Intracellular Enzyme-Instructed Self-Assembly of Peptides (IEISAP) for Biomedical Applications
Abstract
1. Introduction
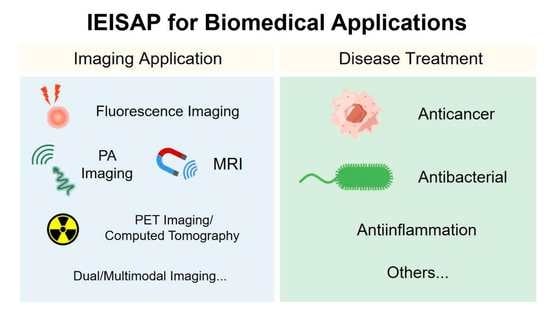
2. IEISAP for Imaging Applications
2.1. Fluorescence Imaging
2.2. Photoacoustic (PA) Imaging
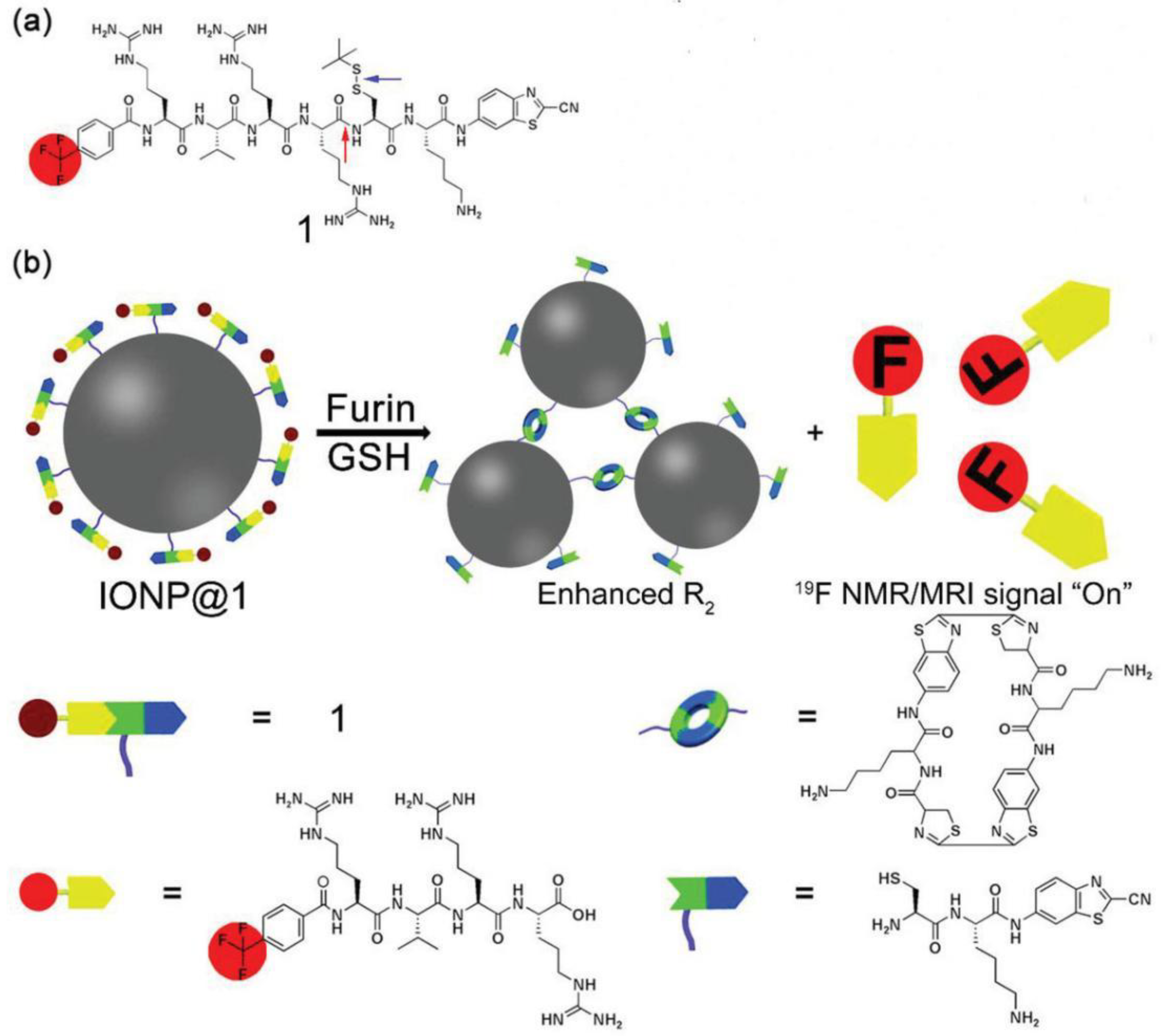
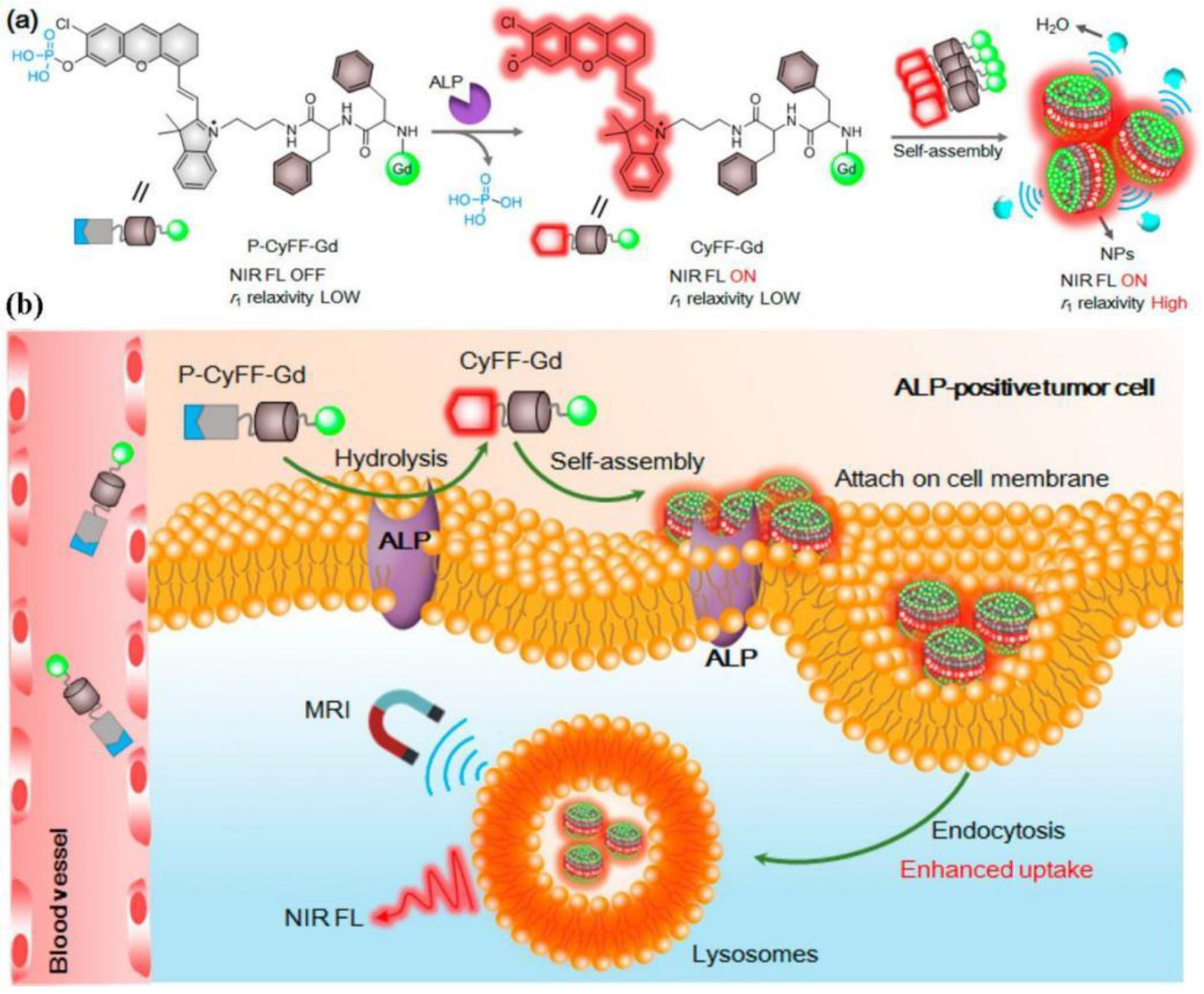
2.3. Magnetic Resonance Imaging (MRI)
2.4. Positron-Emission Tomography (PET) Imaging
2.5. Computed Tomography (CT)
2.6. Dual/Multimodal Imaging
3. Disease Treatments
3.1. Cancer Therapy
3.1.1. IEISAP Materials That Kill Cancer Cells Directly
3.1.2. IEISAP Materials for Delivering Anticancer Drugs
3.1.3. IEISAP Materials Used in Combination with Traditional Chemotherapeutic Drugs for Cancer Treatment
3.1.4. IEISAP Materials for Cancer Theranostics
3.2. Antibacterial Treatment
3.3. Antiinflammation Application
3.4. Others
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Whitesides, G.M.; Grzybowski, B. Self-assembly at all scales. Science 2002, 295, 2418–2421. [Google Scholar] [CrossRef] [PubMed]
- Ariga, K.; Nishikawa, M.; Mori, T.; Takeya, J.; Shrestha, L.K.; Hill, J.P. Self-assembly as a key player for materials nanoarchitectonics. Sci. Technol. Adv. Mater. 2019, 20, 51–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, Y.; Li, Z.; Wu, A.; Wei, G. Bottom-up synthesis and sensor applications of biomimetic nanostructures. Materials 2016, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Helbing, C.; Deckert-Gaudig, T.; Firkowska-Boden, I.; Wei, G.; Deckert, V.; Jandt, K.D. Protein handshake on the nanoscale: How albumin and hemoglobin self-assemble into nanohybrid fibers. ACS Nano 2018, 12, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gong, C.; Akakuru, O.U.; Su, Z.; Wu, A.; Wei, G. The design and biomedical applications of self-assembled two-dimensional organic biomaterials. Chem. Soc. Rev. 2019, 48, 5564–5595. [Google Scholar] [CrossRef]
- Gong, C.; Sun, S.; Zhang, Y.; Sun, L.; Su, Z.; Wu, A.; Wei, G. Hierarchical nanomaterials via biomolecular self-assembly and bioinspiration for energy and environmental applications. Nanoscale 2019, 11, 4147–4182. [Google Scholar] [CrossRef] [PubMed]
- Zou, R.; Wang, Q.; Wu, J.; Wu, J.; Schmuck, C.; Tian, H. Peptide self-assembly triggered by metal ions. Chem. Soc. Rev. 2015, 44, 5200–5219. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.H.; Cong, Y.; Qi, G.B.; Ji, L.; Qiao, Z.Y.; Wang, H. Near-infrared laser-driven in situ self-assembly as a general strategy for deep tumor therapy. Nano Lett. 2018, 18, 6577–6584. [Google Scholar] [CrossRef]
- Haburcak, R.; Shi, J.; Du, X.; Yuan, D.; Xu, B. Ligand–receptor interaction modulates the energy landscape of enzyme-instructed self-assembly of small molecules. J. Am. Chem. Soc. 2016, 138, 15397–15404. [Google Scholar] [CrossRef]
- Kim, B.J. Enzyme-instructed self-assembly of peptides: From concept to representative applications. Chem. Asian J. 2022, 17, e202200094. [Google Scholar] [CrossRef]
- Jeena, M.T.; Palanikumar, L.; Go, E.M.; Kim, I.; Kang, M.G.; Lee, S.; Park, S.; Choi, H.; Kim, C.; Jin, S.M.; et al. Mitochondria localization induced self-assembly of peptide amphiphiles for cellular dysfunction. Nat. Commun. 2017, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Li, X.; Guo, K.; Xie, T.Z.; Moorefield, C.N.; Wesdemiotis, C.; Newkome, G.R. Probing a hidden world of molecular self-assembly: Concentration-dependent, three-dimensional supramolecular interconversions. J. Am. Chem. Soc. 2014, 136, 18149–18155. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhan, J.; Yang, Z. Enzyme-instructed self-assembly (EISA) and hydrogelation of peptides. Adv. Mater. 2020, 32, 1805798. [Google Scholar] [CrossRef]
- Rehm, T.H.; Schmuck, C. Ion-pair induced self-assembly in aqueous solvents. Chem. Soc. Rev. 2010, 39, 3597–3611. [Google Scholar] [CrossRef] [PubMed]
- Dreher, M.R.; Simnick, A.J.; Fischer, K.; Smith, R.J.; Patel, A.; Schmidt, M.; Chilkoti, A. Temperature triggered self-assembly of polypeptides into multivalent spherical micelles. J. Am. Chem. Soc. 2008, 130, 687–694. [Google Scholar] [CrossRef]
- Ghosh, A.; Haverick, M.; Stump, K.; Yang, X.; Tweedle, M.F.; Goldberger, J.E. Fine-tuning the pH trigger of self-assembly. J. Am. Chem. Soc. 2012, 134, 3647–3650. [Google Scholar] [CrossRef]
- Zhou, D.; Dong, S.; Kuchel, R.P.; Perrier, S.; Zetterlund, P.B. Polymerization induced self-assembly: Tuning of morphology using ionic strength and pH. Polym. Chem. 2017, 8, 3082–3089. [Google Scholar] [CrossRef]
- Yan, X.; Zhu, P.; Li, J. Self-assembly and application of diphenylalanine-based nanostructures. Chem. Soc. Rev. 2010, 39, 1877–1890. [Google Scholar] [CrossRef]
- Kokkoli, E.; Mardilovich, A.; Wedekind, A.; Rexeisen, E.L.; Garg, A.; Craig, J.A. Self-assembly and applications of biomimetic and bioactive peptide-amphiphiles. Soft Matter 2006, 2, 1015–1024. [Google Scholar] [CrossRef]
- Qi, G.B.; Gao, Y.J.; Wang, L.; Wang, H. Self-assembled peptide-based nanomaterials for biomedical imaging and therapy. Adv. Mater. 2018, 30, 1703444. [Google Scholar] [CrossRef]
- Ren, C.; Wang, Z.; Wang, Q.; Yang, C.; Liu, J. Self-assembled peptide-based nanoprobes for disease theranostics and disease-related molecular imaging. Small Methods 2020, 4, 1900403. [Google Scholar] [CrossRef]
- Miao, Q.; Pu, K. Emerging designs of activatable photoacoustic probes for molecular imaging. Bioconjugate Chem. 2016, 27, 2808–2823. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liang, G.; Xu, B. Enzymatic control of the self-assembly of small molecules: A new way to generate supramolecular hydrogels. Soft Matter 2007, 3, 515–520. [Google Scholar] [CrossRef]
- Zhou, J.; Du, X.; Xu, B. Regulating the rate of molecular self-assembly for targeting cancer cells. Angew. Chem. Int. Ed. 2016, 55, 5770–5775. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ju, Q.; Pang, S.; Wei, N.; Zhang, Y. Recent progress of fluorescent probes for the detection of alkaline phosphatase (ALP): A review. Dyes Pigm. 2021, 194, 109569. [Google Scholar] [CrossRef]
- Zhan, J.; Cai, Y.; He, S.; Wang, L.; Yang, Z. Tandem molecular self-assembly in liver cancer cells. Angew. Chem. Int. Ed. 2018, 57, 1813–1816. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Chen, P.; Xie, M.; Wu, C.; Luo, Y.; Wang, W.; Jiang, J.; Liang, G. Cell environment-differentiated self-assembly of nanofibers. J. Am. Chem. Soc. 2016, 138, 11128–11131. [Google Scholar] [CrossRef]
- Yang, Z.; Liang, G.; Guo, Z.; Guo, Z.; Xu, B. Intracellular hydrogelation of small molecules inhibits bacterial growth. Angew. Chem. Int. Ed. 2007, 46, 8216–8219. [Google Scholar] [CrossRef]
- Ren, C.; Wang, H.; Zhang, X.; Ding, D.; Wang, L.; Yang, Z. Interfacial self-assembly leads to formation of fluorescent nanoparticles for simultaneous bacterial detection and inhibition. Chem. Commun. 2014, 50, 3473–3475. [Google Scholar] [CrossRef]
- Hughes, M.; Debnath, S.; Knapp, C.W.; Ulijn, R.V. Antimicrobial properties of enzymatically triggered self-assembling aromatic peptide amphiphiles. Biomater. Sci. 2013, 1, 1138–1142. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, Y.; Deng, J.; Ding, X.; Lin, D.; Shi, H.; Chen, L.; Lin, D.; Wang, Y.; Vakal, S.; et al. Enzyme-instructed self-assembly of peptide-drug conjugates in tear fluids for ocular drug delivery. J. Control. Release 2022, 344, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Li, L.L.; Ma, H.L.; Qi, G.B.; Zhang, D.; Yu, F.; Hu, Z.; Wang, H. Pathological-condition-driven construction of supramolecular nanoassemblies for bacterial infection detection. Adv. Mater. 2016, 28, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Ren, H.; Rao, J. A biocompatible condensation reaction for controlled assembly of nanostructures in living cells. Nat. Chem. 2010, 2, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Liang, G.; Ma, M.L.; Rao, J. Controlling intracellular macrocyclization for the imaging of protease activity. Angew. Chem. Int. Ed. 2011, 50, 2275–2279. [Google Scholar] [CrossRef]
- Yang, Z.; Liang, G.; Ma, M.; Gao, Y.; Xu, B. In vitro and in vivo enzymatic formation of supramolecular hydrogels based on self-assembled nanofibers of a β-amino acid derivative. Small 2007, 3, 558–562. [Google Scholar] [CrossRef]
- Li, L.L.; Qiao, Z.Y.; Wang, L.; Wang, H. Programmable construction of peptide-based materials in living subjects: From modular design and morphological control to theranostics. Adv. Mater. 2019, 31, 1804971. [Google Scholar] [CrossRef]
- Abbas, M.; Zou, Q.; Li, S.; Yan, X. Self-assembled peptide- and protein-based nanomaterials for antitumor photodynamic and photothermal therapy. Adv. Mater. 2017, 29, 1605021. [Google Scholar] [CrossRef]
- Shy, A.N.; Kim, B.J.; Xu, B. Enzymatic noncovalent synthesis of supramolecular soft matter for biomedical applications. Matter 2019, 1, 1127–1147. [Google Scholar] [CrossRef]
- Chien, M.P.; Carlini, A.S.; Hu, D.; Barback, C.V.; Rush, A.M.; Hall, D.J.; Orr, G.; Gianneschi, N.C. Enzyme-directed assembly of nanoparticles in tumors monitored by in vivo whole animal imaging and ex vivo super-resolution fluorescence imaging. J. Am. Chem. Soc. 2013, 135, 18710–18713. [Google Scholar] [CrossRef][Green Version]
- Zheng, R.; Yang, J.; Mamuti, M.; Hou, D.Y.; An, H.W.; Zhao, Y.; Wang, H. Controllable self-assembly of peptide-cyanine conjugates in vivo as fine-tunable theranostics. Angew. Chem. Int. Ed. 2021, 60, 7809–7819. [Google Scholar] [CrossRef]
- An, H.W.; Hou, D.; Zheng, R.; Wang, M.D.; Zeng, X.Z.; Xiao, W.Y.; Yan, T.D.; Wang, J.Q.; Zhao, C.H.; Cheng, L.M.; et al. A near-infrared peptide probe with tumor-specific excretion-retarded effect for image-guided surgery of renal cell carcinoma. ACS Nano 2020, 14, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Shuhendler, A.J.; Cui, L.; Tong, L.; Tee, S.S.; Tikhomirov, G.; Felsher, D.W.; Rao, J. Bioorthogonal cyclization-mediated in situ self-assembly of small-molecule probes for imaging caspase activity in vivo. Nat. Chem. 2014, 6, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Miao, Q.; Hai, Z.; Yuan, Y.; Liang, G. Enzymatic hydrogelation-induced fluorescence turn-off for sensing alkaline phosphatase in vitro and in living cells. Anal. Chem. 2015, 87, 6475–6478. [Google Scholar] [CrossRef]
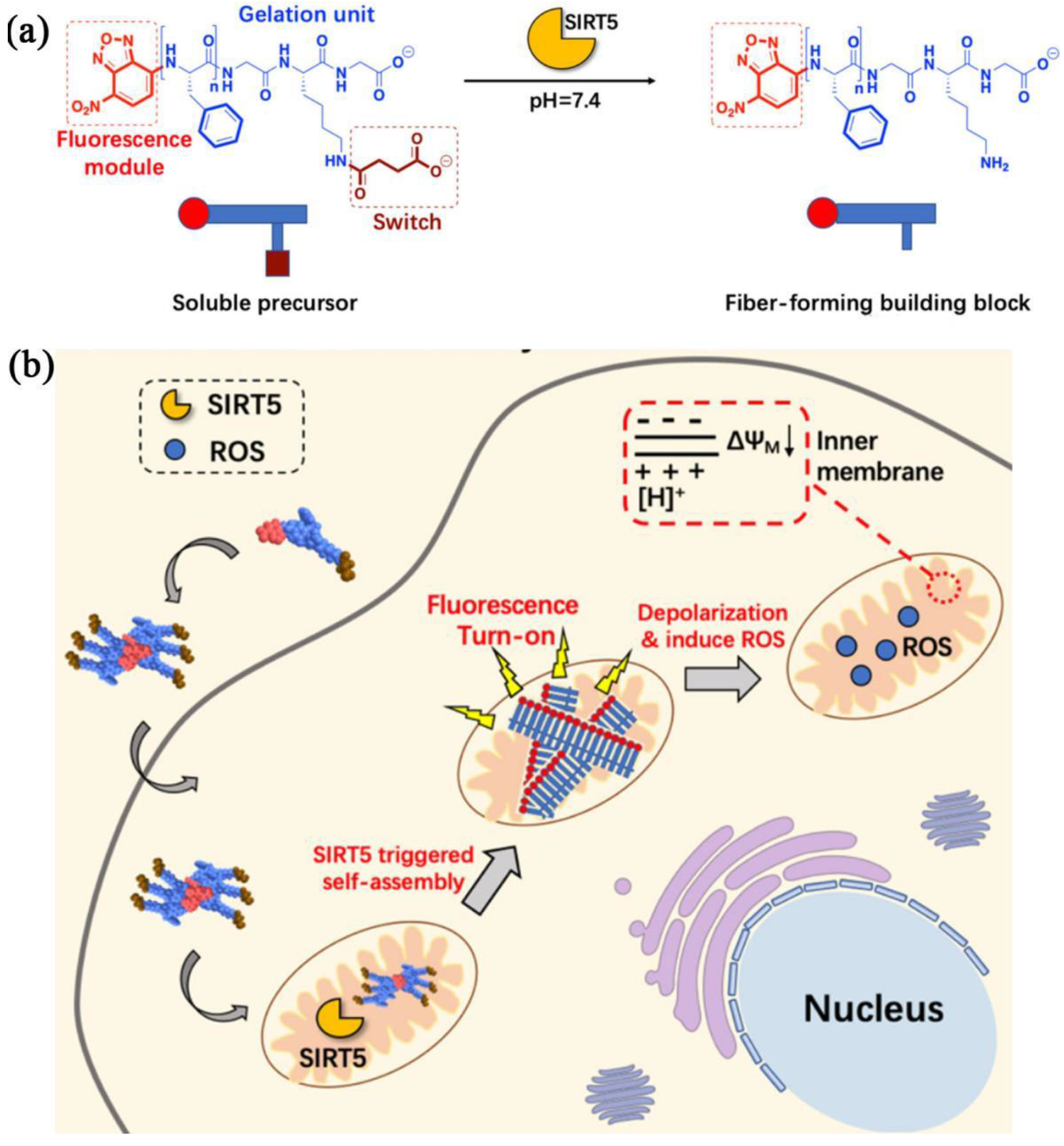
- Yang, L.; Peltier, R.; Zhang, M.; Song, D.; Huang, H.; Chen, G.; Chen, Y.; Zhou, F.; Hao, Q.; Bian, L.; et al. Desuccinylation-triggered peptide self-assembly: Live cell imaging of SIRT5 activity and mitochondrial activity modulation. J. Am. Chem. Soc. 2020, 142, 18150–18159. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Jin, Q.; Wang, Y.; Chen, Y.; Ji, J. The rational design of a gemcitabine prodrug with AIE-based intracellular light-up characteristics for selective suppression of pancreatic cancer cells. Chem. Commun. 2015, 51, 17435–17438. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Shi, J.; Yuan, D.; Xu, B. Imaging enzyme-triggered self-assembly of small molecules inside live cells. Nat. Commun. 2012, 3, 1033. [Google Scholar] [CrossRef]
- Zhou, J.; Du, X.; Li, J.; Yamagata, N.; Xu, B. Taurine boosts cellular uptake of small D-peptides for enzyme-instructed intracellular molecular self-assembly. J. Am. Chem. Soc. 2015, 137, 10040–10043. [Google Scholar] [CrossRef]
- He, H.; Wang, J.; Wang, H.; Zhou, N.; Yang, D.; Green, D.R.; Xu, B. Enzymatic cleavage of branched peptides for targeting mitochondria. J. Am. Chem. Soc. 2018, 140, 1215–1218. [Google Scholar] [CrossRef]
- Wang, H.; Feng, Z.; Del Signore, S.J.; Rodal, A.A.; Xu, B. Active probes for imaging membrane dynamics of live cells with high spatial and temporal resolution over extended time scales and areas. J. Am. Chem. Soc. 2018, 140, 3505–3509. [Google Scholar] [CrossRef]
- Zhou, J.; Du, X.; Berciu, C.; He, H.; Shi, J.; Nicastro, D.; Xu, B. Enzyme-instructed self-assembly for spatiotemporal profiling of the activities of alkaline phosphatases on live cells. Chem 2016, 1, 246–263. [Google Scholar] [CrossRef]
- Ji, S.; Gao, H.; Mu, W.; Ni, X.; Yi, X.; Shen, J.; Liu, Q.; Bao, P.; Ding, D. Enzyme-instructed self-assembly leads to the activation of optical properties for selective fluorescence detection and photodynamic ablation of cancer cells. J. Mater. Chem. B 2018, 6, 2566–2573. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Kwok, R.T.K.; Liu, J.; Xing, B.; Tang, B.Z.; Liu, B. Real-time monitoring of cell apoptosis and drug screening using fluorescent light-up probe with aggregation-induced emission characteristics. J. Am. Chem. Soc. 2012, 134, 17972–17981. [Google Scholar] [CrossRef] [PubMed]
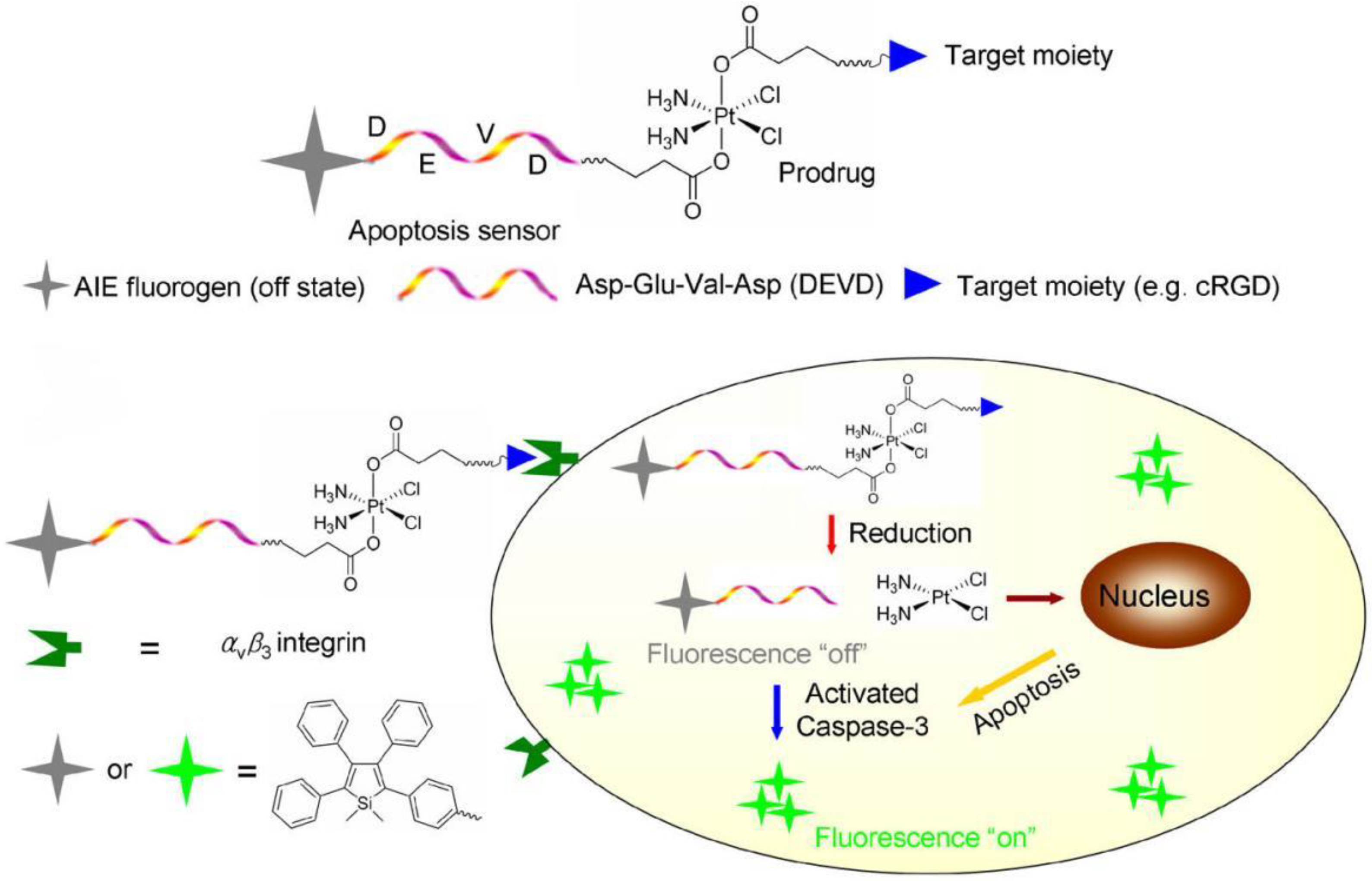
- Yuan, Y.; Kwok, R.T.K.; Tang, B.Z.; Liu, B. Targeted theranostic platinum(IV) prodrug with a built-in aggregation-induced emission light-up apoptosis sensor for noninvasive early evaluation of its therapeutic responses in situ. J. Am. Chem. Soc. 2014, 136, 2546–2554. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhang, C.J.; Gao, M.; Zhang, R.; Tang, B.Z.; Liu, B. Specific light-up bioprobe with aggregation-induced emission and activatable photoactivity for the targeted and image-guided photodynamic ablation of cancer cells. Angew. Chem. Int. Ed. 2015, 54, 1780–1786. [Google Scholar] [CrossRef]
- Zhang, X.; Ren, C.; Hu, F.; Gao, Y.; Wang, Z.; Li, H.; Liu, J.; Liu, B.; Yang, C. Detection of bacterial alkaline phosphatase activity by enzymatic in situ self-assembly of the AIEgen-peptide conjugate. Anal. Chem. 2020, 92, 5185–5190. [Google Scholar] [CrossRef]
- Lin, Y.X.; Qiao, S.L.; Wang, Y.; Zhang, R.X.; An, H.W.; Ma, Y.; Rajapaksha, R.P.Y.J.; Qiao, Z.Y.; Wang, L.; Wang, H. An in situ intracellular self-assembly strategy for quantitatively and temporally monitoring autophagy. ACS Nano 2017, 11, 1826–1839. [Google Scholar] [CrossRef]
- Zhong, Y.; Zhan, J.; Xu, G.; Chen, Y.; Qin, Q.; Liao, X.; Ma, S.; Yang, Z.; Cai, Y. Enzyme-instructed self-assembly enabled monomer–excimer transition to construct higher ordered luminescent supramolecular assembly for activity-based bioimaging. Angew. Chem. Int. Ed. 2021, 60, 8121–8129. [Google Scholar] [CrossRef]
- Zhang, Y.; Ding, Y.; Li, X.; Zhang, Z.; Zhang, X.; Chen, Y.; Yang, Z.; Shi, Y.; Hu, Z.W. Enzyme-instructed self-assembly enabled fluorescence light-up for alkaline phosphatase detection. Talanta 2022, 239, 123078. [Google Scholar] [CrossRef]
- Liang, J.; Kwok, R.T.K.; Shi, H.; Tang, B.Z.; Liu, B. Fluorescent light-up probe with aggregation-induced emission characteristics for alkaline phosphatase sensing and activity study. ACS Appl. Mater. Interfaces 2013, 5, 8784–8789. [Google Scholar] [CrossRef]
- Gao, Y.; Berciu, C.; Kuang, Y.; Shi, J.; Nicastro, D.; Xu, B. Probing nanoscale self-assembly of nonfluorescent small molecules inside live mammalian cells. ACS Nano 2013, 7, 9055–9063. [Google Scholar] [CrossRef]
- Wang, L.; Yang, P.P.; Zhao, X.X.; Wang, H. Self-assembled nanomaterials for photoacoustic imaging. Nanoscale 2016, 8, 2488–2509. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Qi, G.B.; Zhao, Y.X.; Qiao, S.L.; Yang, C.; Wang, H. In situ formation of nanofibers from purpurin18-peptide conjugates and the assembly induced retention effect in tumor sites. Adv. Mater. 2015, 27, 6125–6130. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.X.; Wang, Y.; Qiao, S.L.; An, H.W.; Wang, J.; Ma, Y.; Wang, L.; Wang, H. “In vivo self-assembled” nanoprobes for optimizing autophagy-mediated chemotherapy. Biomaterials 2017, 141, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Zhang, R.; Du, W.; Cheng, L.; Liang, G. Alkaline Phosphatase-triggered self-assembly of near-infrared nanoparticles for the enhanced photoacoustic imaging of tumors. Nano Lett. 2018, 18, 7749–7754. [Google Scholar] [CrossRef]
- Cai, Q.; Fei, Y.; Hu, L.; Huang, Z.; Li, L.L.; Wang, H. Chemotaxis-instructed intracellular Staphylococcus aureus infection detection by a targeting and self-assembly signal-enhanced photoacoustic probe. Nano Lett. 2018, 18, 6229–6236. [Google Scholar] [CrossRef]
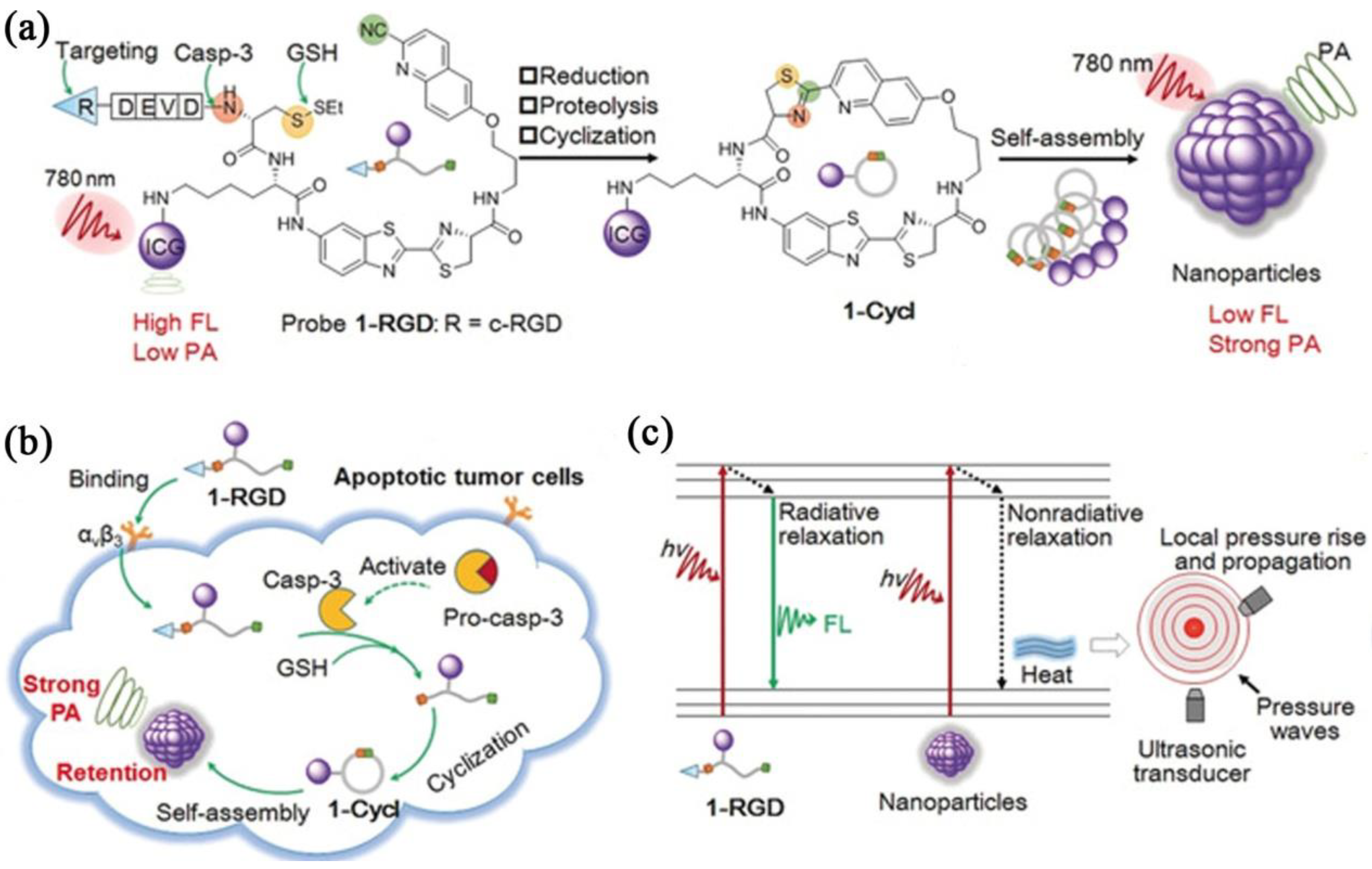
- Wang, Y.; Hu, X.; Weng, J.; Li, J.; Fan, Q.; Zhang, Y.; Ye, D. A photoacoustic probe for the imaging of tumor apoptosis by caspase-mediated macrocyclization and self-assembly. Angew. Chem. Int. Ed. 2019, 58, 4886–4890. [Google Scholar] [CrossRef]
- Dragulescu-Andrasi, A.; Kothapalli, S.R.; Tikhomirov, G.A.; Rao, J.; Gambhir, S.S. Activatable oligomerizable imaging agents for photoacoustic imaging of furin-like activity in living subjects. J. Am. Chem. Soc. 2013, 135, 11015–11022. [Google Scholar] [CrossRef]
- Dong, L.; Qian, J.; Hai, Z.; Xu, J.; Du, W.; Zhong, K.; Liang, G. Alkaline phosphatase-instructed self-assembly of gadolinium nanofibers for enhanced T2-weighted magnetic resonance imaging of tumor. Anal. Chem. 2017, 89, 6922–6925. [Google Scholar] [CrossRef]
- Zhang, J.; Mu, Y.L.; Ma, Z.Y.; Han, K.; Han, H.Y. Tumor-triggered transformation of chimeric peptide for dual-stage-amplified magnetic resonance imaging and precise photodynamic therapy. Biomaterials 2018, 182, 269–278. [Google Scholar] [CrossRef]
- Ye, D.; Shuhendler, A.J.; Pandit, P.; Brewer, K.D.; Tee, S.S.; Cui, L.; Tikhomirov, G.; Rutt, B.; Rao, J. Caspase-responsive smart gadolinium-based contrast agent for magnetic resonance imaging of drug-induced apoptosis. Chem. Sci. 2014, 5, 3845–3852. [Google Scholar] [CrossRef]
- Yuan, Y.; Ding, Z.; Qian, J.; Zhang, J.; Xu, J.; Dong, X.; Han, T.; Ge, S.; Luo, Y.; Wang, Y.; et al. Casp3/7-instructed intracellular aggregation of Fe3O4 nanoparticles enhances T2 MR imaging of tumor apoptosis. Nano Lett. 2016, 16, 2686–2691. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Sun, H.; Ge, S.; Cai, Y.; Yuan, Y.; Hai, Z.; Tao, T.; Hu, J.; Hu, B.; Wang, J.; et al. Furin-controlled Fe3O4 nanoparticle aggregation and 19F signal “turn-on” for precise MR imaging of tumors. Adv. Funct. Mater. 2019, 29, 1903860. [Google Scholar] [CrossRef]
- Zheng, Z.; Sun, H.; Hu, C.; Li, G.; Liu, X.; Chen, P.; Cui, Y.; Liu, J.; Wang, J.; Liang, G. Using “ON/OFF” 19F NMR/magnetic resonance imaging signals to sense tyrosine kinase/phosphatase activity in vitro and in cell lysates. Anal. Chem. 2016, 88, 3363–3368. [Google Scholar] [CrossRef]
- Liu, Y.; Miao, Q.; Zou, P.; Liu, L.; Wang, X.; An, L.; Zhang, X.; Qian, X.; Luo, S.; Liang, G. Enzyme-controlled intracellular self-assembly of 18F nanoparticles for enhanced microPET imaging of tumor. Theranostics 2015, 5, 1058–1067. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, H.; Chen, P.; Wu, H.; Zou, P.; Wu, J.; Liu, Y.; Liang, G. Furin-guided intracellular 68Ga nanoparticle formation enhancing tumor microPET imaging. Anal. Chem. 2019, 91, 14842–14845. [Google Scholar] [CrossRef] [PubMed]
- Xi, D.; Dong, S.; Meng, X.; Lu, Q.; Meng, L.; Ye, J. Gold nanoparticles as computerized tomography (CT) contrast agents. RSC Adv. 2012, 2, 12515–12524. [Google Scholar] [CrossRef]
- Sun, I.C.; Eun, D.K.; Koo, H.; Ko, C.Y.; Kim, H.S.; Yi, D.K.; Choi, K.; Kwon, I.C.; Kim, K.; Ahn, C.H. Tumor-targeting gold particles for dual computed tomography/optical cancer imaging. Angew. Chem. Int. Ed. 2011, 50, 9348–9351. [Google Scholar] [CrossRef]
- Bellat, V.; Ting, R.; Southard, T.L.; Vahdat, L.; Molina, H.; Fernandez, J.; Aras, O.; Stokol, T.; Law, B. Functional peptide nanofibers with unique tumor targeting and enzyme-induced local retention properties. Adv. Funct. Mater. 2018, 28, 1803969. [Google Scholar] [CrossRef]
- Mao, W.; Kim, H.S.; Son, Y.J.; Kim, S.R.; Yoo, H.S. Doxorubicin encapsulated clicked gold nanoparticle clusters exhibiting tumor-specific disassembly for enhanced tumor localization and computerized tomographic imaging. J. Control. Release 2018, 269, 52–62. [Google Scholar] [CrossRef]
- Yan, R.; Hu, Y.; Liu, F.; Wei, S.; Fang, D.; Shuhendler, A.J.; Liu, H.; Chen, H.Y.; Ye, D. Activatable NIR fluorescence/MRI bimodal probes for in vivo imaging by enzyme-mediated fluorogenic reaction and self-assembly. J. Am. Chem. Soc. 2019, 141, 10331–10341. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Huang, Z.; Liu, D.; Chen, J.; Gao, Y. Enzyme-instructed supramolecular self-assembly with anticancer activity. Adv. Mater. 2019, 31, 1804814. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Y.; Shi, J.; Li, J.; Yuan, D.; Alberti, K.A.; Xu, Q.; Xu, B. Pericellular hydrogel/nanonets inhibit cancer cells. Angew. Chem. Int. Ed. 2014, 53, 8104–8107. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Feng, Z.; Wang, Y.; Zhou, R.; Yang, Z.; Xu, B. Integrating enzymatic self-assembly and mitochondria targeting for selectively killing cancer cells without acquired drug resistance. J. Am. Chem. Soc. 2016, 138, 16046–16055. [Google Scholar] [CrossRef] [PubMed]
- Li, L.L.; Qiao, S.L.; Liu, W.J.; Ma, Y.; Wan, D.; Pan, J.; Wang, H. Intracellular construction of topology-controlled polypeptide nanostructures with diverse biological functions. Nat. Commun. 2017, 8, 1276. [Google Scholar] [CrossRef]
- Feng, Z.; Wang, H.; Chen, X.; Xu, B. Self-assembling ability determines the activity of enzyme-instructed self-assembly for inhibiting cancer cells. J. Am. Chem. Soc. 2017, 139, 15377–15384. [Google Scholar] [CrossRef]
- Du, X.; Zhou, J.; Wang, H.; Shi, J.; Kuang, Y.; Zeng, W.; Yang, Z.; Xu, B. In situ generated D-peptidic nanofibrils as multifaceted apoptotic inducers to target cancer cells. Cell Death Dis. 2017, 8, e2614. [Google Scholar] [CrossRef]
- Feng, Z.; Han, X.; Wang, H.; Tang, T.; Xu, B. Enzyme-instructed peptide assemblies selectively inhibit bone tumors. Chem 2019, 5, 2442–2449. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, C.; Chang, J.; Yang, C.; Liu, J.; Fan, S.; Ren, C. Enzyme-instructed self-assembly of a novel histone deacetylase inhibitor with enhanced selectivity and anticancer efficiency. Biomater. Sci. 2019, 7, 1477–1485. [Google Scholar] [CrossRef]
- Zheng, D.; Chen, Y.; Ai, S.; Zhang, R.; Gao, Z.; Liang, C.; Cao, L.; Chen, Y.; Hong, Z.; Shi, Y.; et al. Tandem molecular self-assembly selectively inhibits lung cancer cells by inducing endoplasmic reticulum stress. Research 2019, 2019, 4803624. [Google Scholar] [CrossRef]
- Zheng, D.; Liu, J.; Xie, L.; Wang, Y.; Ding, Y.; Peng, R.; Cui, M.; Wang, L.; Zhang, Y.; Zhang, C.; et al. Enzyme-instructed and mitochondria-targeting peptide self-assembly to efficiently induce immunogenic cell death. Acta Pharm. Sin. B 2022, 12, 2740–2750. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Zheng, D.; Chen, Y.; Zhang, Z.; Yang, Z. Selective degradation of PD-L1 in cancer cells by enzyme-instructed self-assembly. Adv. Funct. Mater. 2021, 31, 2102505. [Google Scholar] [CrossRef]
- Li, J.; Shi, J.; Medina, J.E.; Zhou, J.; Du, X.; Wang, H.; Yang, C.; Liu, J.; Yang, Z.; Dinulescu, D.M.; et al. Selectively inducing cancer cell death by intracellular enzyme-instructed self-assembly (EISA) of dipeptide derivatives. Adv. Healthc. Mater. 2017, 6, 1601400. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Du, X.; Yamagata, N.; Xu, B. Enzyme-instructed self-assembly of small D-peptides as a multiple-step process for selectively killing cancer cells. J. Am. Chem. Soc. 2016, 138, 3813–3823. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Feng, Z.; Wu, D.; Fritzsching, K.J.; Rigney, M.; Zhou, J.; Jiang, Y.; Schmidt-Rohr, K.; Xu, B. Enzyme-regulated supramolecular assemblies of cholesterol conjugates against drug-resistant ovarian cancer cells. J. Am. Chem. Soc. 2016, 138, 10758–10761. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Gao, J.; Mu, G.; Zhang, Y.; Huang, F.; Zhang, W.; Ren, C.; Yang, C.; Liu, J. Selectively enhancing radiosensitivity of cancer cells via in situ enzyme-instructed peptide self-assembly. Acta Pharm. Sin. B 2020, 10, 2374–2383. [Google Scholar] [CrossRef]
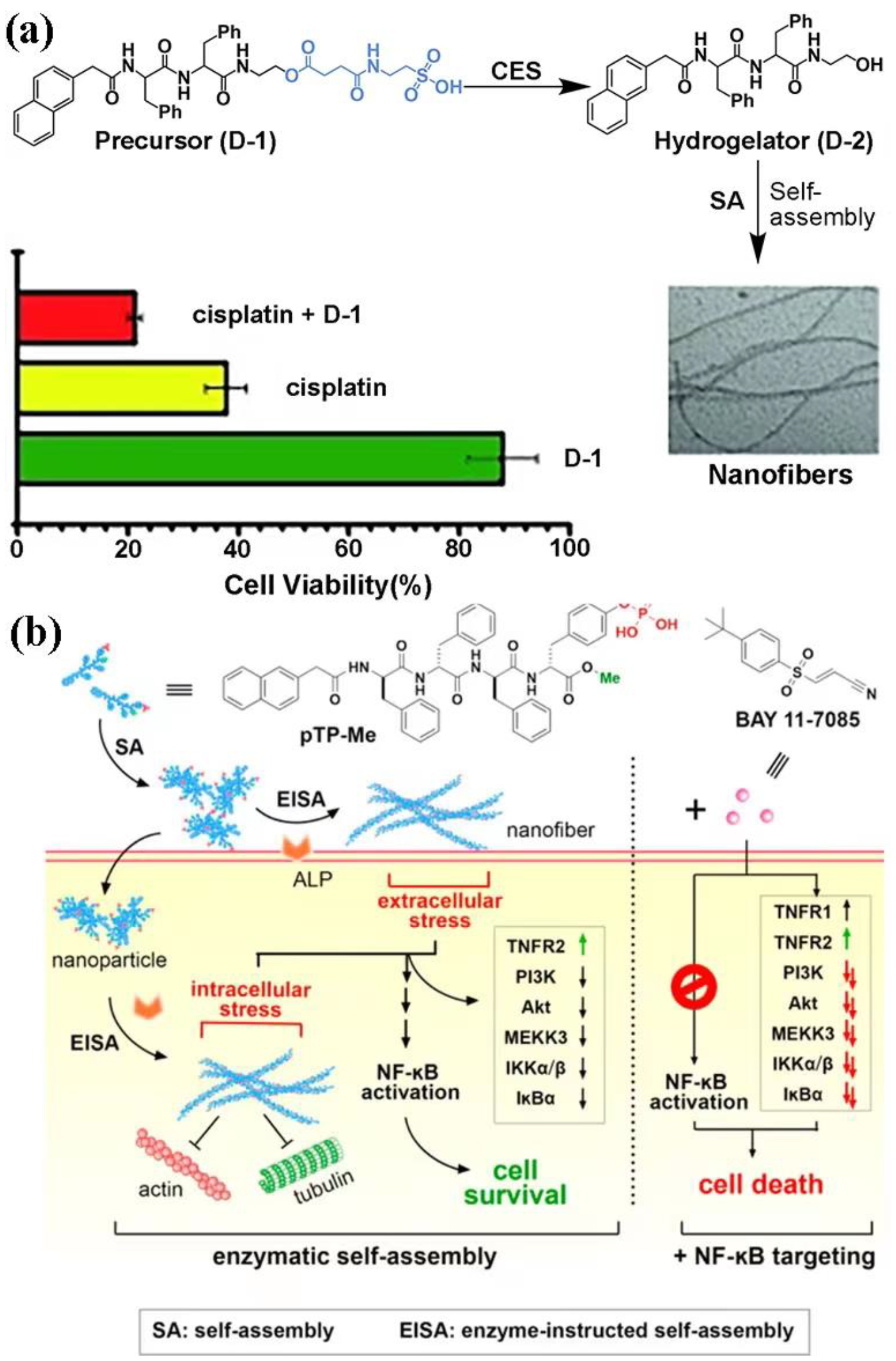
- Zhou, J.; Du, X.; Chen, X.; Wang, J.; Zhou, N.; Wu, D.; Xu, B. Enzymatic self-assembly confers exceptionally strong synergism with NF-κB targeting for selective necroptosis of cancer cells. J. Am. Chem. Soc. 2018, 140, 2301–2308. [Google Scholar] [CrossRef]
- Tanaka, A.; Fukuoka, Y.; Morimoto, Y.; Honjo, T.; Koda, D.; Goto, M.; Maruyama, T. Cancer cell death induced by the intracellular self-assembly of an enzyme-responsive supramolecular gelator. J. Am. Chem. Soc. 2015, 137, 770–775. [Google Scholar] [CrossRef]
- Yang, Z.; Xu, K.; Guo, Z.; Guo, Z.; Xu, B. Intracellular enzymatic formation of nanofibers results in hydrogelation and regulated cell death. Adv. Mater. 2007, 19, 3152–3156. [Google Scholar] [CrossRef]
- Feng, Z.; Wang, H.; Zhou, R.; Li, J.; Xu, B. Enzyme-instructed assembly and disassembly processes for targeting downregulation in cancer cells. J. Am. Chem. Soc. 2017, 139, 3950–3953. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, J.; Qi, X.; Li, S.; Liu, G.; Siddhanta, S.; Barman, I.; Song, X.; McMahon, M.T.; Bulte, J.W.M. Furin-mediated intracellular self-assembly of olsalazine nanoparticles for enhanced magnetic resonance imaging and tumour therapy. Nat. Mater. 2019, 18, 1376–1383. [Google Scholar] [CrossRef]
- Kim, B.J.; Fang, Y.; He, H.; Xu, B. Trypsin-instructed self-assembly on endoplasmic reticulum for selectively inhibiting cancer cells. Adv. Healthc. Mater. 2021, 10, 2000416. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Zhang, Y.; Yang, Z.; Gao, J.; Shi, Y. Enzyme-instructed self-assembly (EISA) assists the self-assembly and hydrogelation of hydrophobic peptides. J. Mater. Chem. B 2022, 10, 3242–3247. [Google Scholar] [CrossRef]
- Yuan, Y.; Wang, L.; Du, W.; Ding, Z.; Zhang, J.; Han, T.; An, L.; Zhang, H.; Liang, G. Intracellular self-assembly of taxol nanoparticles for overcoming multidrug resistance. Angew. Chem. Int. Ed. 2015, 54, 9700–9704. [Google Scholar] [CrossRef]
- Gao, Y.; Kuang, Y.; Guo, Z.F.; Guo, Z.; Krauss, I.J.; Xu, B. Enzyme-instructed molecular self-assembly confers nanofibers and a supramolecular hydrogel of taxol derivative. J. Am. Chem. Soc. 2009, 131, 13576–13577. [Google Scholar] [CrossRef]
- Li, J.; Gao, Y.; Kuang, Y.; Shi, J.; Du, X.; Zhou, J.; Wang, H.; Yang, Z.; Xu, B. Dephosphorylation of D-peptide derivatives to form biofunctional, supramolecular nanofibers/hydrogels and their potential applications for intracellular imaging and intratumoral chemotherapy. J. Am. Chem. Soc. 2013, 135, 9907–9914. [Google Scholar] [CrossRef]
- Huang, A.; Ou, C.; Cai, Y.; Wang, Z.; Li, H.; Yang, Z.; Chen, M. In situ enzymatic formation of supramolecular nanofibers for efficiently killing cancer cells. RSC Adv. 2016, 6, 32519–32522. [Google Scholar] [CrossRef]
- Tian, R.; Wang, H.; Niu, R.; Ding, D. Drug delivery with nanospherical supramolecular cell penetrating peptide–taxol conjugates containing a high drug loading. J. Colloid Interface Sci. 2015, 453, 15–20. [Google Scholar] [CrossRef]
- Callmann, C.E.; Barback, C.V.; Thompson, M.P.; Hall, D.J.; Mattrey, R.F.; Gianneschi, N.C. Therapeutic enzyme-responsive nanoparticles for targeted delivery and accumulation in tumors. Adv. Mater. 2015, 27, 4611–4615. [Google Scholar] [CrossRef]
- Liang, C.; Yan, X.; Zhang, R.; Xu, T.; Zheng, D.; Tan, Z.; Chen, Y.; Gao, Z.; Wang, L.; Li, X.; et al. Enhanced cellular uptake and nuclear accumulation of drug-peptide nanomedicines prepared by enzyme-instructed self-assembly. J. Control. Release 2020, 317, 109–117. [Google Scholar] [CrossRef]
- Liu, H.; Li, Y.; Lyu, Z.; Wan, Y.; Li, X.; Chen, H.; Chen, H.; Li, X. Enzyme-triggered supramolecular self-assembly of platinum prodrug with enhanced tumor-selective accumulation and reduced systemic toxicity. J. Mater. Chem. B 2014, 2, 8303–8309. [Google Scholar] [CrossRef]
- Kim, J.K.; Anderson, J.; Jun, H.W.; Repka, M.A.; Jo, S. Self-assembling peptide amphiphile-based nanofiber gel for bioresponsive cisplatin delivery. Mol. Pharm. 2009, 6, 978–985. [Google Scholar] [CrossRef]
- Gao, G.; Jiang, Y.W.; Zhan, W.; Liu, X.; Tang, R.; Sun, X.; Deng, Y.; Xu, L.; Liang, G. Trident molecule with nanobrush–nanoparticle–nanofiber transition property spatially suppresses tumor metastasis. J. Am. Chem. Soc. 2022, 144, 11897–11910. [Google Scholar] [CrossRef]
- Huang, P.; Gao, Y.; Lin, J.; Hu, H.; Liao, H.S.; Yan, X.; Tang, Y.; Jin, A.; Song, J.; Niu, G.; et al. Tumor-specific formation of enzyme-instructed supramolecular self-assemblies as cancer theranostics. ACS Nano 2015, 9, 9517–9527. [Google Scholar] [CrossRef]
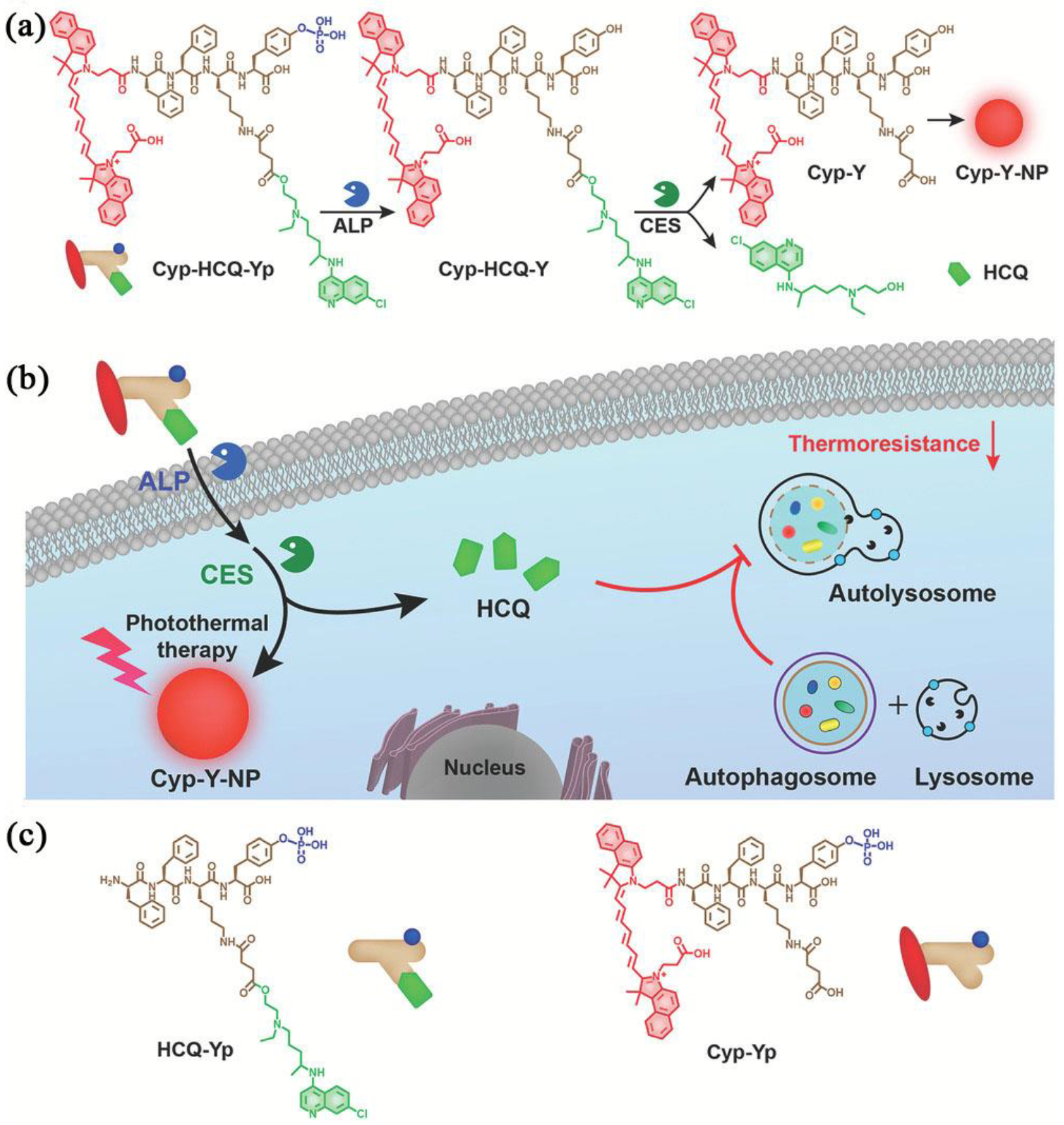
- Gao, G.; Sun, X.; Liu, X.; Jiang, Y.W.; Tang, R.; Guo, Y.; Wu, F.G.; Liang, G. Intracellular nanoparticle formation and hydroxychloroquine release for autophagy-inhibited mild-temperature photothermal therapy for tumors. Adv. Funct. Mater. 2021, 31, 2102832. [Google Scholar] [CrossRef]
- Yang, S.; Yao, D.; Wang, Y.; Yang, W.; Zhang, B.; Wang, D. Enzyme-triggered self-assembly of gold nanoparticles for enhanced retention effects and photothermal therapy of prostate cancer. Chem. Commun. 2018, 54, 9841–9844. [Google Scholar] [CrossRef]
- Li, J.; Kuang, Y.; Shi, J.; Zhou, J.; Medina, J.E.; Zhou, R.; Yuan, D.; Yang, C.; Wang, H.; Yang, Z.; et al. Enzyme-instructed intracellular molecular self-assembly to boost activity of cisplatin against drug-resistant ovarian cancer cells. Angew. Chem. Int. Ed. 2015, 54, 13307–13311. [Google Scholar] [CrossRef]
- Chen, C.; Song, Z.; Zheng, X.; He, Z.; Liu, B.; Huang, X.; Kong, D.; Ding, D.; Tang, B.Z. AIEgen-based theranostic system: Targeted imaging of cancer cells and adjuvant amplification of antitumor efficacy of paclitaxel. Chem. Sci. 2017, 8, 2191–2198. [Google Scholar] [CrossRef]
- Qi, G.B.; Zhang, D.; Liu, F.H.; Qiao, Z.Y.; Wang, H. An “on-site transformation” strategy for treatment of bacterial infection. Adv. Mater. 2017, 29, 1703461. [Google Scholar] [CrossRef]
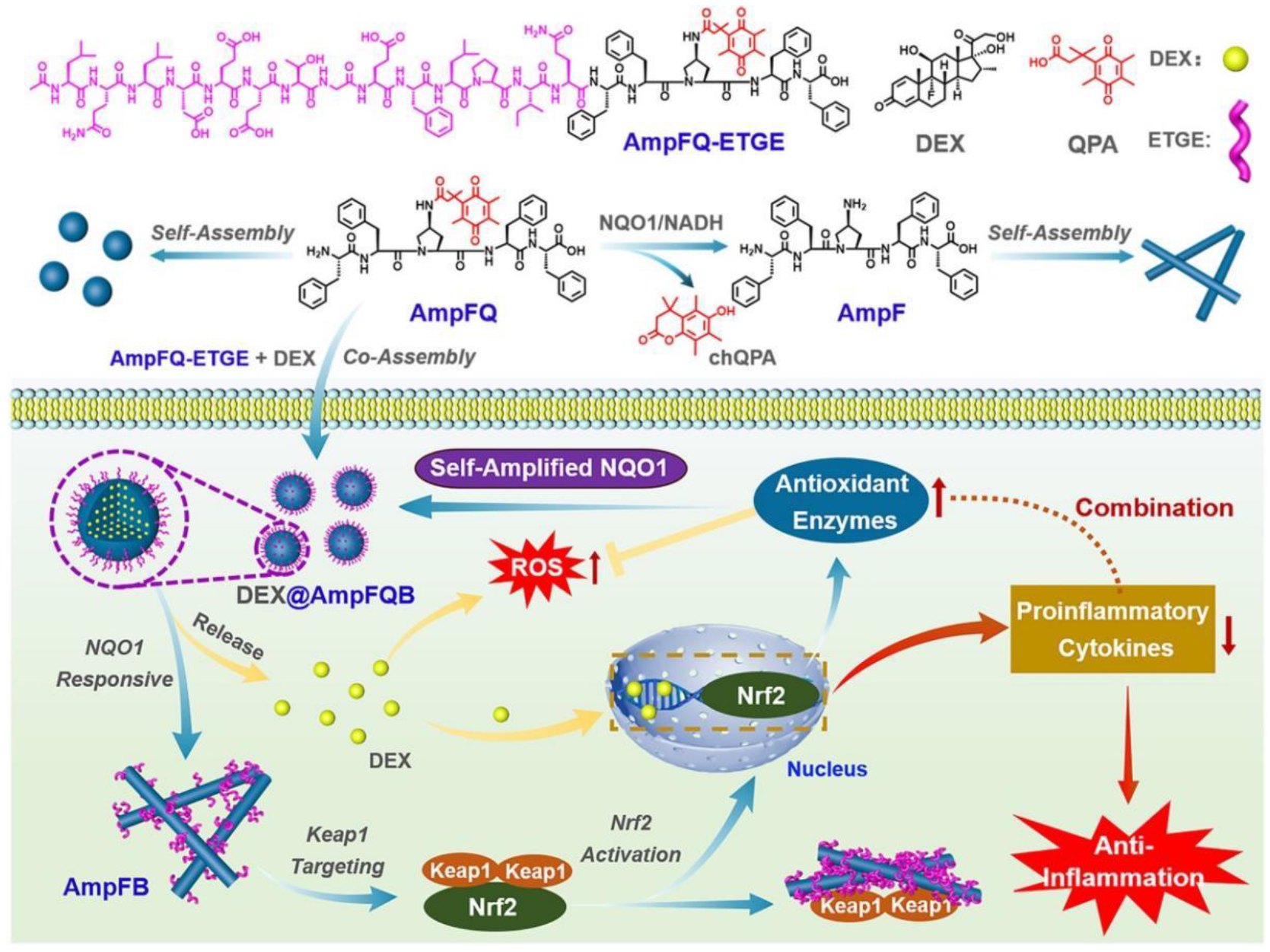
- Tang, W.; Yang, J.; Zhao, Z.; Lian, Z.; Liang, G. Intracellular coassembly boosts the anti-inflammation capacity of dexamethasone. Nanoscale 2017, 9, 17717–17721. [Google Scholar] [CrossRef]
- Song, Y.; Li, M.; Song, N.; Liu, X.; Wu, G.; Zhou, H.; Long, J.; Shi, L.; Yu, Z. Self-amplifying assembly of peptides in macrophages for enhanced inflammatory treatment. J. Am. Chem. Soc. 2022, 144, 6907–6917. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.M.; Carlini, A.S.; Chien, M.P.; Sonnenberg, S.; Luo, C.; Braden, R.L.; Osborn, K.G.; Li, Y.; Gianneschi, N.C.; Christman, K.L. Enzyme-responsive nanoparticles for targeted accumulation and prolonged retention in heart tissue after myocardial infarction. Adv. Mater. 2015, 27, 5547–5552. [Google Scholar] [CrossRef] [PubMed]

| Peptide | Enzyme | Structures | Cell Death | Cancer Cells | Mechanism | Properties | Ref. |
|---|---|---|---|---|---|---|---|
| dNapFFYp | ALP | Hydrogel/nanonet | Apoptosis | HeLa, MES-SA, and MES-SA/Dx5 | Blocking cellular mass exchange | High selectivity and accumulation in cancer cells | [83] |
| NBD-FFYpK-TPP | ALP | Nanofiber | Apoptosis | Saos2 | Causing mitochondrial dysfunction to release cytochrome c | Mitochondria-targeting; no drug resistance | [84] |
| QKVPHVGQK/QKAPGVGQK | TGase | Hydrogel | Apoptosis | HeLa, MCF-7, and SH-SY5Y | Preventing the diffusion and assembly of actin in the cytoplasm and damaging the existing actin filaments | Increased accumulation and retention effects | [85] |
| D-Tetrapeptides | ALP | Nanofiber | Apoptosis | Saos2 | Significant rearrangement of cytoskeletal proteins and plasma membranes | – | [86] |
| dNapFFYp | ALP | Nanofiber | Apoptosis | HeLa, MES-SA/Dx5, T98G, and A2780-cis | Presenting autocrine proapoptotic ligands to their cognate receptors in a juxtacrine manner, as well as directly clustering the death receptors | Killing multidrug-resistant cancer cells | [87] |
| Nap-ffypeMe2 | ALP | Nanoparticle and/or nanofibril * | Apoptosis | Saos-2 | Countering immunosuppression in the tumor microenvironment | Immunotherapy; inhibiting metastatic osteosarcoma | [88] |
| NapGDFDFpYSV | ALP | Nanofiber | Apoptosis | HeLa and A549 | Inhibiting histone deacetylase andimproving the accumulation of acetylated histone | Enhanced cellular uptake and high selectivity | [89] |
| GFFYG | ALP and reductase | Nanofiber | Apoptosis | A549 | Mitochondrial membrane disruption that results in increased ROS, cytochrome C release, and endoplasmic reticulum stress | Inhibiting lung cancer cells | [90] |
| F-pY-T | ALP | Nanoparticle | Apoptosis | CT26 | Inducing mitochondrial oxidative stress that leads to immunogenic cell death | Mitochondria-targeting and cancer immunotherapy | [91] |
| NYSKPTDRQYHF | ALP | Nanofiber | Apoptosis | 4T1 | Selective degradation of PD-L1 in cancer cells | Selective degradation of PD-L1 | [92] |
| L- and D-dipeptide and taurine conjugates | CES | Nanofiber | Apoptosis or necroptosis | HCC1937, SKOV3, and A2780-cis | Disrupting the dynamics of actin filaments | High selectivity to cancer cells; killing resistant cancer cells | [93] |
| Phosphorylated D-tetrapeptide (NapDFDFDYDY) | ALP | Nanofiber | Apoptosis and necroptosis | Saos2 | Interactions between nanofibers and the death receptors | – | [94] |
| D-Phosphotyrosine conjugated with cholesterol | ALP | Nanoparticle | Apoptosis and necroptosis | HeLa, A2780, and A2780-cis | Interacting with actin filaments and microtubules and affecting lipid rafts | Killing drug-resistant ovarian cancer cells | [95] |
| Nap-GFFpYSV | ALP | Nanofiber | Apoptosis | HeLa and A549 | Damaging DNA and arresting cell cycles | Selectively enhancing radiosensitivity of cancer; combinatorial chemo-photodynamic therapy | [96] |
| Nap-FFFYp | ALP | Nanofiber | Necroptosis | Saos-2 | Dramatically disrupting cytoskeletons and acitvating NF-κB | NF-κB targeting; low dosage | [97] |
| GKGSFGFTG | ATG4B | – | Autophagy | MCF-7 | Autophagy-mediated chemotherapy using doxorubicin | Chemotherapy | [63] |
| Nap-FF-NHCH2CH2OH | MMP-7 | – | – | HeLa | Cellular stress | – | [98] |
| Nap-FF-NHCH2CH2OH | Esterase | – | – | HeLa | Cellular stress | – | [99] |
| Nap-ffy | CES and ALP | Nanofiber | – | Hep G2 and OVSAHO | Targeting downregulation in cancer cells | Treating metastatic cancers | [100] |
| Olsalazine-RVRR | Furin | nanoparticle | – | HCT116 | Inhibiting DNA methylation | Minimal effusion | [101] |
| KYDKKKKDG(Nap-ffky) | Trypsin | Nanofiber | Apoptosis and necroptosis | OVSAHO | Inducing endoplasmic reticulum stress | Targeting endoplasmic reticulum | [102] |
| Nap-GFFpYSV (precursor 1) and Nap-GFFpYIGSR (precursor 2) | ALP | Nanofiber and hydrogel | – | A549 and HeLa | – | – | [103] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, F.; Jia, C.; Wu, F.-G. Intracellular Enzyme-Instructed Self-Assembly of Peptides (IEISAP) for Biomedical Applications. Molecules 2022, 27, 6557. https://doi.org/10.3390/molecules27196557
Lin F, Jia C, Wu F-G. Intracellular Enzyme-Instructed Self-Assembly of Peptides (IEISAP) for Biomedical Applications. Molecules. 2022; 27(19):6557. https://doi.org/10.3390/molecules27196557
Chicago/Turabian StyleLin, Fengming, Chenyang Jia, and Fu-Gen Wu. 2022. "Intracellular Enzyme-Instructed Self-Assembly of Peptides (IEISAP) for Biomedical Applications" Molecules 27, no. 19: 6557. https://doi.org/10.3390/molecules27196557
APA StyleLin, F., Jia, C., & Wu, F.-G. (2022). Intracellular Enzyme-Instructed Self-Assembly of Peptides (IEISAP) for Biomedical Applications. Molecules, 27(19), 6557. https://doi.org/10.3390/molecules27196557