C-12), 31.4 (C-7), 31.4 (C-18), 22.8 (C-11), 22.0 (C-2), 19.1 (C-19), 18.6 (C-6), 17.1 (C-16), 17.1 (C-17); HRMS m/z calcd for C27H39O2ClNa ([M + Na]+) 453.2531, found 453.2533 (Figures S58–S60).
Data for 5f: white solid, yield: 53%, m.p. 63–65 °C; IR cm−1: 2358, 1455, 1089, 1012, 800, 668; 1H NMR (600 MHz, CDCl3): δ 7.44 (d, J = 8.4 Hz, 2H, H-Ph), 7.16 (d, J = 8.4 Hz, 2H, H-Ph), 4.42 (d, J = 12.6 Hz, 1H, PhCH2), 4.35 (d, J = 12.6 Hz, 1H, PhCH2), 3.59–3.54 (m, 2H, H-20), 2.29–2.03 (m, 6H), 1.95–1.91 (m, 1H), 1.76–1.57 (m, 5H), 1.51–1.39 (m, 4H), 1.29–1.25 (m, 1H), 1.18–1.13 (m, 1H), 1.08–1.03 (m, 1H), 0.94–0.91 (m, 6H, H-16, 17), 0.87 (s, 3H, H-19), 0.76 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 137.9 (C-Ph), 135.0 (C-Ph), 131.5 (C-9), 129.2 (C-8), 126.2 (C-Ph), 121.3 (C-Ph), 74.3 (C-20), 72.8 (PhCH2), 72.2 (C-13), 51.2 (C-5), 41.8 (C-10), 41.5 (C-3), 40.4 (C-14), 35.9 (C-1), 33.9 (C-4), 33.5 (C-15), 33.4 (C-12), 32.4 (C-7), 31.4 (C-18), 22.8 (C-11), 22.0 (C-2), 19.1 (C-19), 18.6 (C-6), 17.1 (C-16), 17.1 (C-17); HRMS m/z calcd for C27H39O2BrNa ([M + Na]+) 497.2026, found 497.2028 (Figures S61–S63).
Data for 5g: colorless oil, yield: 74%; IR cm−1: 2928, 1466, 1091; 1H NMR (600 MHz, CDCl3): δ 7.16 (d, J = 7.8 Hz, 2H, H-Ph), 7.12 (d, J = 7.8 Hz, 2H, H-Ph), 4.43 (d, J = 12.0 Hz, 1H, PhCH2), 4.36 (d, J = 12.0 Hz, 1H, PhCH2), 3.57–3.54 (m, 2H, H-20), 2.32 (s, 3H, CH3), 2.29–2.03 (m, 6H), 1.94–1.91 (m, 1H), 1.75–1.57 (m, 5H), 1.50–1.37 (m, 4H), 1.27–1.25 (m, 1H), 1.17–1.12 (m, 1H), 1.08–1.01 (m, 1H), 0.94–0.91 (m, 6H, H-16, 17), 0.86 (s, 3H, H-19), 0.76 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 137.0 (C-Ph), 135.1 (C-Ph), 129.1 (C-Ph), 129.0 (C-9), 128.0 (C-Ph), 127.6 (C-8), 73.9 (C-20), 73.3 (PhCH2), 72.2 (C-13), 51.2 (C-5), 41.9 (C-10), 41.5 (C-3), 40.4 (C-14), 35.8 (C-1), 34.0 (C-4), 33.5 (C-15), 33.4 (C-12), 32.4 (C-7), 31.4 (C-18), 22.7 (C-11), 22.0 (C-2), 21.2 (CH3), 19.1 (C-19), 18.6 (C-6), 17.1 (C-16), 17.1 (C-17); HRMS m/z calcd for C28H42O2Na ([M + Na]+) 433.3077, found 433.3076 (Figures S64–S66).
Data for 5h: colorless oil, yield: 43%; IR cm−1: 2941, 1455, 1386, 1090; 1H NMR (600 MHz, CDCl3): δ 7.07 (d, J = 7.2 Hz, 1H, H-Ph), 7.04 (s, 1H, H-Ph), 7.00 (d, J = 7.2 Hz, 1H, H-Ph), 4.41 (d, J = 12.1 Hz, 1H, PhCH2), 4.34 (d, J = 12.0 Hz, 1H, PhCH2), 3.57–3.54 (m, 2H, H-20), 2.34–2.27 (m, 1H), 2.24 (s, 3H, CH3), 2.23 (s, 3H, CH3), 2.18–2.04 (m, 5H), 1.95–1.91 (m, 1H), 1.76–1.58 (m, 5H), 1.51–1.38 (m, 4H), 1.28–1.25 (m, 1H), 1.17–1.12 (m, 1H), 1.07–1.02 (m, 1H), 0.95–0.92 (m, 6H, H-16, 17), 0.86 (s, 3H, H-19), 0.76 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 136.3 (C-Ph), 135.6 (C-Ph), 135.1 (C-Ph), 129.6 (C-9), 129.0 (C-Ph), 126.1 (C-Ph), 125.1 (C-8), 73.9 (C-20), 73.3 (PhCH2), 72.2 (C-13), 51.2 (C-5), 41.9 (C-10), 41.5 (C-3), 40.5 (C-14), 35.8 (C-1), 34.1 (C-4), 33.5 (C-15), 33.5 (C-12), 32.5 (C-7), 31.4 (C-18), 22.8 (C-11), 22.0 (C-2), 19.8 (C-19), 19.6 (CH3), 19.1 (CH3), 18.6 (C-6), 17.2 (C-16), 17.1 (C-17); HRMS m/z calcd for C29H44O2Na ([M + Na]+) 447.3234, found 447.3233 (Figures S67–S69).
3.1.4. Synthesis of Compound 6a–c
To a solution of 3a or 3d (0.3 mmol) and pyridine (1.2 mmol) in DMF (10 mL) at 0 °C was added acetyl chloride (0.6 mmol). The mixture was warmed up to room temperature and stirred for 2–2.5 h. When the reaction was complete, pure water (30 mL) was added to the reaction, which was extracted with ethyl acetate (3 × 30 mL). The combined organic phase was dried over anhydrous Na2SO4, filtered, concentrated under reduced pressure, and purified by silica gel column chromatography eluting with petroleum ether/ethyl acetate to afford compounds 6a (75%), 6b (65%), or 6c (15%).
Data for 6a: white solid, yield: 75%, m.p. 118–120 °C; IR cm−1: 1726, 1361, 1272, 1226, 1203, 1130; 1H NMR (600 MHz, CDCl3): δ 3.59 (s, 3H, OCH3), 2.57–2.52 (m, 2H), 2.39 (d, J = 17.4 Hz, 1H), 2.34–2.30 (m, 1H), 2.28–2.24 (m, 1H), 2.21–1.97 (m, 4H), 1.90 (s, 3H, COCH3), 1.84–1.76 (m, 1H), 1.74–1.68 (m, 2H), 1.62–1.57 (m, 1H), 1.55–1.51 (m, 1H), 1.40–1.35 (m, 2H), 1.22–1.17 (m, 1H), 0.95–0.90 (m, 1H), 0.89–0.88 (m, 9H, H-16, 17, 19), 0.68 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 176.1 (C-20), 170.7 (COCH3), 129.6 (C-9), 129.3 (C-8), 85.9 (C-13), 52.4 (OCH3), 51.3 (C-5), 48.6 (C-10), 42.0 (C-3), 36.2 (C-14), 34.8 (C-1), 33.9 (C-4), 32.1 (C-15), 32.1 (C-12), 32.1 (C-7), 31.8 (C-18), 22.3 (COCH3), 21.7 (C-11), 20.3 (C-2), 20.1 (C-19), 18.3 (C-6), 17.5 (C-16), 17.2 (C-17); HRMS m/z calcd for C23H36O4Na ([M + Na]+) 399.2506, found 399.2507 (Figures S70–S72).
Data for 6b: colorless oil, yield: 65%; IR cm−1: 2957, 1748, 1687, 1458, 1127, 906; 1H NMR (600 MHz, CDCl3): δ 7.36–7.28 (5H, H-Ph), 5.13 (d, J = 12.6 Hz, 1H, PhCH2), 4.98 (d, J = 12.6 Hz, 1H, PhCH2), 2.61 (d, J = 16.8 Hz, 1H), 2.54–2.50 (m, 1H), 2.43 (d, J = 12.6 Hz, 1H), 2.35–2.27 (m, 2H), 2.23 (d, J = 16.8 Hz, 1H), 2.17–2.13 (m, 1H), 2.10–1.98 (m, 2H), 1.85–1.63 (m, 6H), 1.64–1.59 (m, 1H), 1.54–1.52 (m, 1H), 1.40–1.37 (m, 2H), 1.22–1.17 (m, 1H), 0.97–0.92 (m, 1H), 0.89–0.87 (m, 9H, H-16, 17, 19), 0.66 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3): δ 175.3 (C-20), 170.7 (COCH3), 136.3 (C-Ph), 129.8 (C-9), 129.6 (C-8), 128.6 × 2 (C-Ph), 128.0 (C-Ph), 127.8 (C-Ph) × 2, 85.9 (C-13), 66.2 (PhCH2), 52.3 (C-5), 48.8 (C-10), 42.0 (C-3), 36.3 (C-14), 34.7 (C-1), 34.0 (C-4), 32.0 (C-15), 31.9 (C-12), 31.6 (C-7), 31.6 (C-18), 28.2 (COCH3), 22.3 (C-11), 21.7 (C-2), 20.2 (C-19), 18.3 (C-6), 17.5 (C-16), 17.2 (C-17); HRMS m/z calcd for C29H40O4Na ([M + Na]+): 475.2810, found 475.2811 (Figures S73–S75).
Data for 6c: colorless oil, yield: 15%; IR cm−1: 2359, 1716, 1704, 1206; 1H NMR (600 MHz, CDCl3): δ 3.62 (s, 3H, OCH3), 3.33–3.27 (m, 2H, COCH2COCH3), 2.57 (d, J = 13.8 Hz, 1H), 2.52–2.48 (m, 1H), 2.43 (d, J = 16.8 Hz, 1H), 2.36–2.15 (m, 7H), 2.09–1.97 (m, 2H), 1.83–1.63 (m, 4H), 1.54–1.50 (m, 1H), 1.40–1.33 (m, 2H), 1.22–1.17 (m, 1H), 0.95–0.92 (m, 1H), 0.91–0.89 (m, 9H, H-16, 17, 19), 0.68 (s, 3H, H-18); 13C NMR (150 MHz, CDCl3):δ 201.2 (COCH2COCH3), 175.9 (C-20), 166.5 (COCH2COCH3), 129.9 (C-9), 129.2 (C-8), 88.0 (C-13), 52.5 (C-5), 51.4 (OCH3), 51.3 (COCH2COCH3), 48.6 (C-10), 42.0 (C-3), 36.2 (C-14), 34.8 (C-1), 33.8 (C-4), 32.1 (C-15), 32.1 (C-12), 31.8 (C-7), 30.2 (C-18), 28.2 (C-25), 21.7 (C-11), 20.3 (C-2), 20.1 (C-19), 18.2 (C-6), 17.5 (C-16), 17.2 (C-17);HRMS m/z calcd for C25H38O5Na ([M + Na]+): 441.2611, found 441.2612 (Figures S76–S78).
3.2. Pathogens and Culture Conditions
The microbial pathogen strains used for bioassays were methicillin-resistant Staphylococcus aureus (ATCC 43300), Escherichia coli (ATCC 25922), Streptococcus mutans (ATCC 35668), Streptococcus sobrinus (ATCC 33478), Candida albicans (ATCC 10231), Cryptococcus neoformans (ATCC 66031), T. rubrum (ATCC-MYA4438) and T. mentagrophyte (ATCC9533). The bacteria strains S. mutans and S. sobrinus (ATCC 33478) were obtained from the Prince Philip Dental Hospital, Hong Kong University, Hong Kong.
The strains of S. aureus and E. coli were grown at 37 °C on Tryptic Soy media (TSA, TSB; BD Biosciences, CA, USA). Yeast malt (YM; BD Biosciences, CA, USA) media were used for cultivating C. albicans at 30 °C. T. rubrum, T. mentagrophyte, and C. neoformans were grown at 30 °C on Sabouraud dextrose media (SDA; BD Biosciences). S. mutans and S. sobrinus were started from the frozen BHI-glycerol stocks and growth at 37 °C with shaking. Strains grown in liquid media were cultivated on an orbital shaker at 200 rpm.
3.3. In Vitro Antifungal Assay
To evaluate the antifungal activity against filamentous fungi, T. rubrum and T. mentagrophytes were separately cultured for 2 weeks at 28 °C on SDA to produce conidia. A mixed suspension of conidia and hyphae fragments was obtained by covering the fungal colonies with sterile saline (0.85%) and gently rubbing the colonies with the inoculation loop. Then, the suspension was filtered with four layers of sterile lens paper to remove the hyphae and centrifuged at 1000× g for 10 min to collect the conidia. Conidia were washed twice by agitation in sterile saline. The concentration of conidia or spore was adjusted with sterile saline to 1 × 104 cells/mL by hemocytometer counts. The antifungal susceptibility testing was performed as outlined in document M38-A2 and previous research, with minor changes [16,17,18,19]. The medium used was RPMI 1640 with L-glutamine buffered to pH 7.0 with 0.165 M morpholinepropanesulfonic acid (MOPS) and was supplemented with 2% glucose (m/v). The 195 μL of prepared conidia or spore suspension was seeded on 96-well plates that had been previously added with 5 μL of tested agents in each well, and three replicates were used for each treatment. Miconazole was used as a positive control in the assay. The 96-well plates were then incubated at 28 ± 2 °C for 7 days. The optical density (OD) reading was measured by a microplate reader at 510 nm. The fungal growth inhibition is determined using the formula:
Antifungal susceptibility testing of yeasts was performed by using a micro-broth dilution assay. The compounds were dissolved in DMSO at a stock concentration of 2–10 mg/mL and kept at 4 °C for the bioassays. Exponentially growing cultures of each strain were prepared from overnight cultures, and cultures were adjusted to the OD value of about 0.5 at 600 nm. Cultures were then diluted 1:1000 in broth (C. neoformans was directly used at OD600 = 0.05) and added to a 96-well plate (195 µL/well). Miconazole (Dalian Meilun, Dalian, China; 10 µg mL−1) was used as a positive control for C. albicans, and C. neoformans. Plates were read at 600 nm after incubation for 48 h. Inhibition was calculated by subtracting the absorbance of the blank wells, dividing by the average value for the DMSO-only wells, and multiplying by 100.
3.4. In Vitro Antibacterial Assay
Compounds were tested for planktonic microbial growth inhibition using the above micro-broth dilution assay [16]. The compounds and the standard drug were prepared in DMSO. Exponentially growing cultures of S. aureus, E. coli, S. mutans, and S. sobrinus were prepared from overnight cultures, and cultures were adjusted to the OD value of about 0.5 at 600 nm. Cultures were then diluted 1:1000 in broth and added to a 96-well plate (195 µL/well). Tetracycline (Sigma, St. Louis, MO, USA; 10 µg mL−1 in DMSO) was used as a standard drug. Plates were read at 600 nm after incubation for 24 h. Inhibition was calculated by the above calculation formula.
4. Conclusions
A series of novel lophanic acid derivatives have been prepared and evaluated for their antifungal and antibacterial activities. Among the derivatives, 3d is the only compound that showed > 70% inhibitory effects against three fungal and bacterial strains (T. mentagrophytes, C. neoformans, and MRSA), and 3b, 5c, and 5h were found to be able to inhibit the microbial growth of MRSA by over 90%. Through a structure-activity relationship analysis, we observed the presence of a C-20 carboxylic group and a free hydroxyl group at C-13 is essential to retain broad antimicrobial activities for the lophanic acid derivatives (e.g., 3a, 3b, 3d, and 3i). Without the C-20 carboxylic group, the inhibitory effects of the lophanic acid derivatives against T. rubrum and C. neoformans were much weakened (e.g., 5c and 5h). Our present study determined that the C-20 carboxylic group could be the key position for a structural modification to obtain lophanic acid analogs with broad-spectrum antimicrobial activity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27206836/s1, Figures S1–S78: 1H NMR and 13C NMR spectrum of the compounds 3a–6c.
Author Contributions
Data curation, X.Y., X.S., Y.Z. and Y.Y.; formal analysis, J.Y.; funding acquisition: X.S., L.P. and H.Z.; methodology, X.Y. and Y.L.; project administration, X.S.; supervision, L.P.; validation, X.Y.; writing—original draft, X.Y. and X.S.; writing—review and editing, X.S. and H.Z. All authors have read and agreed to the published version of the manuscript.
Funding
The work described in this study was financially supported by the Research Grant Council of the Hong Kong Special Administrative Region, China (Project No. HKBU12102219), the Tip-top Talent Foundation [grant number KY (2021)034]; National Key R&D Program of China (2019YFC1712501); Guizhou Provincial Natural Science Foundation (QKH-J(2020)1Y070); the University stability support program of Shenzhen (20200813201847001); Guangdong Basic and Applied Basic Research Foundation (2020A1515111169); the Technology Fund of Guizhou Administration of Traditional Chinese Medicine [grant number QZYY-2022-019].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are available from the authors.
References
- Ilkit, M.; Durdu, M. Tinea pedis: The etiology and global epidemiology of a common fungal infection. Crit. Rev. Microbiol. 2015, 41, 374–388. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Chow, M.; Daniel, C.R.; Aly, R. Treatments of tinea pedis. Dermatol. Clin. 2003, 21, 431–462. [Google Scholar] [CrossRef]
- Sahoo, A.K.; Mahajan, R. Management of tinea corporis, tinea cruris, and tinea pedis: A comprehensive review. Indian Dermatol. Online J. 2016, 7, 77–86. [Google Scholar] [PubMed]
- Saarikoski, T.; Saari, T.I.; Hagelberg, N.M.; Backman, J.T.; Neuvonen, P.J.; Scheinin, M.; Olkkola, K.T.; Laine, K. Effects of terbinafine and itraconazole on the pharmacokinetics of orally administered tramadol. Eur. J. Clin. Pharmacol. 2015, 71, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Masih, A.; Khurana, A.; Singh, P.K.; Gupta, M.; Hagen, F.; Meis, J.F.; Chowdhary, A. High terbinafine resistance in Trichophyton interdigitale isolates in Delhi, India harbouring mutations in the squalene epoxidase gene. Mycoses 2018, 61, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta 2013, 1830, 3670–3695. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.L.; Cui, G.H.; Guo, J.; Tang, J.F.; Duan, L.X.; Lin, H.X.; Shen, Y.; Chen, T.; Zhang, H.B.; Huang, L.Q. Functional Diversification of Kaurene Synthase-Like Genes in Isodon rubescens. Plant Physiol. 2017, 174, 943–955. [Google Scholar] [CrossRef] [PubMed]
- Pelot, K.A.; Hagelthorn, L.M.; Addison, J.B.; Zerbe, P. Biosynthesis of the oxygenated diterpene nezukol in the medicinal plant Isodon rubescens is catalyzed by a pair of diterpene synthases. PLoS ONE 2017, 12, e0176507. [Google Scholar] [CrossRef]
- Sun, H.D.; Huang, S.X.; Han, Q.B. Diterpenoids from Isodon species and their biological activities. Nat. Prod. Rep. 2006, 23, 673–698. [Google Scholar] [CrossRef]
- Liu, M.; Wang, W.F.; Sun, H.D.; Pu, J.X. Diterpenoids from Isodon species: An update. Nat. Prod. Rep. 2017, 34, 1090–1140. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Pan, L.; Li, Q.; Pu, J.; Yao, P.; Zhu, M.; Banas, J.A.; Zhang, H.; Sun, H. Rubesanolides C-E: Abietane diterpenoids isolated from Isodon rubescens and evaluation of their anti-biofilm activity. Org. Biomol. Chem. 2012, 10, 5039–5044. [Google Scholar] [CrossRef] [PubMed]
- Li, J.X.; Li, Q.J.; Guan, Y.F.; Song, X.; Liu, Y.H.; Zhang, J.J.; Li, W.F.; Du, J.; Zhu, M.; Banas, J.A.; et al. Discovery of antifungal constituents from the Miao medicinal plant Isodon flavidus. J. Ethnopharmacol. 2016, 191, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Lu, Z.F.; Zhang, H.J.; Sun, H.D. Diterpenoids from Isodon lophanthoides. Fitoterapia 2000, 71, 360–364. [Google Scholar] [CrossRef]
- Li, Q.J.; Zhao, C.L.; Ku, C.F.; Zhu, Y.; Zhu, X.J.; Zhang, J.J.; Deyrup, S.T.; Pan, L.T.; Zhang, H.J. Two new bioactive diterpenes identified from Isodon interruptus. Bioorganic Chem. 2020, 95, 103512. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Gaascht, F.; Schmidt-Dannert, C.; Salomon, C.E. Discovery of Antifungal and Biofilm Preventative Compounds from Mycelial Cultures of a Unique North American Hericium sp. Fungus. Molecules 2020, 25, 963. [Google Scholar] [CrossRef]
- Wong-Deyrup, S.W.; Song, X.; Ng, T.W.; Liu, X.B.; Zeng, J.G.; Qing, Z.X.; Deyrup, S.T.; He, Z.D.; Zhang, H.J. Plant-derived isoquinoline alkaloids that target ergosterol biosynthesis discovered by using a novel antifungal screening tool. Biomed. Pharmacother. 2021, 137, 111348. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Hu, H.; Hu, Z.; Zhong, X.; Guan, Y.; Zhao, Y.; Wang, L.; Ye, L.; Ming, L.; Riaz Rajoka, M.S.; et al. Sanguinarine, Isolated from Macleaya cordata, Exhibits Potent Antifungal Efficacy against Candida albicans through Inhibiting Ergosterol Synthesis. Front. Microbiol. 2022, 13, 908461. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wei, Y.X.; Lai, K.M.; He, Z.D.; Zhang, H.J. In vivo antifungal activity of dipyrithione against Trichophyton rubrum on guinea pig dermatophytosis models. Biomed. Pharmacother. 2018, 108, 558–564. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).