Analysis of Polar Lipids in Hemp (Cannabis sativa L.) By-Products by Ultra-High Performance Liquid Chromatography and High-Resolution Mass Spectrometry
Abstract
:1. Introduction
2. Results
2.1. Extraction, Fractionation, and Solid-Phase Extraction (SPE)
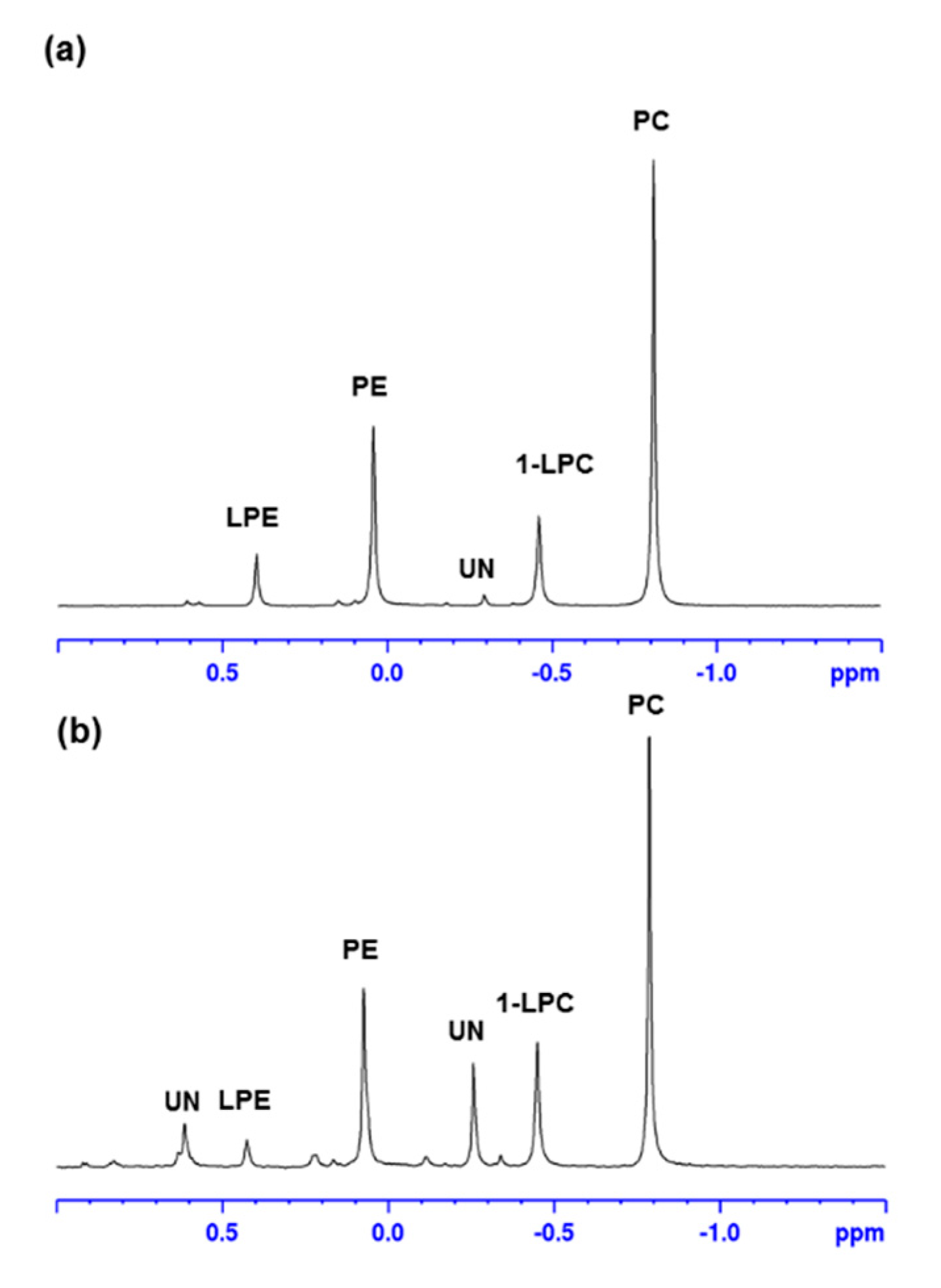
2.2. H NMR Analysis of SPE Fractions
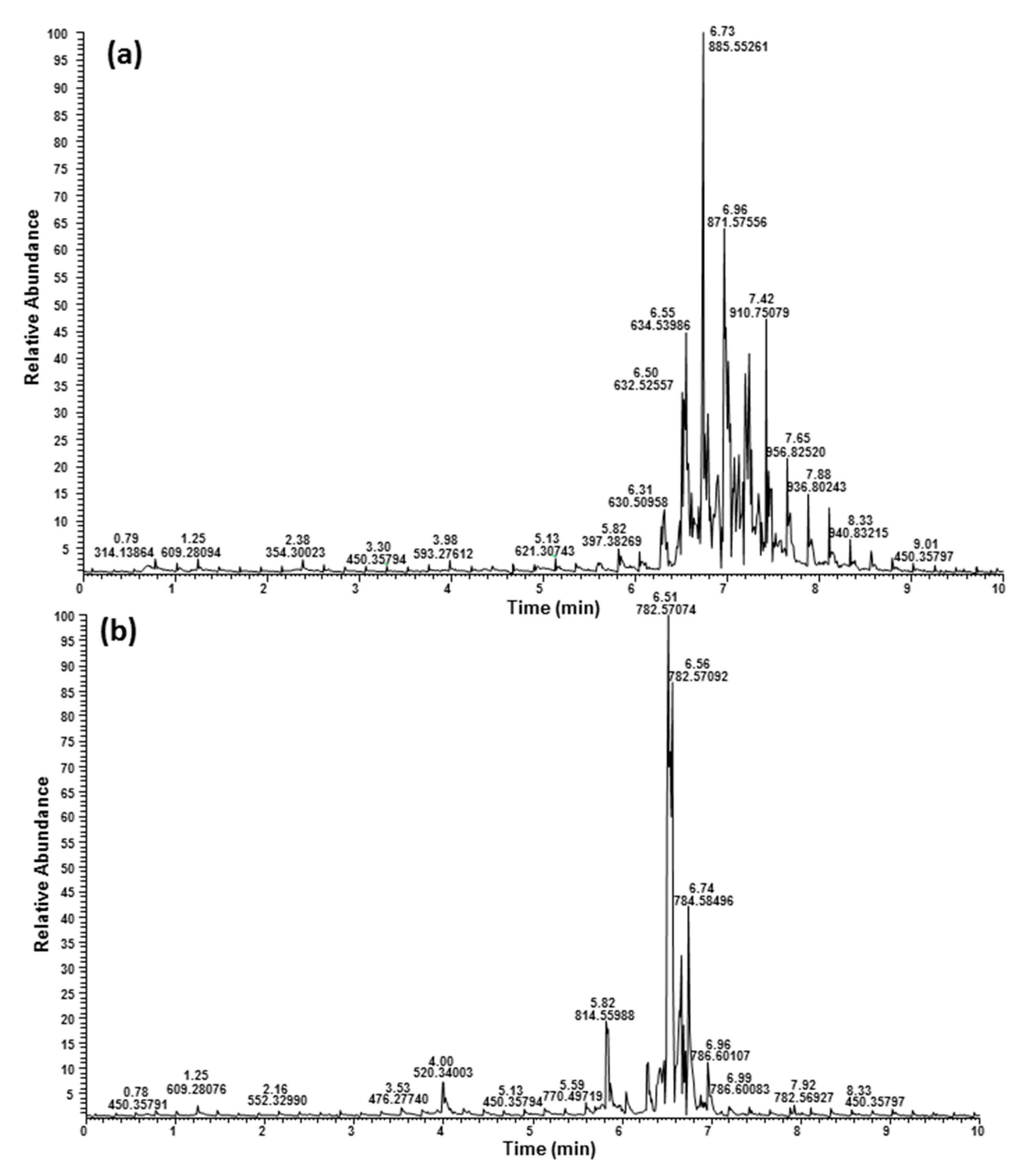
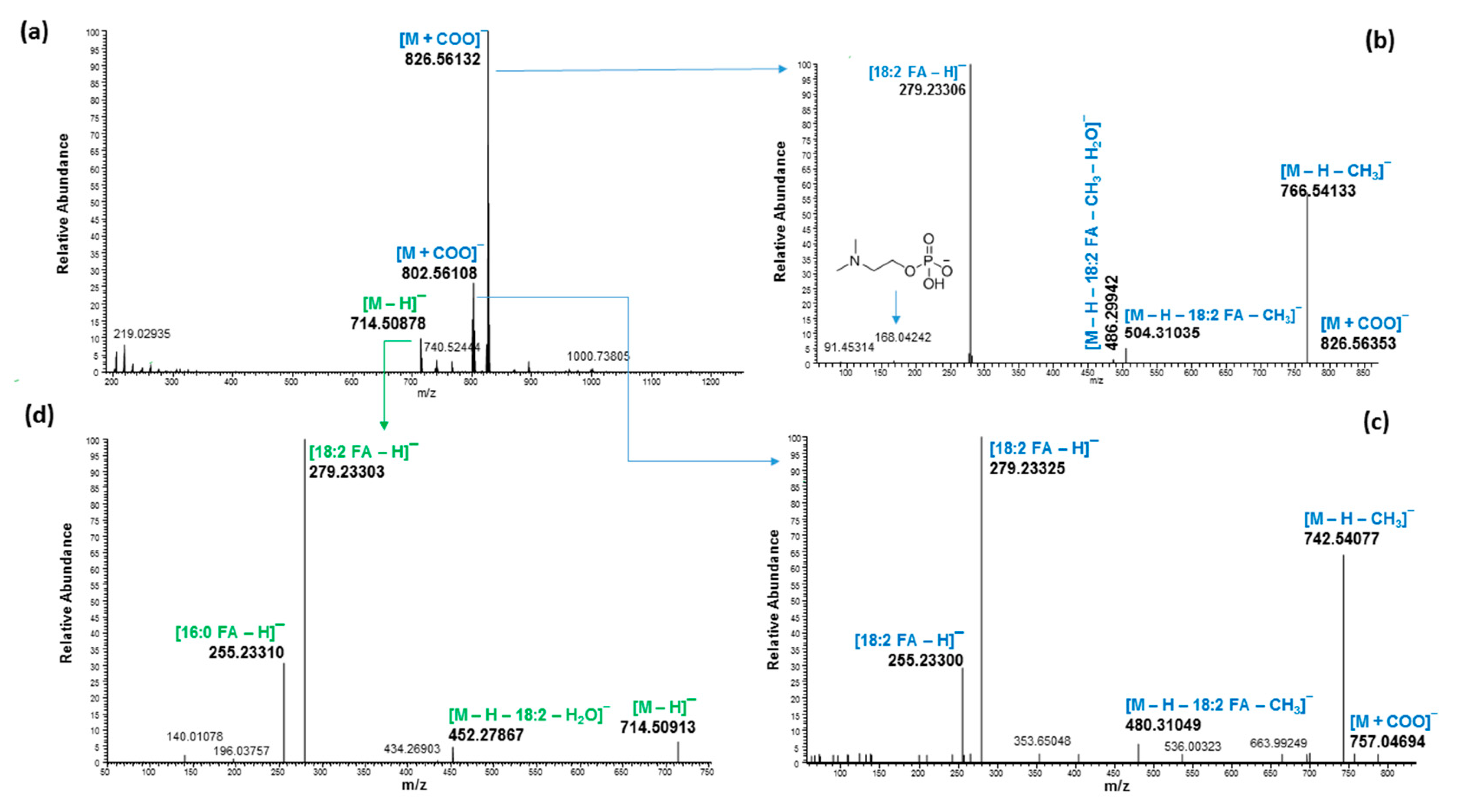
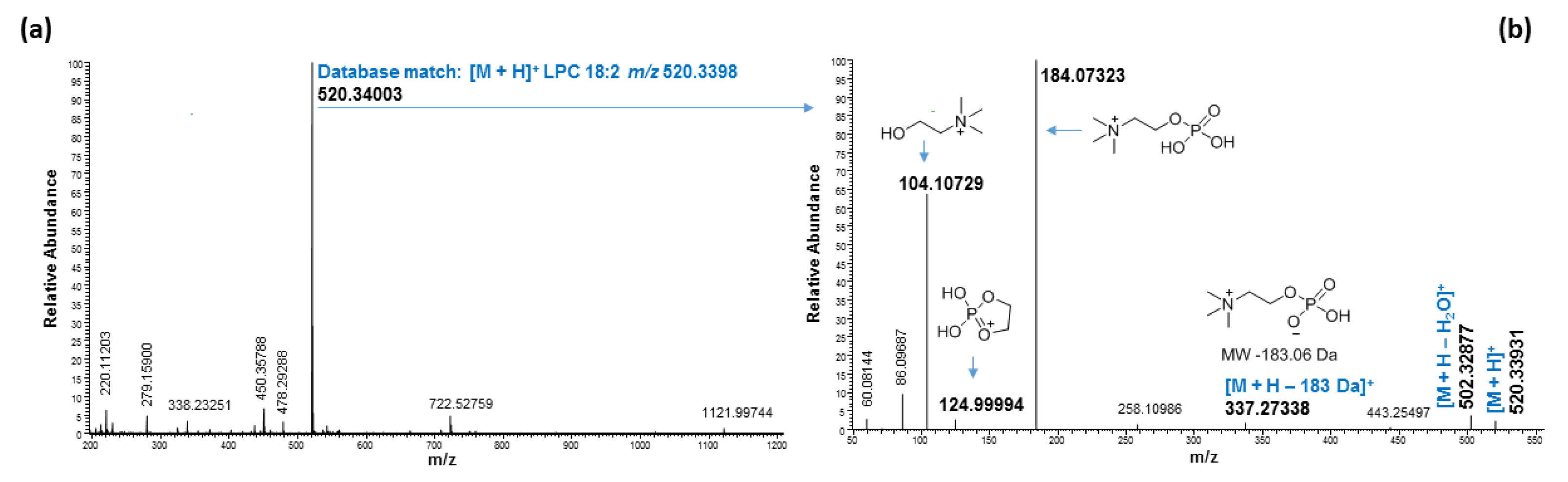
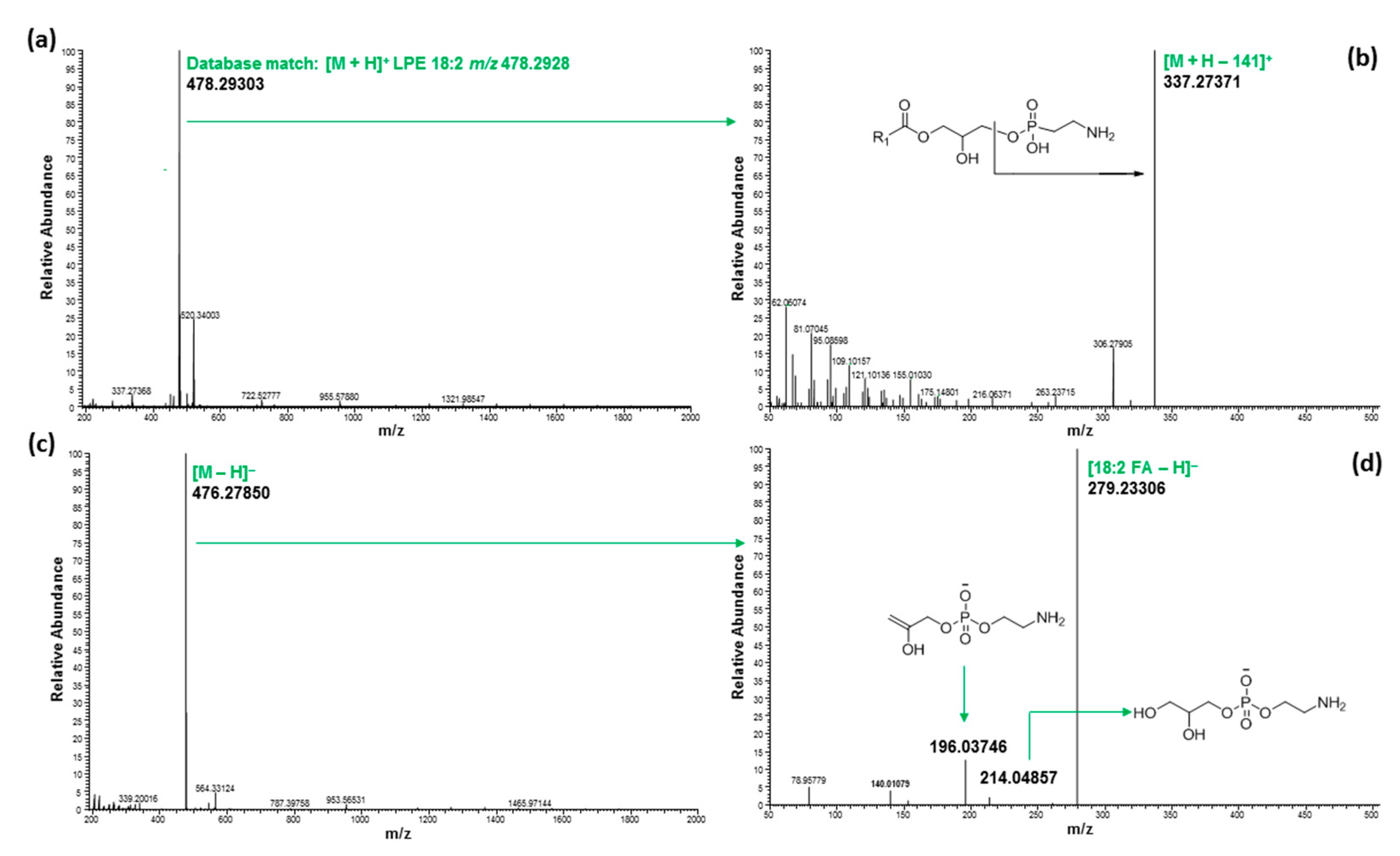
2.3. Ultrahigh-Performance Liquid Chromatography and High-Resolution Mass Spectrometry (UHPLC/HRMS) Analysis
2.4. Fatty Acid Analysis of SPE Fractions
2.5. P NMR and HPLC Purification of Methanol Fraction
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. General
5.2. Research Material
5.3. Polar Lipid Extraction, Fractionation, and Solid-Phase Extraction (SPE)
5.4. UHPLC/HRMS Analysis
5.5. Fatty Acid Analysis
5.6. HPLC Purification of Major PCs
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Cherney, J.H.; Small, E. Industrial hemp in North America: Production politics and potential. Agronomy 2016, 6, 58. [Google Scholar] [CrossRef]
- Bonini, S.A.; Premoli, M.; Tambaro, S.; Kumar, A.; Maccarinelli, G.; Memo, M.; Mastinu, A. Cannabis sativa: A comprehensive ethnopharmacological review of a medicinal plant with a long history. J. Ethnopharmacol. 2018, 227, 300–315. [Google Scholar] [CrossRef] [PubMed]
- Rupasinghe, H.P.V.; Davis, A.; Kumar, S.K.; Murry, B.; Dheljazkov, V.D. Industrial hemp (Cannabis sativa subsp. sativa) as an emerging source for value-added functional food ingredients and nutraceuticals. Molecules 2020, 25, 4078. [Google Scholar] [CrossRef]
- Crescnte, G.; Piccolella, S.; Esposito, A.; Scognamiglio, M.; Fiorentino, A.; Pacifico, S. Chemical composition and nutraceutical properties of hempseed: An ancient food with actual functional value. Phytochem. Rev. 2018, 17, 733–749. [Google Scholar] [CrossRef]
- Oseyko, M.; Sova, N.; Lutsenko, M.; Kalyna, V. Chemical aspects of the composition of industrial hemp products. Food Technol. 2019, 8, 544–558. [Google Scholar] [CrossRef]
- Wang, Q.; Xiong, Y.L. Processing, nutrition, and functionality of hempseed protein: A review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 936–952. [Google Scholar] [CrossRef] [PubMed]
- Nasrollahzadeh, F.; Roman, L.; Swaraj, V.J.S.; Ragavan, K.V.; Vidal, N.P.; Dutcher, J.R.; Martinez, J.M. Hemp (Cannabis sativa L.) protein concentrates from wet and dry industrial fractionation: Molecular properties, nutritional composition, and anisotropic structuring. Food Hydrocoll. 2022, 131, 107755. [Google Scholar] [CrossRef]
- Fathordoobady, F.; Singh, A.; Kitts, D.D.; Singh, A.P. Hemp (Cannabis sativa L.) extract: Anti-microbial properties, methods of extraction, and potential oral delivery. Food Rev. Int. 2019, 35, 664–684. [Google Scholar] [CrossRef]
- Alonso-Esteban, J.I.; Pinela, J.; Ćirić, A.; Calhelha, R.C.; Soković, M.; Ferreira, I.C.F.R.; Barros, L.; Torija-Isasa, E.; de CortesSánchez-Mata, M. Chemical composition and biological activities of whole and dehulled hemp (Cannabis sativa L.) seeds. Food Chem. 2022, 374, 131754. [Google Scholar] [CrossRef]
- Lupescu, M. Industrial hemp production tread and regulation. In Global Agriculture Information Network; GAIN Report Number: CA19030; USDA Foreign Agriculture Service: Ottawa, ON, Canada, 2019. [Google Scholar]
- Ohta, K.; Mizushina, Y.; Hirata, N.; Takemura, M.; Sugawara, F.; Matsukage, A.; Yoshida, S.; Sakaguchi, K. Sulfoquinovosyldiacylglycerol, KM043, a new potent inhibitor of eukaryotic DNA polymerases and HIV-reverse transcriptasetype 1 from a marine red alga, Gigartina tenella. Chem. Pharm. Bull. 1998, 46, 684–686. [Google Scholar] [CrossRef] [Green Version]
- Sun, N.; Chen, J.; Wang, D.; Lin, S. Advance in food-derived phospholipids: Sources, molecular species and structure as well as their biological activities. Trends Food Sci. Technol. 2018, 80, 199–211. [Google Scholar] [CrossRef]
- Christensen, L.P. Galactolipids as potential health promoting compounds in vegetable foods. Recent Pat. Food Nutr. Agric. 2009, 1, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Povedano, M.M.; Callado, C.S.-C.; Priego-Capote, F.; Ferreiro-Vera, C. Untargeted characterization of extracts from Cannabis sativa L. cultivars by gas and liquid chromatography coupled to mass spectrometry in high resolution mode. Talanta 2020, 208, 120384. [Google Scholar] [CrossRef] [PubMed]
- Cerrato, A.; Capriotti, A.L.; Montone, C.M.; Aita, S.E.; Cannazza, G.; Citti, C.; Piovesana, S.; Aldo, L. Analytical methodologies for lipodomics in hemp plant. In Mass Spectrometry-Based Lipidomics: Methods and Protocols, Methods in Molecular Biology; Hsu, F.-F., Ed.; Springer Nature: Berlin, Germany, 2021; Volume 2306, pp. 257–273. [Google Scholar]
- Arena, P.; Rigano, F.; Guarnaccia, P.; Dugo, P.; Mondello, L.; Trovato, E. Elucidation of lipid composition of hemp (Cannabis sativa L.) products by means of gas chromatography and ultra-high performance liquid chromatography coupled to mass spectrometry detection. Molecules 2022, 27, 3358. [Google Scholar] [CrossRef] [PubMed]
- Banskota, A.H.; Jones, A.; Hui, J.P.M.; Stefanova, R. Triacylglycerols and other lipids from hemp by-products. Molecules 2022, 27, 2339. [Google Scholar] [CrossRef]
- Banskota, A.H.; Tibbetts, S.M.; Jones, A.; Stefanova, R.; Behnke, J. Biochemical characterization and in vitro digestibility of protein isolated from hemp (Cannabis sativa L.) by-products for salmonoid feed applications. Molecules 2022, 27, 4794. [Google Scholar] [CrossRef]
- Leizer, C.; Ribnicky, D.; Poulev, A.; Dushenkov, S.; Raskin, I. The composition of hemp seed oil and its potential as an important source of nutrition. J. Nutraceuticals Funct. Med. Foods 2000, 2, 35–53. [Google Scholar] [CrossRef]
- Pojić, M.; Mišan, A.; Sakač, M.; Hadnađev, T.D.; Šarić, B.; Milovanović, I.; Hadnađev, M. Characterization of byproducts originating from hemp oil processing. J. Agric. Food Chem. 2014, 62, 12436–12442. [Google Scholar] [CrossRef]
- Aladić, K.; Jarni, K.; Barbir, T.; Vidović, S.; Vladić, J. Supercritical CO2 extraction of hemp (Cannabis sativa L.) seed oil. Ind. Crops Prod. 2015, 76, 472–478. [Google Scholar] [CrossRef]
- Oomah, B.D.; Bussonb, M.; Godfrey, D.V.; Drovera, J.C.G. Characteristics of hemp (Cannabis sativa L.) seed oil. Food Chem. 2002, 76, 33–43. [Google Scholar] [CrossRef]
- Ellison, C.; Moreno, T.; Catchpole, O.; Fenton, T.; Lagutin, K.; MacKenzie, A.; Mitchell, K.; Scott, D. extraction of hemp seed using near-critical CO2, propane and dimethyl ether. J Supercrit Fluids 2021, 173, 105218. [Google Scholar] [CrossRef]
- Buré, C.; Solgadi, A.; Yen-Nicolaÿ, S.; Bardeau, T.; Libong, D.; Abreu, S.; Chaminade, P.; Subra-Paternault, P.; Cansell, M. Electrospray mass spectrometry as a tool to characterize phospholipid composition in plant cakes. Eur. J. Lipid. Technol. 2016, 118, 1282–1292. [Google Scholar] [CrossRef]
- Antonelli, M.; Benedetti, B.; Cannazza, G.; Cerrato, A.; Citti, C.; Montone, C.M.; Piovesana, S.; Laganà, A. New insights in hemp chemical composition: A comprehensive polar lipidome characterization by combining solid phase enrichment, high-resolution mass spectrometry, and cheminformatics. Anal. Bioanal. Chem. 2020, 412, 413–423. [Google Scholar] [CrossRef]
- Ryckebosch, E.; Muylaert, K.; Foubert, I. Optimization of an analytical procedure for extraction of lipids from microalgae. J. Am. Oil Chem. Soc. 2012, 89, 189–198. [Google Scholar] [CrossRef]
- Garcia, R.; Pires, A.; Martins, N.; Carvalho, T.; Burke, A.J.; Cabrita, M.J. Assessment of the triacylglycerol fraction of olive oil by 1D-NMR spectrometry; exploring the usefulness of DEPT tool on the peak assignments of 13C NMR spectra. Eur. Food Res. Tech. 2019, 245, 2479–2488. [Google Scholar] [CrossRef]
- Melo, T.; Alves, E.; Azevedo, V.; Martins, A.S.; Neves, B.; Domingues, P.; Calado, R.; Abreu, M.H.; Domingus, M.R. Lipidomics as a new approach for the bioprossing of marine macroalgae–unraveling the polar lipid and fatty acid composition of Chondrus crispus. Algal Res. 2015, 8, 181–191. [Google Scholar] [CrossRef]
- MacDougall, K.M.; McNichol, J.; McGinn, P.J.; O’Leary, S.J.B.; Melanson, J.E. Triacylglycerol profiling of microalgae strains for biofuel feedstock by liquid chromatography-high-resolution mass spectrometry. Anal. Bioannal. Chem. 2011, 401, 2609–2616. [Google Scholar] [CrossRef] [PubMed]
- Godzien, J.; Ciborowski, M.; Martínez-Alcázar, M.P.; Samczuk, P.; Kretowski, A.; Barbas, C. Rapid and reliable identification of phospholipids for untargeted metabolomics with LC-ESI-QTOF-MS/MS. J. Proteome Res. 2015, 14, 3204–3216. [Google Scholar] [CrossRef]
- Pi, J.; Wu, X.; Feng, Y. Fragmentation patterns of five types of phospholipids by ultra-high-performance liquid chromatography electrospray ionization quadrupole time-of-flight tandem mass spectrometry. Anal. Methods 2016, 8, 1319–1332. [Google Scholar] [CrossRef]
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS online tools for lipid research. Nucl. Acids Res. 2007, 35, W606–W612. [Google Scholar] [CrossRef] [Green Version]
- Hsu, F.-F.; Turk, J. Electrospray ionization with low energy collosionally activated dissociation Tandem mass spectrometry of glycerophospholipids: Mechanisms of fragmentation and structure characterization. J. Chromatogr B Analyt. Technol. Biomed. Life Sci. 2009, 877, 2673–2695. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.H.; Gage, D.A.; Sweeley, C.C. Characterization of diacylglycerylphosphocholine molecular species by FAB-CAD-MS/MS: A general method not sensitive to the nature of the fatty acyl groups. J. Am. Soc. Mass Spectrom. 1992, 3, 71–78. [Google Scholar] [CrossRef]
- Fang, J.; Barcelona, M.J. Structural determination and quantative analysis of bacterial phospholipids using liquid chromatography/electrospray ionization/mass spectrometry. J. Microbiol. Methods 1998, 33, 23–35. [Google Scholar] [CrossRef]
- Han, X.; Gross, R.W. Structural determination of lysophospholipid regioisomers by electrospray ionization Tandem mass spectrometry. J. Am. Chem. Soc. 1996, 118, 451–457. [Google Scholar] [CrossRef]
- Lee, J.Y.; Min, H.K.; Moon, M.H. Simultaneous profiling of lysophospholipids and phospholipids from human plasma by nanoflow liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2011, 400, 2953–2961. [Google Scholar] [CrossRef]
- Calvano, C.D.; Bianco, M.; Ventura, G.; Losito, I.; Palmisano, F.; Cataldi, T.R.I. Analysis of phospholipids, lysophospholipids, and their linked fatty acyl chains in yellow lupin seeds (Lupinus luteus L.) by liquid chromatography and tandem mass spectrometry. Molecules 2020, 25, 805. [Google Scholar] [CrossRef]
- Monakhova, Y.B.; Diehl, B.W.K. Automated multicomponent phospholipid analysis using 31P NMR spectroscopy: Example of vegetable lecithin and krill oil. Anal. Bioanal. Chem. 2018, 410, 7891–7900. [Google Scholar] [CrossRef] [PubMed]
- AOAC Official Method 991.39; Fatty Acids in Encapsulated Fish Oils and Fish Oil Methyl and Ethyl Esters; AOAC Int.: Gaithersburg, AL, USA, 2000; Chapter 41; pp. 26–28.


| Sample/Fraction | Hemp Cake—Yield (%) | Hemp Seed Hulls—Yield (%) |
|---|---|---|
| EtOH extract-CHCl3 soluble part 1 | 1.5 | 1.6 |
| SPE-Chloroform Fraction 2 | 51.3 | 63.4 |
| SPE-Acetone Fraction 2 | 22.1 | 24.7 |
| SPE-Methanol Fraction 2 | 26.4 | 11.3 |
| HSCA | HSHU | RT (min) | Measured (m/z) | Calculated (m/z) | Error (ppm) | Name: DB 1 | Formula | Ion | Fatty Acid Identity |
|---|---|---|---|---|---|---|---|---|---|
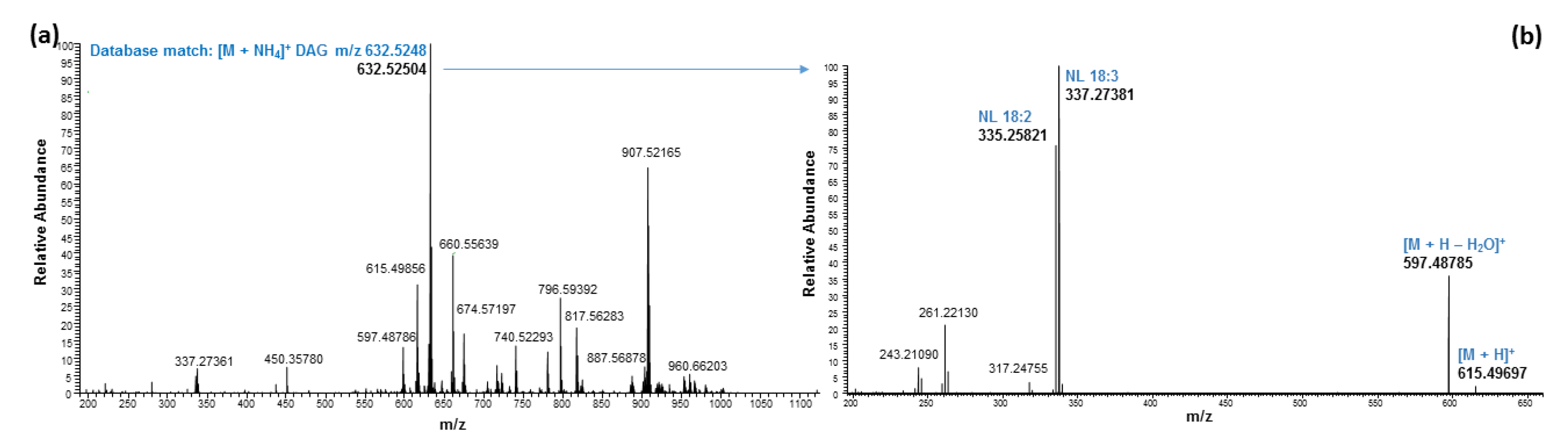
| 6.31 | 630.50958 | 630.50920 | 0.60 | DAG 36:6 | C39H64O5 | [M + NH4]+ | 18:3/18:3 | ||
| 6.47 | 632.52521 | 632.52485 | 0.57 | DAG 36:5 | C39H66O5 | [M + NH4]+ | 18:3/18:2 | ||
| 6.58 | 660.55634 | 660.55615 | 0.29 | DAG 38:5 | C41H70O5 | [M + NH4]+ | 18:3/20:2 | ||
| 6.68 | 634.54065 | 634.54050 | 0.24 | DAG 36:4 | C39H68O5 | [M + NH4]+ | 182:18:2, 18:3/18:1 2 | ||
| 6.81 | 636.55658 | 636.55615 | 0.67 | DAG 36:3 | C39H70O5 | [M + NH4]+ | 18:2/18:1, 18:3/18:0 2 | ||
| 7.00 | 638.57248 | 638.57180 | 1.06 | DAG 36:2 | C39H72O5 | [M + NH4]+ | 18:2/18:0; 18:1/18:1 |
 Low abundance→High abundance 1 Search results from LipidMAPS database (https://www.lipidmaps.org (accessed on 7 September 2022)). 2 Minor component based on the intensity of fragment ion observed for fatty acyl group.
Low abundance→High abundance 1 Search results from LipidMAPS database (https://www.lipidmaps.org (accessed on 7 September 2022)). 2 Minor component based on the intensity of fragment ion observed for fatty acyl group.| HSCA | HSHU | RT (min) | Measured (m/z) | Calculated (m/z) | Error (ppm) | Name: DB 1 | Formula | Ion | FA Identity (R1/R2) |
|---|---|---|---|---|---|---|---|---|---|
| Lysophosphatidylcholines (LPCs) | |||||||||
| 2.42 | 552.32977 | 552.32960 | 0.32 | LPS 20:1 | C26H50NO9P | [M + H]+ | 20:1/0:0 | ||
| 3.35 | 518.32256 | 518.32412 | −3.00 | LPC 18:3 | C26H48NO7P | [M + H]+ | 18:3/0:0 | ||
| 4.00 | 520.34003 | 520.33977 | 0.51 | LPC 18:2 | C26H50NO7P | [M + H]+ | 18:2/0:0 | ||
| 4.20 | 496.33982 | 496.33977 | 0.11 | LPC 16:0 | C24H50NO7P | [M + H]+ | 16:0/0:0 | ||
| 4.50 | 522.35555 | 522.35542 | 0.26 | LPC 18:1 | C26H52NO7P | [M + H]+ | 18:1/0:0 | ||
| 5.00 | 524.37082 | 524.37107 | −0.47 | LPC 18:0 | C26H54NO7P | [M + H]+ | 18:0/0:0 | ||
| Lysophosphatidylethanolamines (LPEs) | |||||||||
| 3.53 | 476.27746 | 476.27717 | 0.61 | LPE 18:3 | C23H42NO7P | [M + H]+ | 18:3/0:0 | ||
| 3.95 | 478.29303 | 478.29281 | 0.46 | LPE 18:2 | C23H44NO7P | [M + H]+ | 18:2/0:0 | ||
| 4.20 | 454.29294 | 454.29281 | 0.29 | LPE 16:0 | C21H44NO7P | [M + H]+ | 16:0/0:0 | ||
| 4.40 | 480.3087 | 480.30847 | 0.48 | LPE 18:1 | C23H46NO7P | [M + H]+ | 18:1/0:0 | ||
| 4.82 | 482.3243 | 482.32411 | 0.39 | LPE 18:0 | C23H48NO7P | [M + H]+ | 18:0/0:0 | ||
| Phosphatidylethanolamines (PEs) | |||||||||
| 6.14 | 736.49145 | 736.49118 | 0.37 | PE 36:6 | C41H70NO8P | [M + H]+ | 18:3/18:3 | ||
| 6.28 | 738.50757 | 738.50683 | 1.00 | PE 36:5 | C41H72NO8P | [M + H]+ | 18:3/18:2 | ||
| 6.40 | 740.52307 | 740.52248 | 0.80 | PE 36:4 | C41H74NO8P | [M + H]+ | 18:2/18:2 | ||
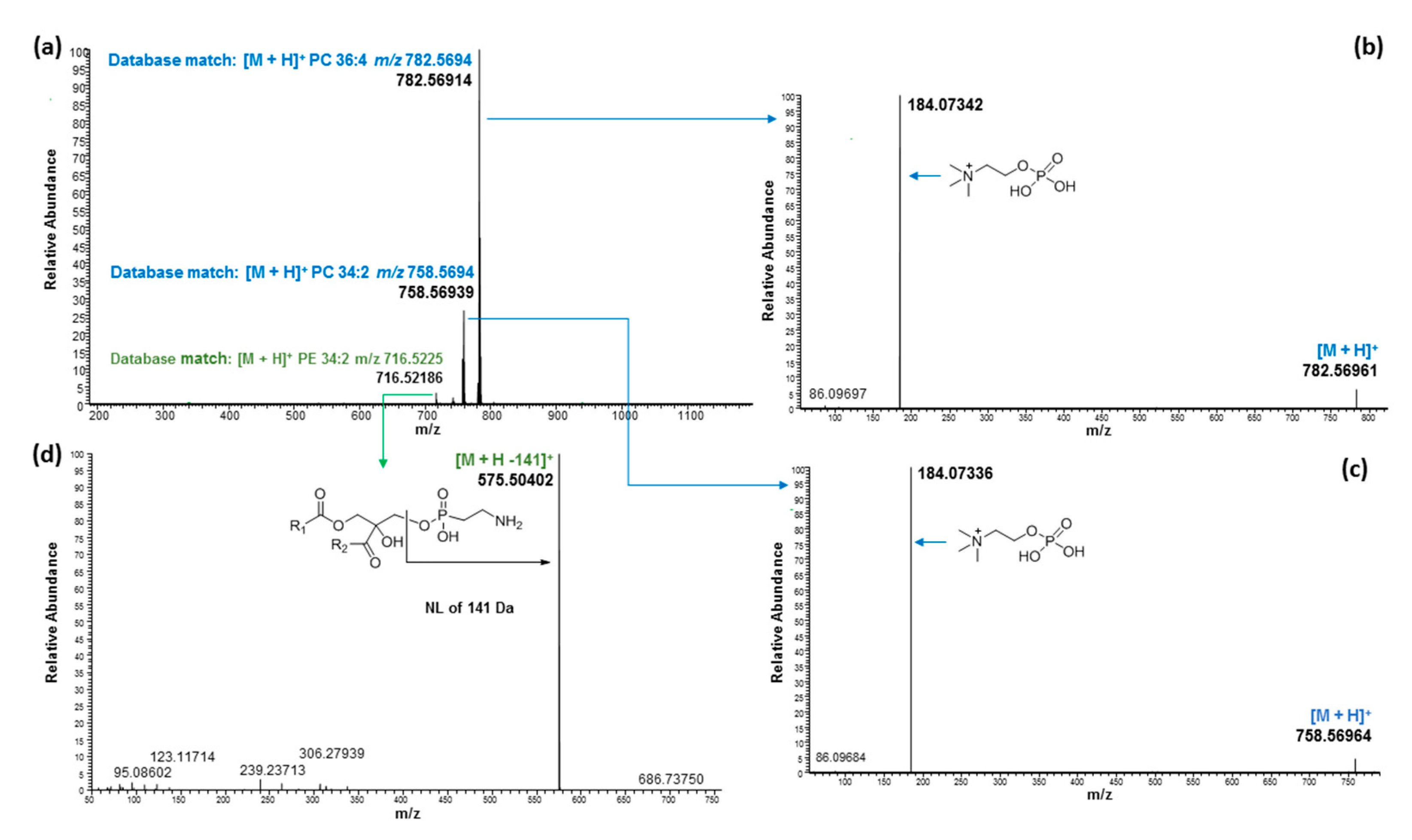
| 6.49 | 716.52250 | 716.52248 | 0.03 | PE 34:2 | C39H74NO8P | [M + H]+ | 16:0/18:2 | ||
| 6.51 | 742.53979 | 742.53813 | 2.23 | PE 36:3 | C41H76NO8P | [M + H]+ | 18:1/18:2; 18:0/18:3 2 | ||
| 6.70 | 744.55438 | 744.55378 | 0.80 | PE 36:2 | C41H78NO8P | [M + H]+ | 18:0/18:2; 18:1/18:1 2 | ||
| Phosphatidylcholines (PCs) | |||||||||
| 6.28 | 778.53900 | 778.53813 | 1.12 | PC 36:6 | C44H76NO8P | [M + H]+ | 18:3/18:3 | ||
| 6.45 | 780.55459 | 780.55378 | 1.04 | PC 36:5 | C44H78NO8P | [M + H]+ | 18:3/18:2 | ||
| 6.60 | 782.57114 | 782.56943 | 2.18 | PC 36:4 | C44H80NO8P | [M + H]+ | 18:2/18:2; 18:3/18:1 2 | ||
| 6.60 | 756.55577 | 756.55378 | 2.63 | PC 34:3 | C42H78NO8P | [M + H]+ | 16:0/18:3 | ||
| 6.66 | 758.57012 | 758.56943 | 0.91 | PC 34:2 | C42H80NO8P | [M + H]+ | 16:0/18:2 | ||
| 6.78 | 784.58642 | 784.58508 | 1.71 | PC 36:3 | C44H82NO8P | [M + H]+ | 18:0/18:3; 18:1/18:2 | ||
| 6.95 | 786.60112 | 786.60073 | 0.49 | PC 36:2 | C44H84NO8P | [M + H]+ | 18:0/18:2; 18:1/18:1 | ||
| 6.96 | 760.58553 | 760.58508 | 0.59 | PC 34:1 | C42H82NO8P | [M + H]+ | 16:0/18:1 | ||
| 7.00 | 812.61623 | 812.61638 | −0.19 | PC 38:3 | C46H86NO8P | [M + H]+ | 20:0/18:3 | ||
| 7.20 | 814.63265 | 814.63203 | 0.76 | PC 38:2 | C46H88NO8P | [M + H]+ | 20:0/18:2 | ||
 Low abundance→High abundance, 1 Search results from LipidMAPS database (https://www.lipidmaps.org (accessed on 7 September 2022)). 2 Minor component based on the intensity of fragment ion observed for fatty acyl group.
Low abundance→High abundance, 1 Search results from LipidMAPS database (https://www.lipidmaps.org (accessed on 7 September 2022)). 2 Minor component based on the intensity of fragment ion observed for fatty acyl group.| Fatty Acid (FA) | HSCA-CHCl3 | HSCA-Acetone | HSCA-MeOH | HSHU-CHCl3 | HSHU-Acetone | HSHU-MeOH |
|---|---|---|---|---|---|---|
| Myristic acid (C14:0) | 1.2 ± 0.2 (0.2) | 1.7 ± 0.1 (1.6) | 1.4 ± 0.7 (0.2) | 0.4 ± 0.7 (0.0) | 0.6 ± 0.5 (0.2) | 1.0 ± 0.2 (0.2) |
| Myristoleic acid (C14:1) | - | 2.5 ± 0.2 (1.7) | - | - | 1.1 ± 0.2 (0.5) | |
| Pentadecanoic acid (C15:0) | - | - | - | - | - | - |
| cis-10-Pentadecenoic acid (C15:1) | - | - | - | - | - | - |
| Palmitic acid (C16:0) | 67.6 ± 1.7 (10.1) | 23.1 ± 2.4 (15.6) | 99.7 ± 4.3 (15.5) | 72.2 ± 10.2 (8.8) | 48.8 ± 0.9 (14.7) | 111.9 ± 2.1 (18.6) |
| Palmitoleic acid (C16:1 n-7) | 1.0 ± 0.1 (0.2) | 1.0 ± 0.3 (0.7) | 1.2 ± 0.3 (0.2) | 2.1 ± 1.5 (0.3) | 10.8 ± 0.6 (3.3) | 1.1 ± 0.2 (0.2) |
| C16:2 n-4 | - | - | - | - | 0.9 ± 0.8 (0.3) | - |
| C17:0 (Heptadecanoic acid) | 0.9 ± 0.0 (0.1) | - | 1.4 ± 0.1 (0.2) | - | - | 1.4 ± 0.0 (0.2) |
| C16:3 n-4 | - | - | - | - | - | - |
| cis-10-Heptadecenoic acid (C17:1) | - | - | - | - | - | - |
| C16:4 n-1 | - | - | - | - | - | - |
| Stearic acid (C18:0) | 19.5 ± 0.5 (2.9) | 4.9 ± 0.7 (3.3) | 27.7 ± 0.7 (4.0) | 21.2 ± 2.1 (2.6) | 11.5 ± 0.2 (3.5) | 22.4 ± 0.4 (3.7) |
| Oleic acid (C18:1 n-9) | 47.8 ± 0.6 (7.1) | 8.3 ± 2.0 (5.6) | 20.9 ± 0.5 (3.3) | 81.5 ± 13.9 (9.9) | 34.3 ± 0.7 (10.4) | 26.2 ± 1.2 (4.4) |
| cis-Vaccenic acid (C18:1 n-7) | 7.1 ± 0.1 (1.1) | 2.3 ± 0.4 (1.6) | 11.6 ± 0.3 (1.8) | - | - | 12.5 ± 0.6 (2.1) |
| Linoleic acid (C18:2 n-6) | 355.1 ± 4.8 (53.0) | 60.9 ± 10.3 (41.1) | 367.1 ± 8.7 (57.2) | 432.8 ± 99.1 (52.2) | 136.5 ± 3.0 (41.3) | 325.0 ± 10.5 (54.1) |
| γ-linolenic acid (C18:3 n-6) | 21.7 ± 0.2 (3.2) | 3.7 ± 0.7 (2.5) | 12.6 ± 0.3 (2.0) | 0.5 ± 0.9 (0.1) | 49.0 ± 1.0 (4.8) | 11.2 ± 0.4 (1.9) |
| α-linolenic acid (C18:3 n-3) | 126.6 ± 1.7 (18.9) | 31.1 ± 4.0 (21.0) | 87.2 ± 3.4 (13.6) | 134.7 ± 36.8 (16.3) | 4.0 ± 0.1 (1.2) | 74.6 ± 3.4 (12.4) |
| Stearidonic acid (C18:4 n-3) | 7.8 ± 0.1 (1.2) | 1.5 ± 0.3 (1.0) | 2.7 ± 0.0 (0.4) | 10.2 ± 2.4 (1.2) | 4.1 ± 0.1 (1.2) | 1.8 ± 0.1 (0.3) |
| Arachidic acid (C20:0) | 6.4 ± 0.2 (0.9) | 1.7 ± 0.2 (1.2) | 3.8 ± 0.2 (0.6) | 6.9 ± 2.8 (0.8) | 1.4 ± 1.4 (0.4) | 4.6 ± 0.2 (0.8) |
| cis-11-Eicosenoic acid (C20:1 n-9) | 2.9 ± 0.0 (0.4) | - | 2.4 ± 0.1 (0.4) | 4.4 ± 0.4 (0.5) | - | 3.0 ± 0.1 (0.5) |
| cis-11,14-Eicosadienoic acid (C20:2) | 1.0 ± 0.0 (0.1) | - | 1.7 ± 0.1 (0.3) | - | - | 1.8 ± 0.1 (0.3) |
| cis-8,11,14-Eicosatrienoic acid (C20:3 n-6) | - | - | - | - | - | - |
| Henicosanoic acid (C21:0) | - | - | - | 0.9 ± 1.6 (0.1) | - | - |
| cis-8,11,14-Eicosatrienoic acid (C20:3 n-3) | - | - | - | - | - | - |
| Arachidonic acid (C20:4 n-6) | - | - | - | - | - | - |
| Eicosapentaenoic acid (C20:5 n-3) | - | - | - | - | - | - |
| Behenic acid (C22:0) | 3.4 ± 0.1 (0.5) | 1.5 ± 0.2 (1.0) | 1.5 ± 0.1 (0.2) | 4.3 ± 0.0 (0.1) | 2.4 ± 1.6 (0.6) | 2.2 ± 0.2 (0.4) |
| Erucic acid (C22:1 n-9) | - | 1.5 ± 0.1 (1.0) | 1.3 ± 0.1 (0.2) | - | - | - |
| Docosadienoic acid (C22:2 n-3) | - | - | - | - | - | - |
| Lignoceric acid (C24:0) | - | - | - | 2.1 ± 0.0 (0.2) | 1.9 ± 1.6 (0.6) | - |
| Docosahexaenoic acid (C22:6 n-3) | - | - | - | - | - | - |
| Others | - | - | - | 15.5 ± 10.7 (1.9) | 10.5 ± 3.3 (3.2) | - |
| Total | 670.1 (100) | 148.1 (100) | 642.2 (100) | 824.7 (100) | 330.2 (100) | 600.8 (100) |
| ∑ SFA | 99.1 (14.8) | 36.8 (24.4) | 134.7 (21.0) | 107.7 (13.1) | 70.5 (21.3) | 143.5 (23.9) |
| ∑ MUFA | 58.8 (8.8) | 14.2 (9.6) | 36.2 (5.6) | 85.9 (10.4) | 47.7 (14.4) | 42.9 (7.1) |
| ∑ PUFA | 512.2 (76.4) | 97.1 (65.5) | 471.4 (73.4) | 615.6 (74.4) | 201.6 (61.0) | 414.4 (69.0) |
| PL-Class | % PL Fraction—HSCA | % PL Fraction—HSHU |
|---|---|---|
| Lysophosphatidylcholines (LPC) | 14.0 | 14.9 |
| Phosphatidylcholines (PC) | 49.3 | 37.1 |
| Lysophosphatidylethanolamines (LPE) | 6.8 | 3.3 |
| Phosphatidylethanolamines (PE) | 24.2 | 20.6 |
| Unknown | <1.0 | 10.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banskota, A.H.; Jones, A.; Hui, J.P.M.; Stefanova, R.; Burton, I.W. Analysis of Polar Lipids in Hemp (Cannabis sativa L.) By-Products by Ultra-High Performance Liquid Chromatography and High-Resolution Mass Spectrometry. Molecules 2022, 27, 5856. https://doi.org/10.3390/molecules27185856
Banskota AH, Jones A, Hui JPM, Stefanova R, Burton IW. Analysis of Polar Lipids in Hemp (Cannabis sativa L.) By-Products by Ultra-High Performance Liquid Chromatography and High-Resolution Mass Spectrometry. Molecules. 2022; 27(18):5856. https://doi.org/10.3390/molecules27185856
Chicago/Turabian StyleBanskota, Arjun H., Alysson Jones, Joseph P. M. Hui, Roumiana Stefanova, and Ian W. Burton. 2022. "Analysis of Polar Lipids in Hemp (Cannabis sativa L.) By-Products by Ultra-High Performance Liquid Chromatography and High-Resolution Mass Spectrometry" Molecules 27, no. 18: 5856. https://doi.org/10.3390/molecules27185856
APA StyleBanskota, A. H., Jones, A., Hui, J. P. M., Stefanova, R., & Burton, I. W. (2022). Analysis of Polar Lipids in Hemp (Cannabis sativa L.) By-Products by Ultra-High Performance Liquid Chromatography and High-Resolution Mass Spectrometry. Molecules, 27(18), 5856. https://doi.org/10.3390/molecules27185856







