Site-Specific Radioiodination of Oligonucleotides with a Phenolic Element in a Programmable Approach
Abstract
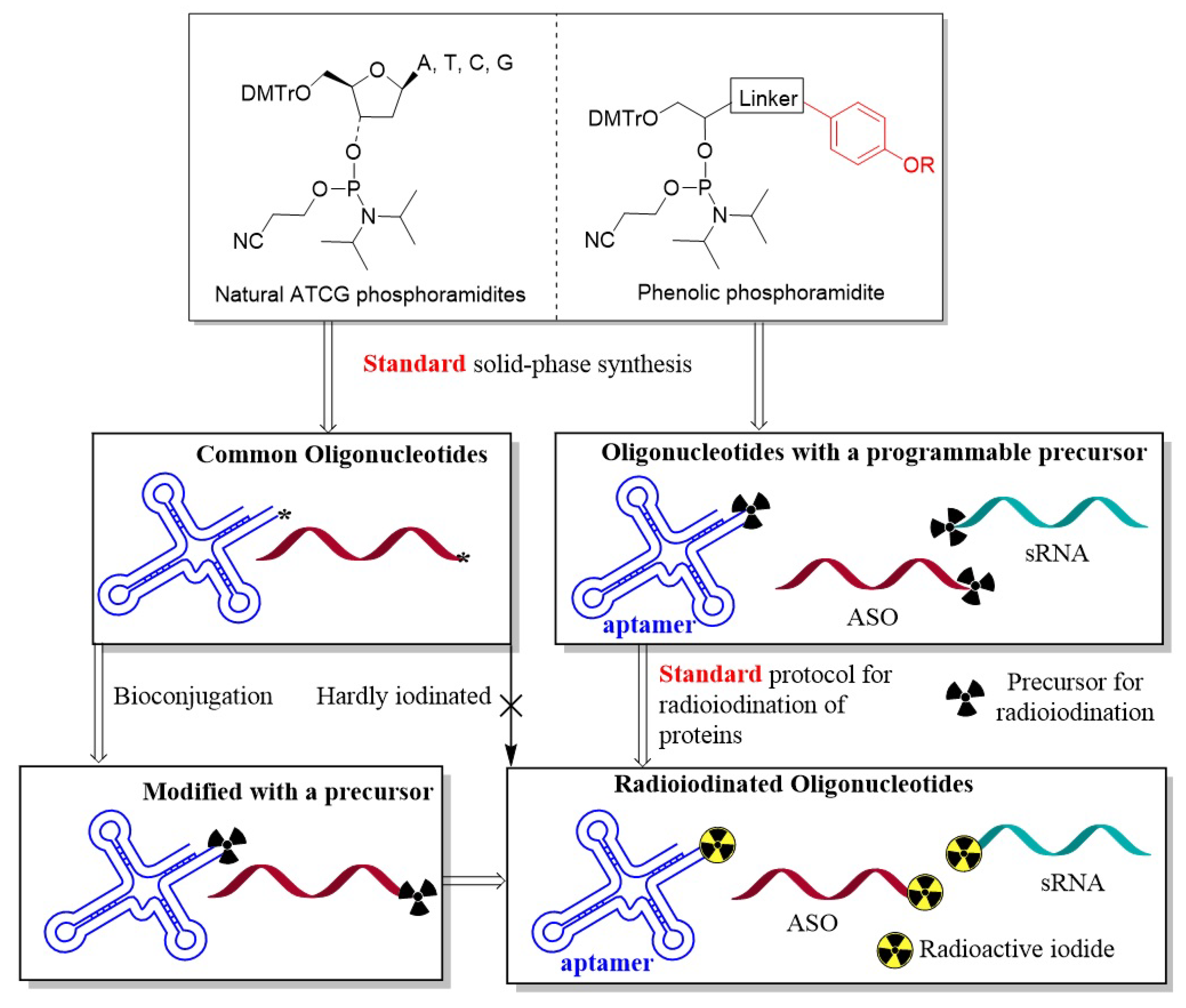
1. Introduction
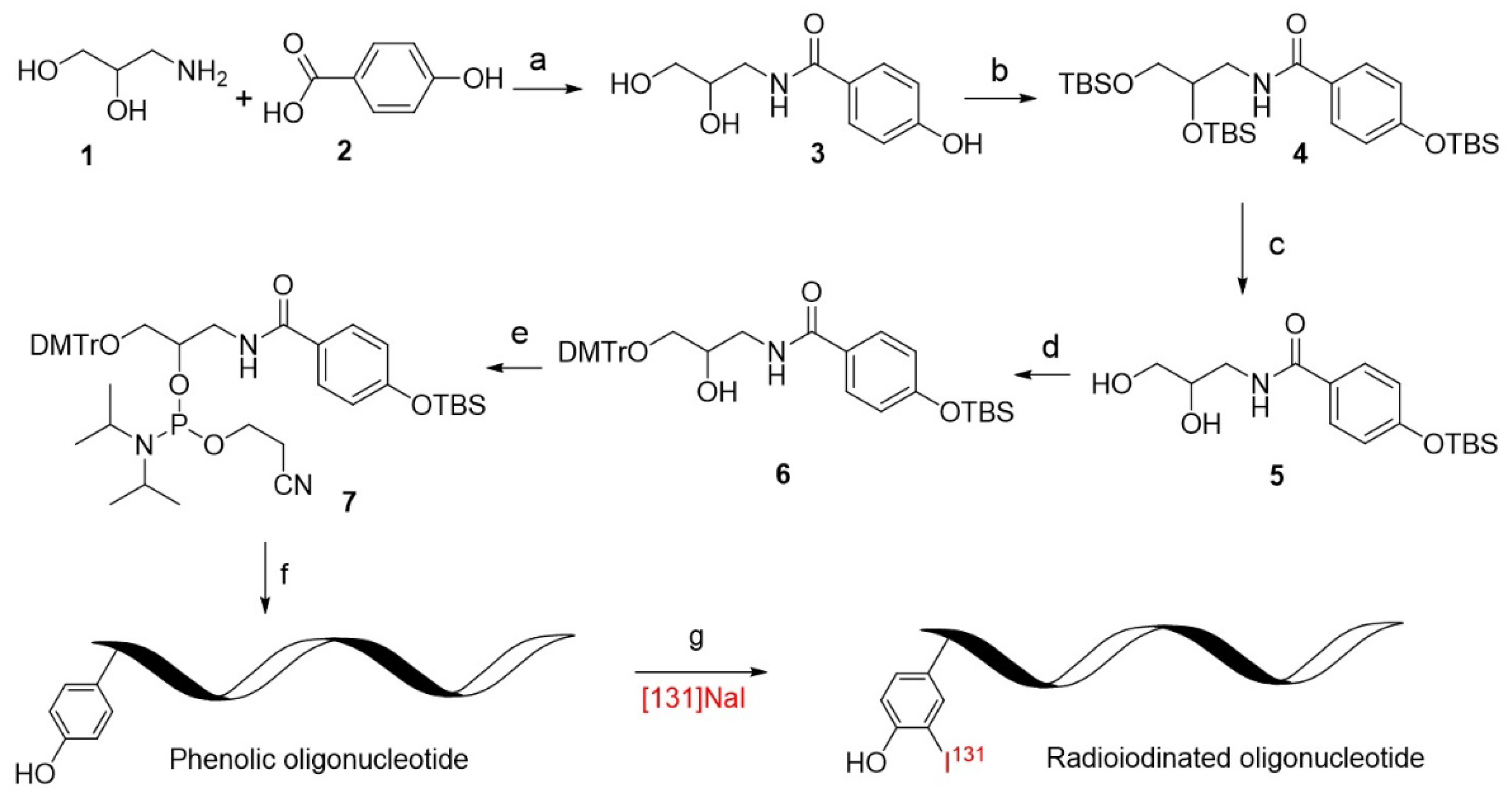
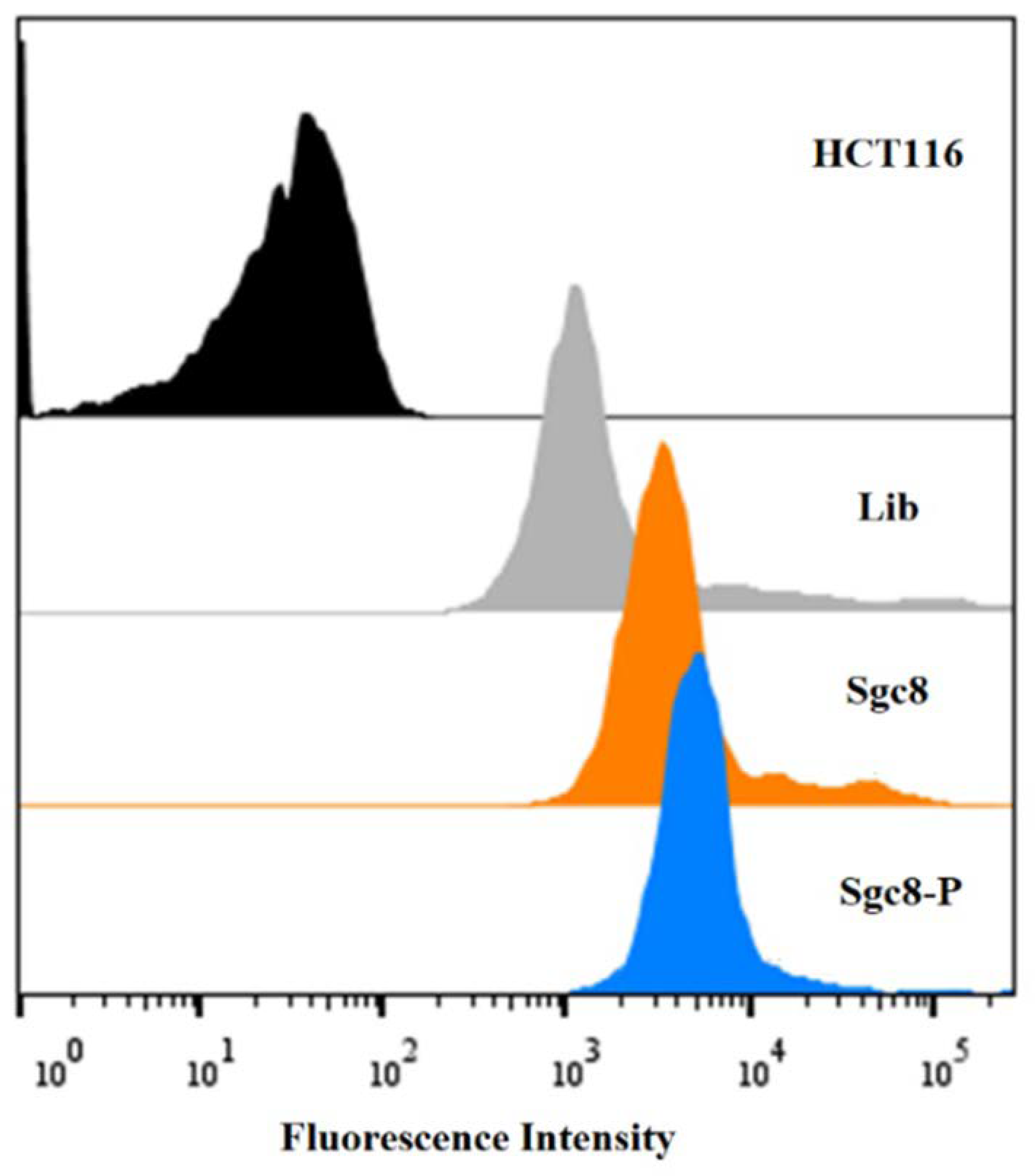
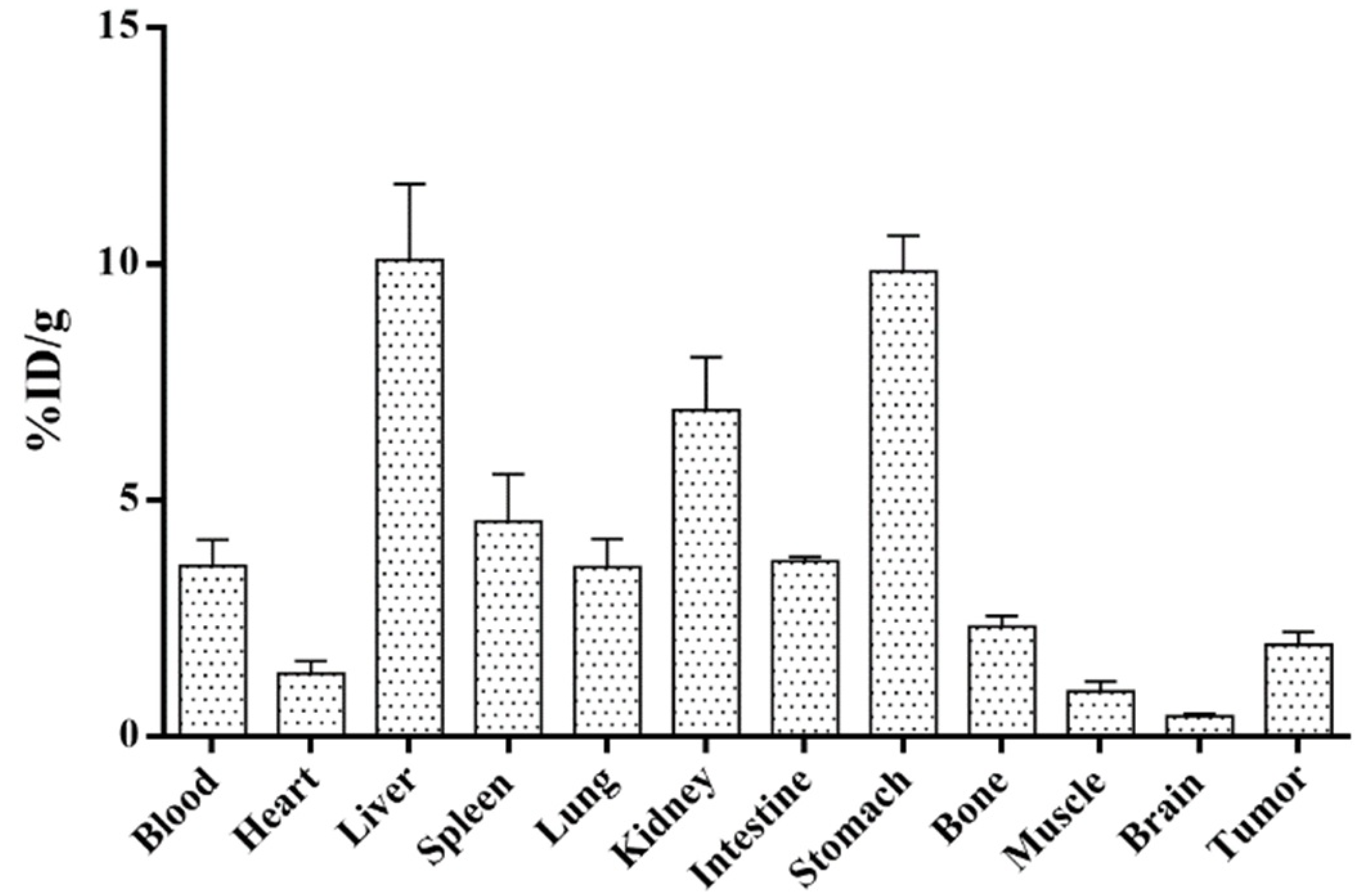
2. Results and Discussion
3. Materials and Methods
3.1. The Preparation of Oligonucleotides
3.2. Radioiodination of the Oligonucleotides
3.3. Biodistribution Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Chen, J.H.; Seeman, N.C. Synthesis from DNA of a Molecule with the Connectivity of a Cube. Nature 1991, 350, 631–633. [Google Scholar] [CrossRef]
- Pianowski, Z.; Gorska, K.; Oswald, L.; Merten, C.A.; Winssinger, N. Imaging of MRNA in Live Cells Using Nucleic Acid-Templated Reduction of Azidorhodamine Probes. J. Am. Chem. Soc. 2009, 131, 6492–6497. [Google Scholar] [CrossRef] [PubMed]
- Bouvier-Muller, A.; Duconge, F. Application of Aptamers for in Vivo Molecular Imaging and Theranostics. Adv. Drug Deliv. Rev. 2018, 134, 94–106. [Google Scholar] [CrossRef]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, J.A.; Witzigmann, D.; Thomson, S.B.; Chen, S.; Leavitt, B.R.; Cullis, P.R.; Meel, R. The Current Landscape of Nucleic Acid Therapeutics. Nat. Nanotechnol. 2021, 16, 630–643. [Google Scholar] [CrossRef]
- Scoles, D.R.; Meera, P.; Schneider, M.D.; Paul, S.; Dansithong, W.; Figueroa, K.P.; Hung, G.; Rigo, F.; Bennett, C.F.; Otis, T.S.; et al. Antisense Oligonucleotide Therapy for Spinocerebellar Ataxia Type 2. Nature 2017, 544, 362–366. [Google Scholar] [CrossRef]
- Fougerolles, A.; Vornlocher, H.P.; Maraganore, J.; Lieberman, J. Interfering with Disease: A Progress Report on SiRNA-Based Therapeutics. Nat. Rev. Drug Discov. 2007, 6, 443–453. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ng, E.W.M.; Shima, D.T.; Calias, P.; Cunningham, E.T.C., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.; Drabkin, H.A.; Lara, P.N.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. Laber: A Phase II Trial of AS1411 (a Novel Nucleolin-Targeted DNA Aptamer) in Metastatic Renal Cell Carcinoma. Investig. New Drugs 2014, 32, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhu, G.; Mei, L.; Xie, Y.; Ma, H.; Ye, M.; Qing, F.L.; Tan, W. Automated Modular Synthesis of Aptamer-Drug Conjugates for Targeted Drug Delivery. J. Am. Chem. Soc. 2014, 136, 2731–2734. [Google Scholar] [CrossRef]
- Rothlisberger, P.; Gasse, C.; Hollenstein, M. Nucleic Acid Aptamers: Emerging Applications in Medical Imaging, Nanotechnology, Neurosciences, and Drug Delivery. Int. J. Mol. Sci. 2017, 18, 2430. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Aruva, M.R.; Zhang, K.; Shanthly, N.; Cardi, C.A.; Thakur, M.L.; Wickstrom, E. PET Imaging of CCND1 MRNA in Human MCF7 Estrogen Receptor Positive Breast Cancer Xenografts with Oncogene-Specific [64Cu]Chelator-Peptide Nucleic Acid-IGF1 Analog Radiohybridization Probes. J. Nucl. Med. 2007, 48, 1699–1707. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Lee, T.S.; Song, I.H.; Cho, Y.L.; Chae, J.R.; Yun, M.; Kang, H.; Lee, J.H.; Lim, J.H.; Cho, W.G.; et al. Kang: Hybridization-Based Aptamer Labeling Using Complementary Oligonucleotide Platform for PET and Optical Imaging. Biomaterials 2016, 100, 143–151. [Google Scholar] [CrossRef]
- Li, J.; Zhang, L.; Li, W.; Lei, C.; Cao, Y.; Wang, Y.; Wang, Z.; Pang, H. Preparation and SPECT/CT Imaging of (177)Lu-Labeled Peptide Nucleic Acid (PNA) Targeting CITED1: Therapeutic Evaluation in Tumor-Bearing Nude Mice. Oncol. Targets Ther. 2020, 13, 487–496. [Google Scholar] [CrossRef]
- Varmira, K.; Hosseinimehr, S.J.; Noaparast, Z.; Abedi, S.M. A HER2-Targeted RNA Aptamer Molecule Labeled with 99mTc for Single-Photon Imaging in Malignant Tumors. Nucl. Med. Biol. 2013, 40, 980–986. [Google Scholar] [CrossRef]
- Jacobson, O.; Weiss, I.D.; Wang, L.; Wang, Z.; Yang, X.; Dewhurst, A.; Ma, Y.; Zhu, G.; Niu, G.; Kiesewetter, D.O.; et al. 18F-Labeled Single-Stranded DNA Aptamer for PET Imaging of Protein Tyrosine Kinase-7 Expression. J. Nucl. Med. 2015, 56, 1780–1785. [Google Scholar] [CrossRef]
- Wang, L.; Jacobson, O.; Avdic, D.; Rotstein, B.H.; Weiss, I.D.; Collier, L.; Chen, X.; Vasdev, N.; Liang, S.H. Ortho-Stabilized (18) F-Azido Click Agents and Their Application in PET Imaging with Single-Stranded DNA Aptamers. Angew. Chem. Int. Ed. Engl. 2015, 54, 12777–12781. [Google Scholar] [CrossRef]
- McCready, V.R. Radioiodine—the Success Story of Nuclear Medicine: 75th Anniversary of the First Use of Iodine-131 in Humans. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 179–182. [Google Scholar] [CrossRef]
- Ferris, T.; Carroll, L.; Jenner, S.O.E. Aboagye: Use of Radioiodine in Nuclear Medicine-A Brief Overview. J. Label. Comp. Radiopharm. 2021, 64, 92–108. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Rosenkrans, Z.T.; Liu, J.; Huang, G.; Luo, Q.Y.; Cai, W. ImmunoPET: Concept, Design, and Applications. Chem. Rev. 2020, 120, 3787–3851. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, M.J.; Bridges, J.W.; Marks, V. Enzyme immunoassay: A review. Ann. Clin. Biochem. 1979, 16, 221–240. [Google Scholar] [CrossRef]
- Vogel, A.; Saborowski, A. Adjuvant (131)I-Metuximab in Hepatocellular Carcinoma: A New Option for an Old Drug? Lancet Gastroenterol. Hepatol. 2020, 5, 517–519. [Google Scholar] [CrossRef]
- Chen, H.; Nan, G.; Wei, D.; Zhai, R.Y.; Huang, M.; Yang, W.W.; Xing, B.C.; Zhu, X.; Xu, H.F.; Wang, X.D.; et al. Hepatic Artery Injection of (131)I-Metuximab Combined with Transcatheter Arterial Chemoembolization for Unresectable Hepatocellular Carcinoma: A Prospective Non-Randomized, Multicenter Clinical Trial. J. Nucl. Med. 2022, 63, 556–559. [Google Scholar] [CrossRef]
- Rollag, M.D.; Myers, D.; Osteryoung, J.; Niswender, G.D. Radioiodination of Protein Hormones Using a Constant Potential Thin-Layer Electrochemical Cell. J. Nucl. Biol. Med. 1975, 19, 80–85. [Google Scholar]
- Bryant, M.L.; Nalewaik, R.P.; Tibbs, V.L.; Todaro, G.J. Comparison of Two Techniques for Protein Isolation and Radioiodination by Tryptic Peptide Mapping. Anal. Biochem. 1979, 96, 84–89. [Google Scholar] [CrossRef]
- Hamilton, R.G.; Berson, S.A.; Button, T.M. Protein Radioiodination in a Radioassay Laboratory: Evaluation of Commercial Na125I Reagents and Related Biohazards. J. Immunoass. 1980, 1, 435–448. [Google Scholar] [CrossRef]
- Arano, Y.; Wakisaka, K.; Ohmomo, Y.; Uezono, T.; Mukai, T.; Motonari, H.; Shiono, H.; Sakahara, H.; Konishi, J. Maleimidoethyl 3-(Tri-n-Butylstannyl)Hippurate: A Useful Radioiodination Reagent for Protein Radiopharmaceuticals to Enhance Target Selective Radioactivity Localization. J. Med. Chem. 1994, 37, 2609–2618. [Google Scholar] [CrossRef]
- Wakisaka, K.; Arano, Y.; Uezono, T.; Akizawa, H.; Ono, M.; Kawai, K.; Ohomomo, Y.; Nakayama, M.; Saji, H. A Novel Radioiodination Reagent for Protein Radiopharmaceuticals with L-Lysine as a Plasma-Stable Metabolizable Linkage to Liberate m-Iodohippuric Acid after Lysosomal Proteolysis. J. Med. Chem. 1997, 40, 2643–2652. [Google Scholar] [CrossRef]
- Shimura, N.; Sogawa, Y.; Kawakita, Y.; Ikekita, M.; Yamazaki, N.; Kojima, S. Radioiodination of Glycoprotein-Conjugated Liposomes by Using the Bolton-Hunter Reagent and Biodistribution in Tumor-Bearing Mice. Nucl. Med. Biol. 2002, 29, 491–496. [Google Scholar] [CrossRef]
- Butler, S.R.; Lam, R.W.; Fisher, D.A. Iodination of thyroliberin by use of Iodogen. Clin. Chem. 1984, 30, 547–548. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.P.; Mountford, P.J.; Baird, A.C.; Heyderman, E.; Richardson, T.C.; Coakley, A.J. An Improved Iodogen Method of Labelling Antibodies with 123I. Nucl. Med. Commun. 1986, 7, 355–362. [Google Scholar] [CrossRef]
- Chen, J.; Wang, M.; Joyce, A.; DeFranco, D.; Kavosi, M.; Xu, X.; O’Hara, D.M. Comparison of Succinimidyl [(125)I]Iodobenzoate with Iodogen Iodination Methods to Study Pharmacokinetics and ADME of Biotherapeutics. Pharm. Res. 2014, 31, 2810–2821. [Google Scholar] [CrossRef] [PubMed]
- Commerford, S.L. Iodination of Nucleic Acids in Vitro. Biochemistry 1971, 10, 1993–2000. [Google Scholar] [CrossRef]
- Commerford, S.L. In Vitro Iodination of Nucleic Acids. Methods Enzymol. 1980, 70, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Kuhnast, B.; Dolle, F.; Terrazzino, S.; Rousseau, B.; Loc’h, C.; Vaufrey, F.; Hinnen, F.; Doignon, I.; Pillon, F.; David, C.; et al. Tavitian: General Method to Label Antisense Oligonucleotides with Radioactive Halogens for Pharmacological and Imaging Studies. Bioconjug. Chem. 2000, 11, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Fontanel, M.; Bazin, H.; Teoule, R. End Attachment of Phenol-Oligonucleotide Conjugates to Diazotized Cellulose. Bioconjug. Chem. 1993, 4, 380–385. [Google Scholar] [CrossRef]
- Dougan, H.; Hobbs, J.B.; Weitz, J.I.; Lyster, D.M. Synthesis and Radioiodination of a Stannyl Oligodeoxyribonucleotide. Nucleic Acids Res. 1997, 25, 2897–2901. [Google Scholar] [CrossRef]
- Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.J.; Tan, W. Aptamers Evolved from Live Cells as Effective Molecular Probes for Cancer Study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Qin, Y.; Liu, D.; Geng, X.; Wang, C.; Ding, D.; Ding, X.; Xia, Q.; Liu, J.; Wang, R.; et al. Site-Specific Radioiodination of Oligonucleotides with a Phenolic Element in a Programmable Approach. Molecules 2022, 27, 6257. https://doi.org/10.3390/molecules27196257
Zhao H, Qin Y, Liu D, Geng X, Wang C, Ding D, Ding X, Xia Q, Liu J, Wang R, et al. Site-Specific Radioiodination of Oligonucleotides with a Phenolic Element in a Programmable Approach. Molecules. 2022; 27(19):6257. https://doi.org/10.3390/molecules27196257
Chicago/Turabian StyleZhao, Haitao, Yu Qin, Dunfang Liu, Xinyao Geng, Cheng Wang, Ding Ding, Xuan Ding, Qian Xia, Jianjun Liu, Ruowen Wang, and et al. 2022. "Site-Specific Radioiodination of Oligonucleotides with a Phenolic Element in a Programmable Approach" Molecules 27, no. 19: 6257. https://doi.org/10.3390/molecules27196257
APA StyleZhao, H., Qin, Y., Liu, D., Geng, X., Wang, C., Ding, D., Ding, X., Xia, Q., Liu, J., Wang, R., & Tan, W. (2022). Site-Specific Radioiodination of Oligonucleotides with a Phenolic Element in a Programmable Approach. Molecules, 27(19), 6257. https://doi.org/10.3390/molecules27196257







