Preparation and Characterization of an Ancient Aminopeptidase Obtained from Ancestral Sequence Reconstruction for L-Carnosine Synthesis
Abstract
1. Introduction
2. Results and Discussion
2.1. Ancestral Sequence Reconstruction of Aminopeptidase
2.2. Cloning, Expression and Purification of LUCA–DmpA
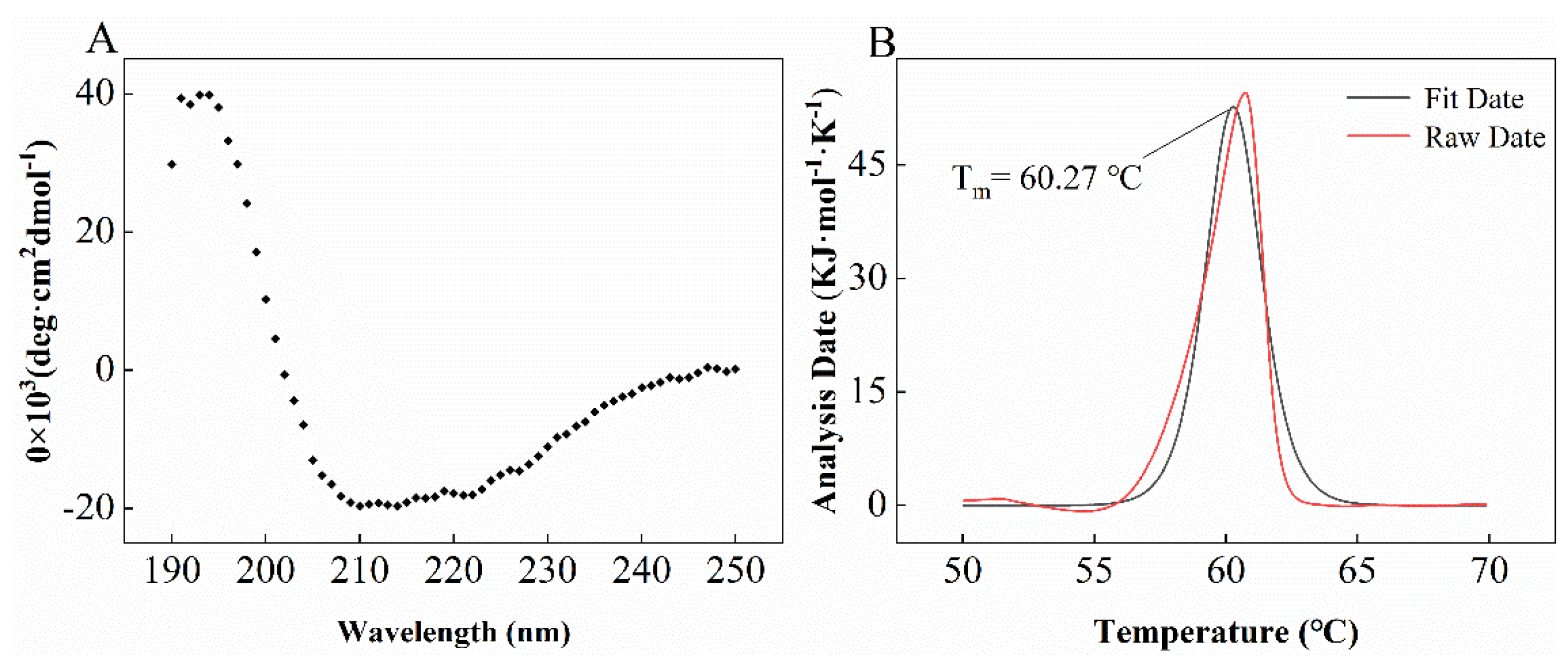
2.3. Characterization of LUCA–DmpA
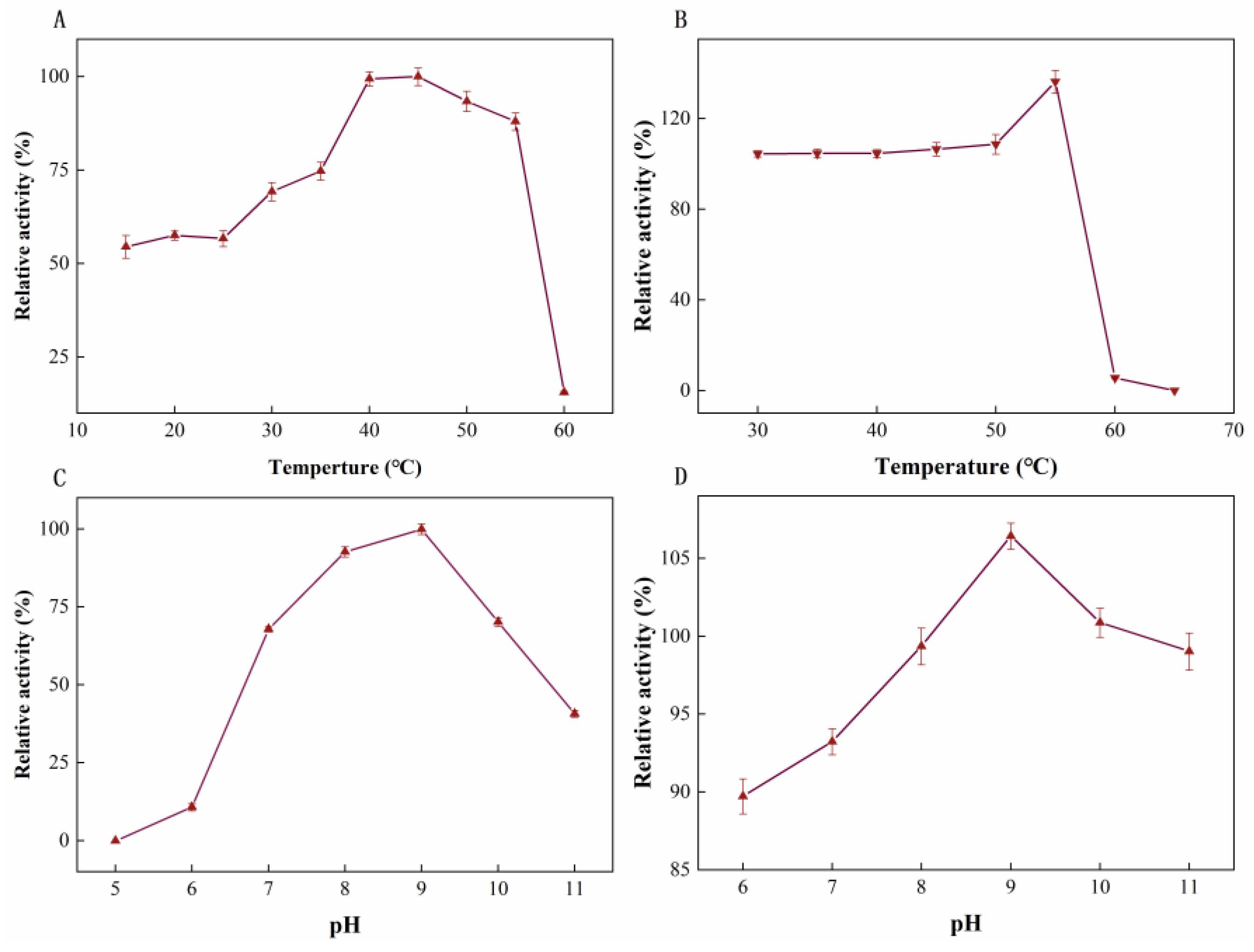
2.4. Effects of pH and Temperature on Stability and Activity of LUCA–DmpA
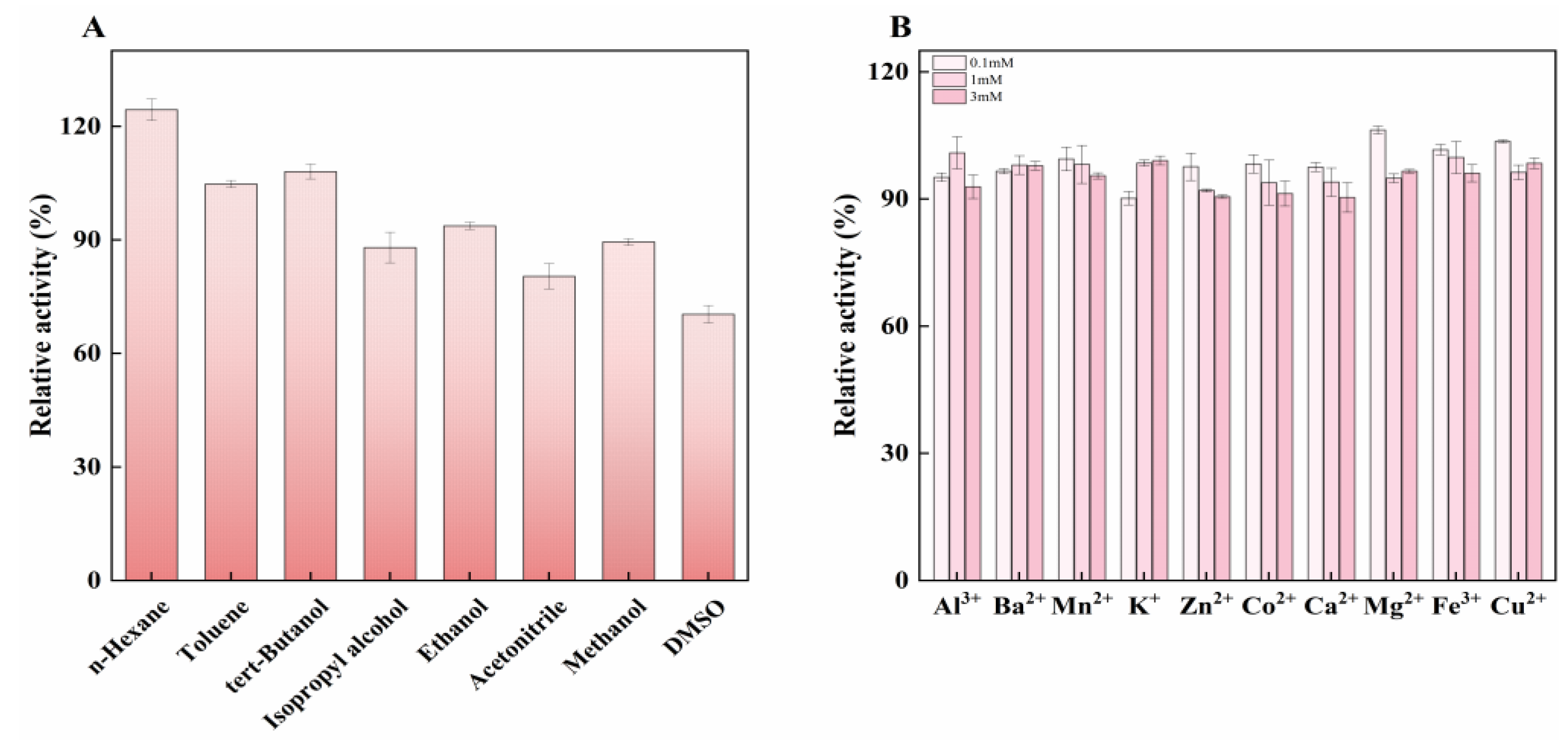
2.5. Effect of Organic Solvents and Metal Ions on Enzyme Activity
2.6. Determination of Kinetic Parameters of LUCA–DmpA
3. Conclusions
4. Materials and Methods
Ancestral Sequence Reconstruction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boldyrev, A.A.; Aldini, G.; Derave, W. Physiology and pathophysiology of carnosine. Physiol. Rev. 2013, 93, 1803–1845. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G.; Fresta, C.G.; Musso, N.; Giambirtone, M.; Grasso, M.; Spampinato, S.F.; Merlo, S.; Drago, F.; Lazzarino, G.; Sortino, M.A. Carnosine prevents Aβ-induced oxidative stress and inflammation in microglial cells: A key role of TGF-β1. Cells 2019, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Hipkiss, A.R.; Baye, E.; de Courten, B. Carnosine and the processes of ageing. Maturitas 2016, 93, 28–33. [Google Scholar] [CrossRef]
- Turner, M.D.; Sale, C.; Garner, A.C.; Hipkiss, A.R. Anti-cancer actions of carnosine and the restoration of normal cellular homeostasis. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2021, 1868, 119117. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, J.-R.; Lee, H.; Jang, S.; Ryu, S.-M.; Kim, H.; Kim, D.; Jang, A.; Yang, S.-R. L-carnosine induces apoptosis/cell cycle arrest via suppression of NF-κB/STAT1 pathway in HCT116 colorectal cancer cells. In Vitro Cell. Dev. Biol.-Anim. 2018, 54, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Matthews, J.J.; Artioli, G.G.; Turner, M.D.; Sale, C. The physiological roles of carnosine and β-alanine in exercising human skeletal muscle. Med. Sci. Sport. Exerc. 2019, 51, 2098–2108. [Google Scholar] [CrossRef]
- Xing, L.; Chee, M.E.; Zhang, H.; Zhang, W.; Mine, Y. Carnosine—A natural bioactive dipeptide: Bioaccessibility, bioavailability and health benefits. J. Food Bioact. 2019, 5, 8–17. [Google Scholar] [CrossRef]
- Chevalot, I.; Arab-Tehrany, E.; Husson, E.; Gerardin, C. Application of Carnosine and Its Functionalised Derivatives. In Industrial Biotechnology of Vitamins, Biopigments, and Antioxidants; Wiley: Weinheim, Germany, 2016. [Google Scholar]
- Tanaka, K.-I.; Kawahara, M. Carnosine and lung disease. Curr. Med. Chem. 2020, 27, 1714–1725. [Google Scholar] [CrossRef]
- Robin, A.; Ghosh, S.; Gabay, B.; Levkov, K.; Golberg, A. Identifying critical parameters for extraction of carnosine and anserine from chicken meat with high voltage pulsed electric fields and water. Innov. Food Sci. Emerg. Technol. 2022, 76, 102937. [Google Scholar] [CrossRef]
- Yin, D.-Y.; Pan, J.; Zhu, J.; Liu, Y.-Y.; Xu, J.-H. A green-by-design bioprocess for L-carnosine production integrating enzymatic synthesis with membrane separation. Catal. Sci. Technol. 2019, 9, 5971–5978. [Google Scholar] [CrossRef]
- Kim, M.; Ko, Y.J.; Jeong, D.W.; Jeong, W.-Y.; Han, S.O. Ecofriendly Synthesis of L-carnosine in Metabolically Engineered Corynebacterium glutamicum by Reinforcing Precursor Accumulation. ACS Synth. Biol. 2021, 10, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Heyland, J.; Antweiler, N.; Lutz, J.; Heck, T.; Geueke, B.; Kohler, H.P.E.; Blank, L.M.; Schmid, A. Simple enzymatic procedure for L-carnosine synthesis: Whole-cell biocatalysis and efficient biocatalyst recycling. Microb. Biotechnol. 2010, 3, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, T.; Robert-Baudouy, J. Bacterial aminopeptidases: Properties and functions. FEMS Microbiol. Rev. 1996, 18, 319–344. [Google Scholar] [CrossRef] [PubMed]
- Nandan, A.; Nampoothiri, K.M. Molecular advances in microbial aminopeptidases. Bioresour. Technol. 2017, 245, 1757–1765. [Google Scholar] [CrossRef] [PubMed]
- Bompard-Gilles, C.; Villeret, V.; Davies, G.J.; Fanuel, L.; Joris, B.; Frère, J.-M.; Van Beeumen, J. A new variant of the Ntn hydrolase fold revealed by the crystal structure of L-aminopeptidase D-ala-esterase/amidase from Ochrobactrum anthropi. Structure 2000, 8, 153–162. [Google Scholar] [CrossRef][Green Version]
- Arima, J.; Morimoto, M.; Usuki, H.; Mori, N.; Hatanaka, T. β-Alanyl peptide synthesis by Streptomyces S9 aminopeptidase. J. Biotechnol. 2010, 147, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, R.; Takakura, Y.; Suzuki, S.; Yokozeki, K. Method for Producing Beta-Alanyl-Amino Acid or Derivative Thereof. U.S. Patent Application No. 12/943,509, 7 April 2011. [Google Scholar]
- Heck, T.; Kohler, H.P.E.; Limbach, M.; Flögel, O.; Seebach, D.; Geueke, B. Enzyme-catalyzed formation of β-peptides: β-peptidyl aminopeptidases BapA and DmpA acting as β-peptide-synthesizing enzymes. Chem. Biodivers. 2007, 4, 2016–2030. [Google Scholar] [CrossRef]
- Heck, T.; Makam, V.S.; Lutz, J.; Blank, L.M.; Schmid, A.; Seebach, D.; Kohler, H.P.E.; Geueke, B. Kinetic Analysis of L-carnosine Formation by β-Aminopeptidases. Adv. Synth. Catal. 2010, 352, 407–415. [Google Scholar] [CrossRef]
- Holinski, A.; Heyn, K.; Merkl, R.; Sterner, R. Combining ancestral sequence reconstruction with protein design to identify an interface hotspot in a key metabolic enzyme complex. Proteins Struct. Funct. Bioinform. 2017, 85, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Xin, Y.; Guo, Z.; Shi, Y.; Zhang, L.; Li, Y.; Gu, Z.; Ding, Z.; Shi, G. Ancestral sequence reconstruction and spatial structure analysis guided alteration of longer-chain substrate catalysis for Thermomicrobium roseum lipase. Enzym. Microb. Technol. 2022, 156, 109989. [Google Scholar] [CrossRef] [PubMed]
- Babkova, P.; Dunajova, Z.; Chaloupkova, R.; Damborsky, J.; Bednar, D.; Marek, M. Structures of hyperstable ancestral haloalkane dehalogenases show restricted conformational dynamics. Comput. Struct. Biotechnol. J. 2020, 18, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- Spence, M.A.; Kaczmarski, J.A.; Saunders, J.W.; Jackson, C.J. Ancestral sequence reconstruction for protein engineers. Curr. Opin. Struct. Biol. 2021, 69, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Merkl, R.; Sterner, R. Ancestral protein reconstruction: Techniques and applications. Biol. Chem. 2016, 397, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Zhou, Y.; Yi, Z.; Zhou, R.; Jin, W.; Zhang, G. Highly thermostable and promiscuous β-1,3-xylanasen designed by optimized ancestral sequence reconstruction. Bioresour. Technol. 2021, 340, 125732. [Google Scholar] [CrossRef]
- Rozi, M.F.A.M.; Rahman, R.N.Z.R.A.; Leow, A.T.C.; Ali, M.S.M. Ancestral sequence reconstruction of ancient lipase from family I. 3 bacterial lipolytic enzymes. Mol. Phylogenetics Evol. 2022, 168, 107381. [Google Scholar] [CrossRef]
- Merz, T.; Heck, T.; Geueke, B.; Mittl, P.R.; Briand, C.; Seebach, D.; Kohler, H.-P.E.; Grütter, M.G. Autoproteolytic and catalytic mechanisms for the β-aminopeptidase BapA—A member of the Ntn hydrolase family. Structure 2012, 20, 1850–1860. [Google Scholar] [CrossRef]
- Asano, Y.; Nakazawa, A.; Kato, Y.; Kondo, K. Properties of a novel D-stereospecific aminopeptidase from Ochrobactrum anthropi. J. Biol. Chem. 1989, 264, 14233–14239. [Google Scholar] [CrossRef]
- Komeda, H.; Asano, Y. A DmpA-homologous protein from Pseudomonas sp. is a dipeptidase specific for β-alanyl dipeptides. FEBS J. 2005, 272, 3075–3084. [Google Scholar] [CrossRef]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.T.; Musil, M.; Stourac, J.; Damborsky, J.; Bednar, D. Fully Automated Ancestral Sequence Reconstruction using FireProtASR. Curr. Protoc. 2021, 1, e30. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Zheng, M.; Wang, Q.; Lu, L.; Zhang, L.; Tong, Y.; Wang, W. Structural and catalytic alteration of sarcosine oxidase through reconstruction with coenzyme-like ligands. J. Mol. Catal. B Enzym. 2016, 133, S250–S258. [Google Scholar] [CrossRef] [PubMed]


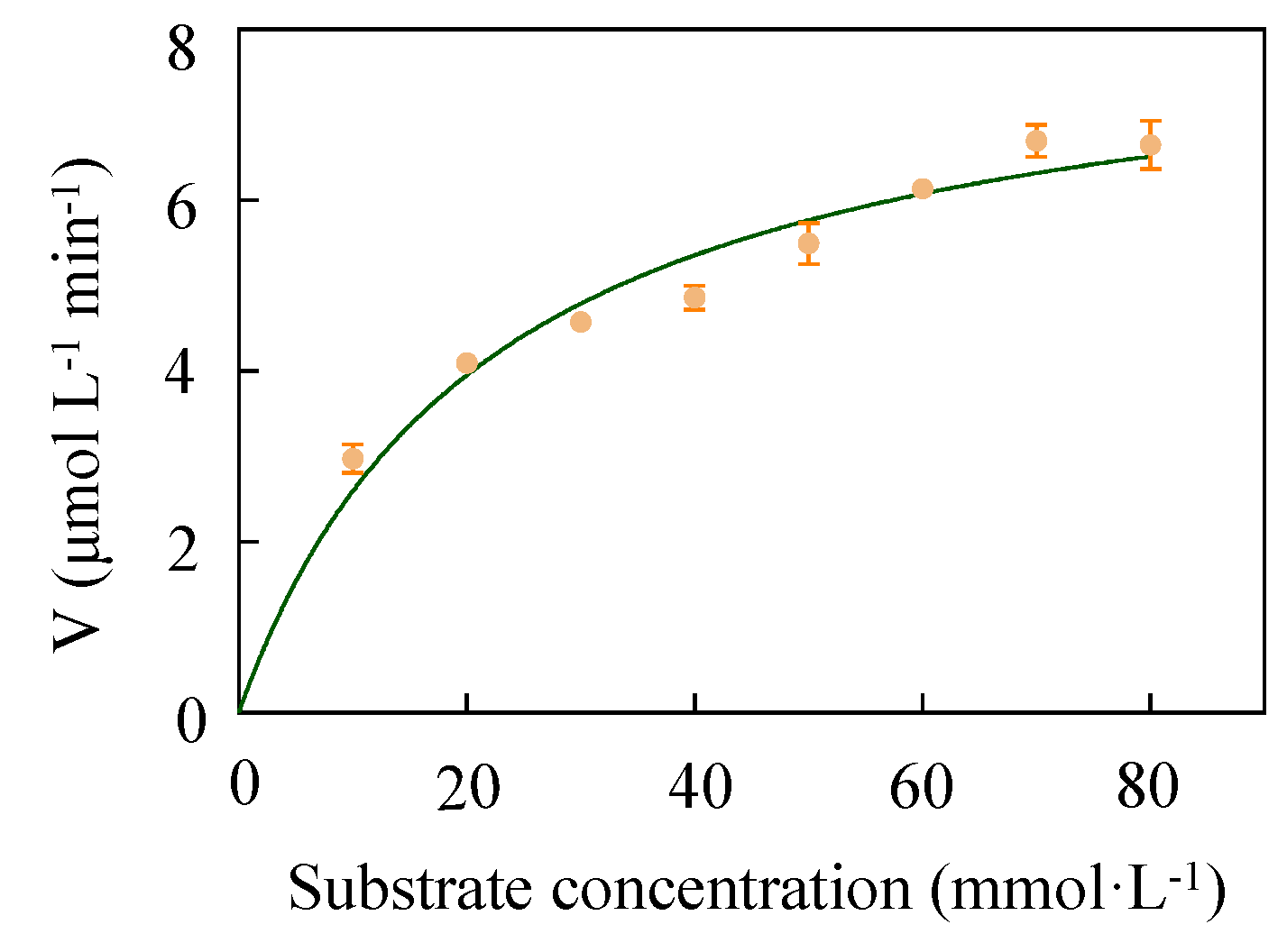
| Kinetic Parameters | LUCA–DmpA | DmpA | BapA |
|---|---|---|---|
| Km (mM) | 22.06 ± 1.02 | 0.48 ± 0.05 | 23.00 ± 8.00 |
| kcat (s−1) | 76.53 ± 2.68 | 12.90 ± 0.59 | 0.87 ± 0.23 |
| kcat/Km (s−1 L mmol−1) | 3.47 | 26.88 | 0.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, F.; Shi, Y.; Fang, Y.; Liu, Z.; Xin, Y.; Gu, Z.; Guo, Z.; Zhang, L. Preparation and Characterization of an Ancient Aminopeptidase Obtained from Ancestral Sequence Reconstruction for L-Carnosine Synthesis. Molecules 2022, 27, 6620. https://doi.org/10.3390/molecules27196620
Liu F, Shi Y, Fang Y, Liu Z, Xin Y, Gu Z, Guo Z, Zhang L. Preparation and Characterization of an Ancient Aminopeptidase Obtained from Ancestral Sequence Reconstruction for L-Carnosine Synthesis. Molecules. 2022; 27(19):6620. https://doi.org/10.3390/molecules27196620
Chicago/Turabian StyleLiu, Fan, Yi Shi, Yakun Fang, Zhenshan Liu, Yu Xin, Zhenghua Gu, Zitao Guo, and Liang Zhang. 2022. "Preparation and Characterization of an Ancient Aminopeptidase Obtained from Ancestral Sequence Reconstruction for L-Carnosine Synthesis" Molecules 27, no. 19: 6620. https://doi.org/10.3390/molecules27196620
APA StyleLiu, F., Shi, Y., Fang, Y., Liu, Z., Xin, Y., Gu, Z., Guo, Z., & Zhang, L. (2022). Preparation and Characterization of an Ancient Aminopeptidase Obtained from Ancestral Sequence Reconstruction for L-Carnosine Synthesis. Molecules, 27(19), 6620. https://doi.org/10.3390/molecules27196620







