Virtual Screening in the Identification of Sirtuins’ Activity Modulators
Abstract
1. Introduction
2. Sirtuins as Targets in Different Pathologies
2.1. SIRT-1 Modulators as Therapeutics
2.1.1. SIRT-1 Modulators in Cancer
2.1.2. SIRT-1 Modulators in Neurodegenerative Disorders
2.1.3. SIRT-1 Modulators in T2D
2.2. SIRT-2 Modulators as Therapeutics
2.2.1. SIRT-2 Modulators in Cancer
2.2.2. SIRT-2 Modulators in Neurodegenerative Disorders
2.2.3. SIRT-2 Modulators in T2D
2.3. SIRT-3 Modulators as Therapeutics
2.3.1. SIRT-3 Modulators in Cancer
2.3.2. SIRT-3 Modulators in Neurodegenerative Disorders
2.3.3. SIRT-3 Modulators in T2D
2.4. SIRT-4 Modulators as Therapeutics
2.4.1. SIRT-4 Modulators in Cancer
2.4.2. SIRT-4 Modulators in Neurodegenerative Disorders
2.4.3. SIRT-4 Modulators in T2D
2.5. SIRT-5 Modulators as Therapeutics
2.5.1. SIRT-5 Modulators in Cancer
2.5.2. SIRT-5 Modulators in Neurodegenerative Disorders
2.5.3. SIRT-5 Modulators in T2D
2.6. SIRT-6 Modulators as Therapeutics
2.6.1. SIRT-6 Modulators in Cancer
2.6.2. SIRT-6 Modulators in Neurodegenerative Disorders
2.6.3. SIRT-6 Modulators in T2D
2.7. SIRT-7 Modulators as Therapeutics
2.7.1. SIRT-7 Modulators in Cancer
2.7.2. SIRT-7 Modulators in Neurodegenerative Disorders
2.7.3. SIRT-7 Modulators in T2D
2.8. Pan-Sirtuin Modulators as Therapeutics
2.8.1. Pan-Sirtuin Modulators in Cancer
2.8.2. Pan-Sirtuin Modulators in Neurodegenerative Disorders
2.8.3. Pan-Sirtuin Modulators in T2D
3. Virtual Screening Strategies Guiding the Discovery of Sirtuins’ Modulators
3.1. Virtual Screening of SIRT-1 Modulators
3.2. Virtual Screening of SIRT-2 Modulators
| Year | Ref. | Title | SIRT(s) | Type of VS | Notes | Selectivity Over Other Isoforms | Experimental Validation | Screened Database (n. of Compounds) | Software(s) | Most Active Compound/Proposed Compound | Activator/Inhibitor | Potency [n. of Proposed Compounds by Computational Study] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2008 | [220] | Oxadizole-carbonylaminothioureas as SIRT-1 and SIRT-2 inhibitors | 1 | SBVS | Pharmacophore based | 1,2 | YES | Maybridge and Leadquest libraries | Unity 4.3.1/Sybyl 7.1 |  oxadizole-carbonylaminothiourea | I | 13 μM (IC50, SIRT-1) |
| 2012 | [52] | Novel acridinedione derivatives: design, synthesis, SIRT-1 enzyme and tumor cell growth inhibition studies. | 1 | SBVS | Target: HM | Not tested | YES | In house database (2500) | Glide, Gold, AutoDock 4.0 | 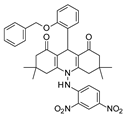 acridinedione derivatives | I | 0.25 μM (IC50) |
| 2014 | [232] | Structure-based drug design of small molecule SIRT-1 modulators to treat cancer and metabolic disorders | 1 | SBVS | Target model for inhibitors: crystal structureTarget model for activators: HM of the allosteric site | Not tested | YES | Asinex (>600000) | Glide 5.0 | 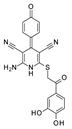 | I, A | 16.35 μM (IC50) |
| 2016 | [234] | Identification of New Inhibitors for Human SIRT-1: An in-silico Approach | 1 | SBVS | Not tested | YES | Drug bank library from ZINC (1716) | AutoDock Vina 1.1.2 |  diphenyl and oxycoumarin derivatives | I | 77.7% inhibition @5μM | |
| 2020 | [59] | Sirtuin 1 Inhibiting Thiocyanates (S1th)-A New Class of Isotype Selective Inhibitors of NAD(+) Dependent Lysine Deacetylases | 1 | SBVS | Iterative in vitro-in silico screenings | 2,3,5 | YES | Small library of previously identified putative SIRT-1 inhibitors | GOLD 5.6 |  Thiocyanates | I | 5.2 μM (IC50) |
| 2021 | [238] | In Silico Design of Novel SIRT-1 Enzyme Activators for the Treatment of Age-related Diseases and Life Span | 1 | SBVS | Not tested | NO | Zinc (150 000) | Information not available |  acebutolol and others | A | NC [7] | |
| 2009 | [222] | Pharmacophore Mapping and Virtual Screening for SIRT-1 Activators | 1 | LBVS | Pharmacophore based | Not tested | NO | Maybridge | HipHop module/CATALYST | isothiazole scaffold benzimidazole scaffold | A | NC† [7] |
| 2014 | [224] | Theoretical approaches to identify the potent scaffold for human SIRT-1 activator: Bayesian modeling and density functional theory | 1 | LBVS | Bayesian model, pharmacophore model | Not tested | NO | Maybridge (60,000), Chembridge (50,000), NCI (200,000), and ChemDiv (700 000) | Discovery Studio v 3.1 | Various | A | NC [16] |
| 2016 | [244] | Ligand-based virtual screening and inductive learning for identification of SIRT-1 inhibitors in natural products | 1 | LBVS | Inductive logic programming | Not tested | NO | Traditional Chinese Medicines-Taiwan Database and Traditional Chinese Medicine Integrated Database (1 444 880) | DMax Chemistry Assistant software | Various | I | NC [3] |
| 2015 | [225] | Ligand and structure-based approaches for the identification of SIRT-1 activators. | 1 | LB-SB | LBVS: Pharmacophore basedSBVS Target model: HM of Sirt-1 | Not tested | NO | ZINC database | DISCOtech, GASP/SYBYL X 1.2 (pharmacophore)Surflex-Dock/SYBYL X 1.2 (docking) | Various | A | NC [2] |
| 2016 | [57] | Energy-Based Pharmacophore and Three-Dimensional Quantitative Structure--Activity Relationship (3D-QSAR) Modeling Combined with Virtual Screening To Identify Novel Small-Molecule Inhibitors of Silent Mating-Type Information Regulation 2 Homologue 1 (SIRT-1). | 1 | LB-SB | LB and SB pharmacophore models combined with docking-based VS | Not tested | YES | ASINEX (5 000 000), in-house (971) | PHASE 3.4/Maestro 9.3 (pharmacophore) Glide 5.8/Maestro 9.4 (docking) | 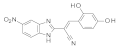 benzimidazole derivative | I | 4.34 μM (IC50) |
| 2019 | [227] | In silico and in vitro identification of candidate SIRT-1 activators from Indonesian medicinal plants compounds database | 1 | LB-SB | LB and SB pharmacophore models | Not tested | YES | Indonesian Herbal Database (1377) | LigandScout 4.2 | 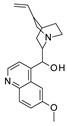 mulberrin, quinine, quinidine, and gartanin | A | 1.14 μM (EC50) |
| 2011 | [239] | Computational screening for active compounds targeting protein sequences: methodology and experimental validation. | 1 | Other | Sequence-based VS | Not tested | YES | SPECS drug-like library (85 000) | LIBSVM | 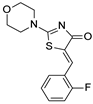 various | I | 5.72 μM (IC50) |
| 2019 | [247] | A prospective compound screening contest identified broader inhibitors for SIRT-1 | 1 | Other | Contest-based approach | Not tested | YES | ENAMINE (2 459 912) | various | 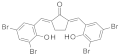 various various | I | 4.1 μM (IC50) |
| 2004 | [255] | An in silico approach to discovering novel inhibitors of human SIRT-2 | 2 | SBVS | Queries: (SB) features calculated on MD generated conformation + Docking | Not tested | YES | Maybridge | Unity 4.3.1/Sybyl v6.8 (feature-based), GOLD v1.2 (docking) |  Phenol derivatives | I | 56.7 μM (IC50) |
| 2006 | [256] | Discovering inhibitors of human SIRT-2: Novel structural scaffolds | 2 | SBVS | Queries: (SB) features calculated on MD generated conformation + Docking | Not tested | YES | Maybridge Screening Collection and LeadQuest libraries | Unity 4.4/Sybyl 6.9 (feature-based), GOLD 2.0 (docking) | 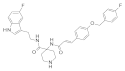 indole derivatives, phenol derivatives | I | 51 μM (IC50) |
| 2010 | [251] | Design of a novel nucleoside analog as potent inhibitor of the NAD+ dependent deacetylase, SIRT-2. | 2 | SBVS | 1 | YES | NCI Diversity Set II | AutoDock 4.0 | 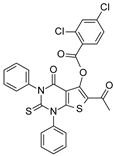 nucleoside analog (thieno [2,3-d]pyrimidine) | I | 8.7 μM (IC50) | |
| 2012 | [257] | Molecular Docking and Dynamics Simulation, Receptor-based Hypothesis: Application to Identify Novel SIRT-2 Inhibitors | 2 | SBVS | SBVS pharmacophoric model based on MD-generated conformations + docking | Not tested | NO | CHEMDIV database (700 000) | Ligand Pharmacophore Mapping/DS (screening), LigandFit/DS and GOLD (docking) | Various | I | NC [21] |
| 2016 | [253] | 5-Benzylidene-hydantoin is a new scaffold for SIRT inhibition: From virtual screening to activity assays. | 2 | SBVS | 1 | YES | Specs library (197 477) | GOLD v5.2 | 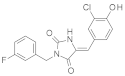 5-Benzylidene-hydantoin | I | 37.7 μM (IC50) | |
| 2008 | [248] | Thiobarbiturates as Sirtuin Inhibitors:Virtual Screening, Free-Energy Calculations, and Biological Testing | 2 | LB-SB | LB: similarity based | 1 | YES | Chembridge database (∼328 000) | MOE (fingerprints) GOLD 3.2 (docking) | 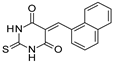 thiobarbiturates | I | 9.1 μM (IC50) |
| 2008 | [105] | Structure-activity studies on splitomicin derivatives as sirtuin inhibitors and computational prediction of binding mode | 2 | LB-SB | LB: similarity based | Not tested | YES | Chembridge (∼328 000) | MOE (fingerprint), GOLD 3.0 (docking) | 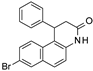 lactame analogues of beta-Phenylsplitomicins | I | 6.4 μM (IC50) |
| 2012 | [259] | Pharmacophore modeling and molecular dynamics simulation to identify the critical chemical features against human SIRT2 inhibitors | 2 | LB-SB | LB: pharmacophore basedSB: VS on MD-derived protein conformation | Not tested | NO | NCI (5672), Maybridge (26 490), Chembridge (17 885) | Discovery Studio v2.5 (pharmacophore), GOLD (SB) | Not reported | I | NC [29] |
| 2012 | [250] | Binding free energy calculations and biological testing of novel thiobarbiturates as inhibitors of the human NAD(+) dependent histone deacetylase SIRT-2 | 2 | LB-SB | LB: similarity-based | Not tested | YES | Chembridge | MOE (fingerprint), GOLD 4.0 (SBVS) | 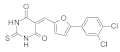 Thiobarbiturates | I | 1.5 μM (IC50) |
| 2019 | [97] | Pharmacophore modeling and virtual screening studies to identify novel selective SIRT-2 inhibitors | 2 | LB-SB | LB: Pharmacophore-based | 1,3,5 | YES | ZINC drug-like database (13 000 000) | Schrodinger Small-Molecule Drug Discovery Suite (pharmacophore) Glide (SBVS) |  various (triazines and dimethylfuran derivatives among others) | I | 84.28% inhibition @300μM |
| 2021 | [264] | Discovery of Potent Natural-Product-Derived SIRT-2 Inhibitors Using Structure- Based Exploration of SIRT-2 Pharmacophoric Space Coupled With QSAR Analyses. | 2 | LB-SB | SBVS pharmacophoric model combined with (SB+LB) QSAR | Not tested | YES | AnalytiCon Discovery database of purified natural products (5637) | DISCOVERY STUDIOv2.5 | 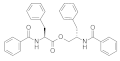 asperphenamate and salvianolic acid B | I | 1.94 μM (IC50) |
| 2021 | [263] | Targeting Epigenetic Regulators Using Machine Learning: Potential SIRT-2 Inhibitors | 2 | LB-SB | LB: Machine learning model | Not tested | NO | ZINC/FDA library | WEKA (ML), AutoDock Vina/PyRx (SBVS) | Various | I | NC [43] |
| 2015 | [271] | Virtual screening approach of sirtuin inhibitors results in two new scaffolds | 3 | SBVS | 1,2 | YES | ZINC database (> 8 000 000) | Glide v5.8 | 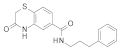 3-oxo-1,4-benzothiazinyl compounds and 4-(1-piperidyliminomethyl)benzene-1,3-dioles | I | 40% inhibition @200 μM | |
| 2013 | [272] | Identification of novel SIRT-3 inhibitor scaffolds by virtual screening | 3 | LB-SB | LB: similarity based | 1,2 | YES | Chembridge EXPRESS-Pick Collection database | Phase 3.2/ Schrodinger (similarity) + Glide 5.7 (SBVS) |  benzoxazoles, furopyrimidines | I | 71% inhibition (200 μM) |
| 2021 | [143] | Structure-Guided Design of a Small-Molecule Activator of SIRT-3 that Modulates Autophagy in Triple Negative Breast Cancer | 3 | SBVS | SB compound optimization | Not tested | YES | ZINC database (> 8 000 000) | Glide | 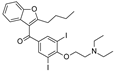 ZINC03830212 | A | EC50 = 3.25 μM |
| 2017 | [273] | Molecular modeling, dynamics studies and density functional theory approaches to identify potential inhibitors of SIRT-4 protein from Homo sapiens: a novel target for the treatment of type 2 diabetes. | 4 | SBVS | Target: HM | Not tested | NO | LifeChem, Specs, ZINC and MayBridge libraries | Glide/Maestro/Schrodinger v10.1 | 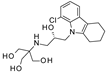 Indole derivative | I | NC [1] |
| 2018 | [274] | Structure-based discovery of new selective small-molecule SIRT-5 inhibitors | 5 | SBVS | SBVS + protein–ligand interaction fingerprint (IFP)-based filter | 2,6 | YES | In-house database (>15,000) | AutoDock Vina | 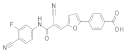 (E)-2-cyano-N-phenyl-3-(5-phenylfuran-2-yl)acrylamide derivatives | I | 5,59 μM (IC50) |
| 2014 | [179] | Discovery of Novel and Selective SIRT-6 Inhibitors | 6 | SBVS | Target: modified structure of Sirt-6 | 1,2 | YES | ASINEX subset of CoCoCo database | Glide v.5.8 | 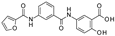 salicylate-based derivative | I | 89 μM (IC50) |
| 2015 | [174] | Quinazolinedione SIRT-6 inhibitors sensitize cancer cells to chemotherapeutics | 6 | LB-SB | LB: pharmacophore, similarity based | 1,2 | YES | CoCoCo database | Instant JChem (Chemaxon) ver. 5.11.5 | 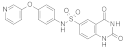 Compound 1 as Quinazolinedione prototype | I | 106 μM (IC50) |
| 2018 | [170] | Identification of a cellularly active SIRT-6 allosteric activator | 6 | SBVS | Allosteric site search + docking | Not tested | YES | Various DB (> 5,000,000) | GLIDE software (Schrödinger suite 2009, v5.5) | 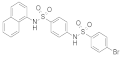 AN-988/40889624 | A | EC50 = 173 μM |
| 2020 | [173] | Discovery of Potent Small-Molecule SIRT-6 Activators: Structure–Activity Relationship and Anti-Pancreatic Ductal Adenocarcinoma Activity | 6 | SBVS | Allosteric site search + docking | Not tested | YES | Specs, ChemDiv, Selleck, and MedChemExpress and in-house database | GOLD software | 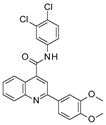 HIT20 | A | 147.60 (% Peptide Demyristoylation Activation @20 μM) |
| 2021 | [275] | Screening of SIRT-6 inhibitors and activators: A novel activator has an impact on breast cancer cells | 6 | LB-SB | LB: pharmacophore, similarity based | 1,2 | YES | ENAMINE (4 103 115), Chembridge (1 022 400), in house library of 1,4-dihydropyridine derivatives (∼100) | MOE (pharmacophore, similarity, docking), Glide/Maestro/ Schrodinger (docking) | 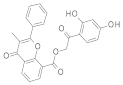 4H-chromen analogs (A), 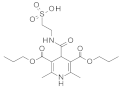 1,4-dihydropyridine (I) | A, I | 80 μM (EC50, activator) 60% @200 μM (IC50, inhibitor) |
| 2022 | [184] | Discovery of SIRT-7 Inhibitor as New Therapeutic Options Against Liver Cancer | 7 | SBVS | Target: protein modeling via fold recognition | 1,6 | YES | Chemdiv database (939319) | AutoDock Vina | 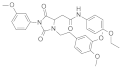 Compound 2800Z | I | Inhibition of SIRT-7 deacetylase activity |
| 2011 | [276] | Structure-based development of novel sirtuin inhibitors | 2,3,5,6 | SBVS | 2,3,5,6 | YES | NCI diversity set (1990) | AutoDock Vina | 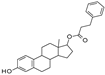 SIRT-2 specific: tetracyclic compounds | I | 4.8 μM (IC50, SIRT-2) | |
| 2017 | [197] | Discovery and Characterization of R/S-N-3-Cyanophenyl-N’-(6-tertbutoxycarbonylamino-3,4-dihydro-2,2-dimethyl-2H-1-benzopyran-4- yl)urea, a New Histone Deacetylase Class III Inhibitor Exerting Antiproliferative Activity against Cancer Cell Lines | 1,2 | SBVS | 1,2,3 | YES | In-house library (17) | AutoDock Vina |  Compound 18 | 6.2 μM (IC50, SIRT-1) 4.2 μM (IC50, SIRT-2) | ||
| 2018 | [277] | Identification of Bichalcones as Sirtuin Inhibitors by Virtual Screening and In Vitro Testing | 1,2 | SBVS | SBVS on different conformations of sirt-1 and sirt-2 | 3 | YES | pan-African Natural Products Library (463) | GOLD, Glide/Maestro/ Schrödinger v5.8 | 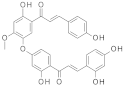 Bichalcones | I | 40.8 μM (IC50, SIRT-1) |
| 2019 | [278] | Structure-based identification of novel sirtuin inhibitors against triple negative breast cancer: An in silico and in vitro study | 1,2,3,4,5,6,7 | SBVS | Not applicable | YES | Plant-derived inhibitors (24), synthetic inhibitors (3) with reported epigenetic modulatory and anticancer potential, PubChem | Glide/Maestro/ Schrödinger | 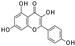 Sulforaphane (SIRT-1,-5), Kaempferol (SIRT-3) Apigenin (SIRT-6) | I | 12,5 μM (IC50) | |
| 2021 | [279] | In silico Repurposing of Drugs for pan-HDAC and pan-SIRT Inhibitors: Consensus Structure-based Virtual Screening and Pharmacophore Modeling Investigations | 1,2,3,5,6 | SBVS | Consensus SBVS on sirt-1,2,3,5,6 with 3 software | Not applicable | NO | (FDA)-approved drugs (1502) | Glide, FRED v3.3.1.2, AutoDock Vina/PyRx v1.1.2 | 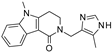 Alosetron, cinacalcet, indacaterol (virtual pan-SIRT I) | I | NC [3] |
| 2008 | [280] | Structure function analysis of Leishmania sirtuin: An ensemble of In silico and biochemical studies | Lm Sir2 | LB-SB | LB: Fingerprint basedSBVS target: LmSir2 HM | 2 | YES | National Cancer Institute (NCI) 3D database (~200 000) | MOE, FlexX/SYBYL 6.9 | 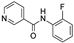 Nicotinamide derivative | I | 1,49 mM (IC50) |
| 2012 | [281] | Anti-Trypanosoma cruzi activity of nicotinamide | Tc Sir2 | LB-SB | LB: fingerprint based (nicotinamide)SBVS target: TcSir2 HM | Not tested | YES | Zinc database | MolDock |  Nicotinamide | I | ~100 μM (IC50) |
| 2014 | [282] | Computational Studies on Sirtuins from Trypanosoma cruzi: Structures, Conformations and Interactions with Phytochemicals | Tc Sir2 | SBVS | Targets: 2 HM of the 2 Tc sirtuins (closed state) | 2,5 | NO | Phytochemicals with antitrypanosomal activity collected by the literature (50) | GOLD v5.1 | 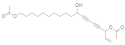 Anacardic acid derivative, aculeatin D, 16-acetoxy-11-hydroxyoctadeca-17-ene12,14-diynylethanoate, vismione D | I | NC [4] |
| 2016 | [283] | In-silico analysis of SIRT-2 from Schistosoma mansoni: structures, conformations, and interactions with inhibitors | Sm Sir2 | SBVS | Target: SmSir2 HM | 2 | NO | ZINC derived database (18 560) | idock | 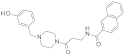 Phenol derivative | I | NC [4] |
3.3. Virtual Screening of SIRT-3 Modulators
3.4. Virtual Screening of SIRT-4 Modulators
3.5. Virtual Screening of SIRT-5 Modulators
3.6. Virtual Screening of SIRT-6 Modulators
3.7. Virtual Screening of SIRT-7 modulators
3.8. Pan-Sirtuin Modulators
3.9. Parasitic Sirtuins
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Finkel, T.; Deng, C.-X.; Mostoslavsky, R. Recent Progress in the Biology and Physiology of Sirtuins. Nature 2009, 460, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Makkar, R.; Behl, T.; Arora, S. Role of HDAC Inhibitors in Diabetes Mellitus. Curr. Res. Transl. Med. 2020, 68, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Sinclair, D.A.; Ellis, J.L.; Steegborn, C. Sirtuin Activators and Inhibitors: Promises, Achievements, and Challenges. Pharmacol. Ther. 2018, 188, 140–154. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, R.L.; Daniels, E.G.; Molenaars, M.; Houtkooper, R.H.; Janssens, G.E. From Molecular Promise to Preclinical Results: HDAC Inhibitors in the Race for Healthy Aging Drugs. EMBO Mol. Med. 2019, 11, e9854. [Google Scholar] [CrossRef] [PubMed]
- Hull, E.E.; Montgomery, M.R.; Leyva, K.J. HDAC Inhibitors as Epigenetic Regulators of the Immune System: Impacts on Cancer Therapy and Inflammatory Diseases. BioMed Res. Int. 2016, 2016, 1–15. [Google Scholar] [CrossRef]
- Finnin, M.S.; Donigian, J.R.; Pavletich, N.P. Structure of the Histone Deacetylase SIRT2. Nat. Struct. Biol. 2001, 8, 621–625. [Google Scholar] [CrossRef]
- Haigis, M.C.; Guarente, L.P. Mammalian Sirtuins—Emerging Roles in Physiology, Aging, and Calorie Restriction. Genes Dev. 2006, 20, 2913–2921. [Google Scholar] [CrossRef]
- Kitada, M.; Ogura, Y.; Monno, I.; Koya, D. Sirtuins and Type 2 Diabetes: Role in Inflammation, Oxidative Stress, and Mitochondrial Function. Front. Endocrinol. 2019, 10, 187. [Google Scholar] [CrossRef]
- Wang, W.; Li, J.; Cai, L. Research Progress of Sirtuins in Renal and Cardiovascular Diseases. Curr. Opin. Nephrol. Hypertens. 2021, 30, 108–114. [Google Scholar] [CrossRef]
- Jurkowska, K.; Szymańska, B.; Knysz, B.; Kuźniarski, A.; Piwowar, A. Sirtuins as Interesting Players in the Course of HIV Infection and Comorbidities. Cells 2021, 10, 2739. [Google Scholar] [CrossRef]
- Leite, J.A.; Ghirotto, B.; Targhetta, V.P.; Lima, J.; Câmara, N.O.S. Sirtuins as Pharmacological Targets in Neurodegenerative and Neuropsychiatric Disorders. Br. J. Pharmacol. 2022, 179, 1496–1511. [Google Scholar] [CrossRef]
- Zhao, E.; Hou, J.; Ke, X.; Abbas, M.N.; Kausar, S.; Zhang, L.; Cui, H. The Roles of Sirtuin Family Proteins in Cancer Progression. Cancers 2019, 11, 1949. [Google Scholar] [CrossRef]
- Mei, Z.; Zhang, X.; Yi, J.; Huang, J.; He, J.; Tao, Y. Sirtuins in Metabolism, DNA Repair and Cancer. J. Exp. Clin. Cancer Res. 2016, 35, 182. [Google Scholar] [CrossRef] [PubMed]
- Bonkowski, M.S.; Sinclair, D.A. Slowing Ageing by Design: The Rise of NAD+ and Sirtuin-Activating Compounds. Nat. Rev. Mol. Cell Biol. 2016, 17, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Mautone, N.; Zwergel, C.; Mai, A.; Rotili, D. Sirtuin Modulators: Where Are We Now? A Review of Patents from 2015 to 2019. Expert Opin. Ther. Pat. 2020, 30, 389–407. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yang, T.; Li, X.; Peng, T.; Hang, H.C.; Li, X.D. Integrative Chemical Biology Approaches for Identification and Characterization of “Erasers” for Fatty-Acid-Acylated Lysine Residues within Proteins. Angew. Chem. Int. Ed. Engl. 2015, 54, 1149–1152. [Google Scholar] [CrossRef] [PubMed]
- Pillai, V.B.; Kanwal, A.; Fang, Y.H.; Sharp, W.W.; Samant, S.; Arbiser, J.; Gupta, M.P. Honokiol, an Activator of Sirtuin-3 (SIRT3) Preserves Mitochondria and Protects the Heart from Doxorubicin-Induced Cardiomyopathy in Mice. Oncotarget 2017, 8, 34082–34098. [Google Scholar] [CrossRef]
- Hebert, A.S.; Dittenhafer-Reed, K.E.; Yu, W.; Bailey, D.J.; Selen, E.S.; Boersma, M.D.; Carson, J.J.; Tonelli, M.; Balloon, A.J.; Higbee, A.J.; et al. Calorie Restriction and SIRT3 Trigger Global Reprogramming of the Mitochondrial Protein Acetylome. Mol. Cell 2013, 49, 186–199. [Google Scholar] [CrossRef]
- Li, S.; Zheng, W. Mammalian Sirtuins SIRT4 and SIRT7. Prog. Mol. Biol. Transl. Sci. 2018, 154, 147–168. [Google Scholar] [CrossRef]
- Bheda, P.; Jing, H.; Wolberger, C.; Lin, H. The Substrate Specificity of Sirtuins. Annu. Rev. Biochem. 2016, 85, 405–429. [Google Scholar] [CrossRef]
- Tan, M.; Peng, C.; Anderson, K.A.; Chhoy, P.; Xie, Z.; Dai, L.; Park, J.; Chen, Y.; Huang, H.; Zhang, Y.; et al. Lysine Glutarylation Is a Protein Posttranslational Modification Regulated by SIRT5. Cell Metab. 2014, 19, 605–617. [Google Scholar] [CrossRef]
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Choi, B.H.; He, B.; Chen, W.; et al. Sirt5 Is a NAD-Dependent Protein Lysine Demalonylase and Desuccinylase. Science 2011, 334, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Rardin, M.J.; He, W.; Nishida, Y.; Newman, J.C.; Carrico, C.; Danielson, S.R.; Guo, A.; Gut, P.; Sahu, A.K.; Li, B.; et al. SIRT5 Regulates the Mitochondrial Lysine Succinylome and Metabolic Networks. Cell Metab. 2013, 18, 920–933. [Google Scholar] [CrossRef] [PubMed]
- van Meter, M.; Mao, Z.; Gorbunova, V.; Seluanov, A. SIRT6 Overexpression Induces Massive Apoptosis in Cancer Cells but Not in Normal Cells. Cell Cycle 2011, 10, 3153–3158. [Google Scholar] [CrossRef] [PubMed]
- Liszt, G.; Ford, E.; Kurtev, M.; Guarente, L. Mouse Sir2 Homolog SIRT6 Is a Nuclear ADP-Ribosyltransferase. J. Biol. Chem. 2005, 280, 21313–21320. [Google Scholar] [CrossRef] [PubMed]
- Feldman, J.L.; Dittenhafer-Reed, K.E.; Denu, J.M. Sirtuin Catalysis and Regulation. J. Biol. Chem. 2012, 287, 42419–42427. [Google Scholar] [CrossRef]
- Jiang, H.; Khan, S.; Wang, Y.; Charron, G.; He, B.; Sebastian, C.; Du, J.; Kim, R.; Ge, E.; Mostoslavsky, R.; et al. SIRT6 Regulates TNF-Alpha Secretion through Hydrolysis of Long-Chain Fatty Acyl Lysine. Nature 2013, 496, 110–113. [Google Scholar] [CrossRef]
- Bosch-Presegue, L.; Vaquero, A. The Dual Role of Sirtuins in Cancer. Genes Cancer 2011, 2, 648–662. [Google Scholar] [CrossRef]
- Ehrenberg, A.J.; Khatun, A.; Coomans, E.; Betts, M.J.; Capraro, F.; Thijssen, E.H.; Senkevich, K.; Bharucha, T.; Jafarpour, M.; Young, P.N.E.; et al. Relevance of Biomarkers across Different Neurodegenerative Diseases. Alzheimers Res. Ther. 2020, 12, 56. [Google Scholar] [CrossRef]
- Luo, G.; Jian, Z.; Zhu, Y.; Zhu, Y.; Chen, B.; Ma, R.; Tang, F.; Xiao, Y. Sirt1 Promotes Autophagy and Inhibits Apoptosis to Protect Cardiomyocytes from Hypoxic Stress. Int. J. Mol. Med. 2019, 43, 2033–2043. [Google Scholar] [CrossRef]
- Raynes, R.; Brunquell, J.; Westerheide, S.D. Stress Inducibility of SIRT1 and Its Role in Cytoprotection and Cancer. Genes Cancer 2013, 4, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Oberdoerffer, P.; Michan, S.; McVay, M.; Mostoslavsky, R.; Vann, J.; Park, S.-K.; Hartlerode, A.; Stegmuller, J.; Hafner, A.; Loerch, P.; et al. SIRT1 Redistribution on Chromatin Promotes Genomic Stability but Alters Gene Expression during Aging. Cell 2008, 135, 907–918. [Google Scholar] [CrossRef]
- Bai, X.; Yao, L.; Ma, X.; Xu, X. Small Molecules as SIRT Modulators. Mini-Rev. Med. Chem. 2018, 18, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Pacholec, M.; Bleasdale, J.E.; Chrunyk, B.; Cunningham, D.; Flynn, D.; Garofalo, R.S.; Griffith, D.; Griffor, M.; Loulakis, P.; Pabst, B.; et al. SRT1720, SRT2183, SRT1460, and Resveratrol Are Not Direct Activators of SIRT1. J. Biol. Chem. 2010, 285, 8340–8351. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Wang, M.; Zhong, A.; Wang, Y.; Du, J.; Wang, J.; Qi, L.; Bi, Z.; Zhang, P.; Lin, T.; et al. SRT1720 Inhibits the Growth of Bladder Cancer in Organoids and Murine Models through the SIRT1-HIF Axis. Oncogene 2021, 40, 6081–6092. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, R. SRT1720, a SIRT1 Activator, Promotes Tumor Cell Migration, and Lung Metastasis of Breast Cancer in Mice. Oncol. Rep. 2012, 27, 1726–1732. [Google Scholar] [CrossRef][Green Version]
- Zhang, X.; Chen, J.; Sun, L.; Xu, Y. SIRT1 Deacetylates KLF4 to Activate Claudin-5 Transcription in Ovarian Cancer Cells. J. Cell Biochem. 2018, 119, 2418–2426. [Google Scholar] [CrossRef]
- Guida, N.; Laudati, G.; Anzilotti, S.; Secondo, A.; Montuori, P.; di Renzo, G.; Canzoniero, L.M.T.; Formisano, L. Resveratrol via Sirtuin-1 Downregulates RE1-Silencing Transcription Factor (REST) Expression Preventing PCB-95-Induced Neuronal Cell Death. Toxicol. Appl. Pharmacol. 2015, 288, 387–398. [Google Scholar] [CrossRef]
- Liu, T.; Ma, Y.; Zhang, R.; Zhong, H.; Wang, L.; Zhao, J.; Yang, L.; Fan, X. Resveratrol Ameliorates Estrogen Deficiency-Induced Depression- and Anxiety-like Behaviors and Hippocampal Inflammation in Mice. Psychopharmacology 2019, 236, 1385–1399. [Google Scholar] [CrossRef]
- Scuderi, C.; Stecca, C.; Bronzuoli, M.R.; Rotili, D.; Valente, S.; Mai, A.; Steardo, L. Sirtuin Modulators Control Reactive Gliosis in an in Vitro Model of Alzheimer’s Disease. Front. Pharmacol. 2014, 5, 89. [Google Scholar] [CrossRef]
- Kumar, R.; Nigam, L.; Singh, A.P.; Singh, K.; Subbarao, N.; Dey, S. Design, Synthesis of Allosteric Peptide Activator for Human SIRT1 and Its Biological Evaluation in Cellular Model of Alzheimer’s Disease. Eur. J. Med. Chem. 2017, 127, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Cea, M.; Soncini, D.; Fruscione, F.; Raffaghello, L.; Garuti, A.; Emionite, L.; Moran, E.; Magnone, M.; Zoppoli, G.; Reverberi, D.; et al. Synergistic Interactions between HDAC and Sirtuin Inhibitors in Human Leukemia Cells. PLoS ONE 2011, 6, e22739. [Google Scholar] [CrossRef] [PubMed]
- Kalle, A.M.; Mallika, A.; Badiger, J.; Alinakhi; Talukdar, P. Sachchidanand Inhibition of SIRT1 by a Small Molecule Induces Apoptosis in Breast Cancer Cells. Biochem. Biophys. Res. Commun. 2010, 401, 13–19. [Google Scholar] [CrossRef]
- Wang, T.; Li, X.; Sun, S. EX527, a Sirt-1 Inhibitor, Induces Apoptosis in Glioma via Activating the P53 Signaling Pathway. Anticancer Drugs 2020, 31, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cao, L.; Li, Z.; Li, Y. SIRT1 Promotes GLUT1 Expression and Bladder Cancer Progression via Regulation of Glucose Uptake. Hum. Cell 2019, 32, 193–201. [Google Scholar] [CrossRef]
- Asaka, R.; Miyamoto, T.; Yamada, Y.; Ando, H.; Mvunta, D.H.; Kobara, H.; Shiozawa, T. Sirtuin 1 Promotes the Growth and Cisplatin Resistance of Endometrial Carcinoma Cells: A Novel Therapeutic Target. Lab. Investig. 2015, 95, 1363–1373. [Google Scholar] [CrossRef]
- Oon, C.E.; Strell, C.; Yeong, K.Y.; Östman, A.; Prakash, J. SIRT1 Inhibition in Pancreatic Cancer Models: Contrasting Effects in Vitro and in Vivo. Eur. J. Pharmacol. 2015, 757, 59–67. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, B.; Xu, H.; Sun, Y.; Shi, Y.; Luo, Y.; Jia, H.; Wang, F. Suppression of Sirt1 Sensitizes Lung Cancer Cells to WEE1 Inhibitor MK-1775-Induced DNA Damage and Apoptosis. Oncogene 2017, 36, 6863–6872. [Google Scholar] [CrossRef]
- Yousafzai, N.A.; Zhou, Q.; Xu, W.; Shi, Q.; Xu, J.; Feng, L.; Chen, H.; Shin, V.Y.; Jin, H.; Wang, X. SIRT1 Deacetylated and Stabilized XRCC1 to Promote Chemoresistance in Lung Cancer. Cell Death Dis. 2019, 10, 363. [Google Scholar] [CrossRef]
- Peck, B.; Chen, C.-Y.; Ho, K.-K.; di Fruscia, P.; Myatt, S.S.; Coombes, R.C.; Fuchter, M.J.; Hsiao, C.-D.; Lam, E.W.-F. SIRT Inhibitors Induce Cell Death and P53 Acetylation through Targeting Both SIRT1 and SIRT2. Mol. Cancer Ther. 2010, 9, 844–855. [Google Scholar] [CrossRef]
- Kim, H.-B.; Lee, S.-H.; Um, J.-H.; Oh, W.K.; Kim, D.-W.; Kang, C.-D.; Kim, S.-H. Sensitization of Multidrug-Resistant Human Cancer Cells to Hsp90 Inhibitors by down-Regulation of SIRT1. Oncotarget 2015, 6, 36202–36218. [Google Scholar] [CrossRef] [PubMed]
- Alvala, M.; Bhatnagar, S.; Ravi, A.; Jeankumar, V.U.; Manjashetty, T.H.; Yogeeswari, P.; Sriram, D. Novel Acridinedione Derivatives: Design, Synthesis, SIRT1 Enzyme and Tumor Cell Growth Inhibition Studies. Bioorg. Med. Chem. Lett. 2012, 22, 3256–3260. [Google Scholar] [CrossRef]
- Manjulatha, K.; Srinivas, S.; Mulakayala, N.; Rambabu, D.; Prabhakar, M.; Arunasree, K.M.; Alvala, M.; Basaveswara Rao, M.V.; Pal, M. Ethylenediamine Diacetate (EDDA) Mediated Synthesis of Aurones under Ultrasound: Their Evaluation as Inhibitors of SIRT1. Bioorg. Med. Chem. Lett. 2012, 22, 6160–6165. [Google Scholar] [CrossRef]
- Mellini, P.; Kokkola, T.; Suuronen, T.; Salo, H.S.; Tolvanen, L.; Mai, A.; Lahtela-Kakkonen, M.; Jarho, E.M. Screen of Pseudopeptidic Inhibitors of Human Sirtuins 1–3: Two Lead Compounds with Antiproliferative Effects in Cancer Cells. J. Med. Chem. 2013, 56, 6681–6695. [Google Scholar] [CrossRef] [PubMed]
- Oh, W.K.; Cho, K.B.; Hien, T.T.; Kim, T.H.; Kim, H.S.; Dao, T.T.; Han, H.-K.; Kwon, S.-M.; Ahn, S.-G.; Yoon, J.-H.; et al. Amurensin G, a Potent Natural SIRT1 Inhibitor, Rescues Doxorubicin Responsiveness via Down-Regulation of Multidrug Resistance 1. Mol. Pharmacol. 2010, 78, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Muscolini, M.; Castiello, L.; Palermo, E.; Zevini, A.; Ferrari, M.; Olagnier, D.; Hiscott, J. SIRT1 Modulates the Sensitivity of Prostate Cancer Cells to Vesicular Stomatitis Virus Oncolysis. J. Virol. 2019, 93, e00626-19. [Google Scholar] [CrossRef] [PubMed]
- Pulla, V.K.; Sriram, D.S.; Viswanadha, S.; Sriram, D.; Yogeeswari, P. Energy-Based Pharmacophore and Three-Dimensional Quantitative Structure-Activity Relationship (3D-QSAR) Modeling Combined with Virtual Screening to Identify Novel Small-Molecule Inhibitors of Silent Mating-Type Information Regulation 2 Homologue 1 (SIRT1). J. Chem. Inf. Model. 2016, 56, 173–187. [Google Scholar] [CrossRef]
- Zhang, Q.; Zeng, S.X.; Zhang, Y.; Zhang, Y.; Ding, D.; Ye, Q.; Meroueh, S.O.; Lu, H. A Small Molecule Inauhzin Inhibits SIRT1 Activity and Suppresses Tumour Growth through Activation of P53. EMBO Mol. Med. 2012, 4, 298–312. [Google Scholar] [CrossRef]
- Wössner, N.; Alhalabi, Z.; González, J.; Swyter, S.; Gan, J.; Schmidtkunz, K.; Zhang, L.; Vaquero, A.; Ovaa, H.; Einsle, O.; et al. Sirtuin 1 Inhibiting Thiocyanates (S1th)—A New Class of Isotype Selective Inhibitors of NAD+ Dependent Lysine Deacetylases. Front. Oncol. 2020, 10, 657. [Google Scholar] [CrossRef]
- Wan, J.; Deng, L.; Zhang, C.; Yuan, Q.; Liu, J.; Dun, Y.; Zhou, Z.; Zhao, H.; Liu, C.; Yuan, D.; et al. Chikusetsu Saponin V Attenuates H2O2-Induced Oxidative Stress in Human Neuroblastoma SH-SY5Y Cells through Sirt1/PGC-1α/Mn-SOD Signaling Pathways. Can. J. Physiol. Pharmacol. 2016, 94, 919–928. [Google Scholar] [CrossRef]
- Kim, H.-D.; Hesterman, J.; Call, T.; Magazu, S.; Keeley, E.; Armenta, K.; Kronman, H.; Neve, R.L.; Nestler, E.J.; Ferguson, D. SIRT1 Mediates Depression-Like Behaviors in the Nucleus Accumbens. J. Neurosci. 2016, 36, 8441–8452. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, F.; Guan, X. Baicalin Reverse Depressive-like Behaviors through Regulation SIRT1-NF-kB Signaling Pathway in Olfactory Bulbectomized Rats. Phytother. Res. 2019, 33, 1480–1489. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Huang, F.; Jian, C.; Qin, L.; Lu, F.; Wang, Y.; Zhang, Z.; Zhang, Q. Neuroprotective Effect of Salidroside against Central Nervous System Inflammation-Induced Cognitive Deficits: A Pivotal Role of Sirtuin 1-Dependent Nrf-2/HO-1/NF-ΚB Pathway. Phytother. Res. 2019, 33, 1438–1447. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, J. Training for the Future. Nurs. Stand. 1988, 2, 32. [Google Scholar]
- Min, S.-W.; Cho, S.-H.; Zhou, Y.; Schroeder, S.; Haroutunian, V.; Seeley, W.W.; Huang, E.J.; Shen, Y.; Masliah, E.; Mukherjee, C.; et al. Acetylation of Tau Inhibits Its Degradation and Contributes to Tauopathy. Neuron 2010, 67, 953–966. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Mi, M. Resveratrol Attenuates Aβ25–35 Caused Neurotoxicity by Inducing Autophagy Through the TyrRS-PARP1-SIRT1 Signaling Pathway. Neurochem. Res. 2016, 41, 2367–2379. [Google Scholar] [CrossRef]
- Shekhar, S.; Yadav, Y.; Singh, A.P.; Pradhan, R.; Desai, G.R.; Dey, A.B.; Dey, S. Neuroprotection by Ethanolic Extract of Syzygium Aromaticum in Alzheimer’s Disease like Pathology via Maintaining Oxidative Balance through SIRT1 Pathway. Exp. Gerontol. 2018, 110, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Yin, J.-B.; Liu, L.-H.; Guo, J.; Wang, S.-H.; Qu, C.-H.; Wang, C.-X. Protective Role of Dihydromyricetin in Alzheimer’s Disease Rat Model Associated with Activating AMPK/SIRT1 Signaling Pathway. Biosci. Rep. 2019, 39, BSR20180902. [Google Scholar] [CrossRef]
- Bonfili, L.; Cecarini, V.; Cuccioloni, M.; Angeletti, M.; Berardi, S.; Scarpona, S.; Rossi, G.; Eleuteri, A.M. SLAB51 Probiotic Formulation Activates SIRT1 Pathway Promoting Antioxidant and Neuroprotective Effects in an AD Mouse Model. Mol. Neurobiol. 2018, 55, 7987–8000. [Google Scholar] [CrossRef]
- Lee, H.R.; Shin, H.K.; Park, S.Y.; Kim, H.Y.; Lee, W.S.; Rhim, B.Y.; Hong, K.W.; Kim, C.D. Cilostazol Suppresses β-Amyloid Production by Activating a Disintegrin and Metalloproteinase 10 via the Upregulation of SIRT1-Coupled Retinoic Acid Receptor-β. J. Neurosci. Res. 2014, 92, 1581–1590. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Lee, H.; Yoo, H.B.; Choi, J.-S.; Jung, H.-Y.; Yoon, E.J.; Kim, H.; Jung, Y.-H.; Lee, H.-Y.; Kim, Y.K. Efficacy of Cilostazol Administration in Alzheimer’s Disease Patients with White Matter Lesions: A Positron-Emission Tomography Study. Neurotherapeutics 2019, 16, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.R.; Syed, A.; Lukacsovich, T.; Purcell, J.; Barbaro, B.A.; Worthge, S.A.; Wei, S.R.; Pollio, G.; Magnoni, L.; Scali, C.; et al. A Potent and Selective Sirtuin 1 Inhibitor Alleviates Pathology in Multiple Animal and Cell Models of Huntington’s Disease. Hum. Mol. Genet. 2014, 23, 2995–3007. [Google Scholar] [CrossRef] [PubMed]
- Westerberg, G.; Chiesa, J.A.; Andersen, C.A.; Diamanti, D.; Magnoni, L.; Pollio, G.; Darpo, B.; Zhou, M. Safety, Pharmacokinetics, Pharmacogenomics and QT Concentration−effect Modelling of the SirT1 Inhibitor Selisistat in Healthy Volunteers. Br. J. Clin. Pharmacol. 2015, 79, 477–491. [Google Scholar] [CrossRef] [PubMed]
- Süssmuth, S.D.; Haider, S.; Landwehrmeyer, G.B.; Farmer, R.; Frost, C.; Tripepi, G.; Andersen, C.A.; di Bacco, M.; Lamanna, C.; Diodato, E.; et al. An Exploratory Double-Blind, Randomized Clinical Trial with Selisistat, a SirT1 Inhibitor, in Patients with Huntington’s Disease. Br. J. Clin. Pharmacol. 2015, 79, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Shin, B.H.; Lim, Y.; Oh, H.J.; Park, S.M.; Lee, S.-K.; Ahnn, J.; Kim, D.H.; Song, W.K.; Kwak, T.H.; Park, W.J. Pharmacological Activation of Sirt1 Ameliorates Polyglutamine-Induced Toxicity through the Regulation of Autophagy. PLoS ONE 2013, 8, e64953. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Ban, J.-J.; Chung, J.-Y.; Im, W.; Kim, M. Amelioration of Huntington’s Disease Phenotypes by Beta-Lapachone Is Associated with Increases in Sirt1 Expression, CREB Phosphorylation and PGC-1α Deacetylation. PLoS ONE 2018, 13, e0195968. [Google Scholar] [CrossRef]
- Chen, C.; Xia, B.; Tang, L.; Wu, W.; Tang, J.; Liang, Y.; Yang, H.; Zhang, Z.; Lu, Y.; Chen, G.; et al. Echinacoside Protects against MPTP/MPP+-Induced Neurotoxicity via Regulating Autophagy Pathway Mediated by Sirt1. Metab. Brain Dis. 2019, 34, 203–212. [Google Scholar] [CrossRef]
- Rao, S.P.; Sharma, N.; Kalivendi, S.V. Embelin Averts MPTP-Induced Dysfunction in Mitochondrial Bioenergetics and Biogenesis via Activation of SIRT1. Biochim. Et Biophys. Acta (BBA)-Bioenerg. 2020, 1861, 148157. [Google Scholar] [CrossRef]
- Valle, C.; Salvatori, I.; Gerbino, V.; Rossi, S.; Palamiuc, L.; René, F.; Carrì, M.T. Tissue-Specific Deregulation of Selected HDACs Characterizes ALS Progression in Mouse Models: Pharmacological Characterization of SIRT1 and SIRT2 Pathways. Cell Death Dis. 2014, 5, e1296. [Google Scholar] [CrossRef]
- Prozorovski, T.; Ingwersen, J.; Lukas, D.; Göttle, P.; Koop, B.; Graf, J.; Schneider, R.; Franke, K.; Schumacher, S.; Britsch, S.; et al. Regulation of Sirtuin Expression in Autoimmune Neuroinflammation: Induction of SIRT1 in Oligodendrocyte Progenitor Cells. Neurosci. Lett. 2019, 704, 116–125. [Google Scholar] [CrossRef]
- Lim, H.W.; Kang, S.G.; Ryu, J.K.; Schilling, B.; Fei, M.; Lee, I.S.; Kehasse, A.; Shirakawa, K.; Yokoyama, M.; Schnölzer, M.; et al. SIRT1 Deacetylates RORγt and Enhances Th17 Cell Generation. J. Exp. Med. 2015, 212, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Nayagam, V.M.; Wang, X.; Tan, Y.C.; Poulsen, A.; Goh, K.C.; Ng, T.; Wang, H.; Song, H.Y.; Ni, B.; Entzeroth, M.; et al. SIRT1 Modulating Compounds from High-Throughput Screening as Anti-Inflammatory and Insulin-Sensitizing Agents. J. Biomol. Screen. 2006, 11, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Zhou, P.; Xu, X.; Fang, W.; Jia, S.; Liu, W.; Su, X.; Zhang, J.; Wang, H.; Yu, P.; et al. Benzothiazole Derivatives Upregulate SIRT1 and Relevant Genes in High-Fat Fed C57BL/6J Mice. Med. Chem. Res. 2015, 24, 2454–2460. [Google Scholar] [CrossRef]
- Liu, P.; Feng, T.; Zuo, X.; Wang, X.; Luo, J.; Li, N.; Han, X.; Zhu, N.; Xu, S.; Xu, Y.; et al. A Novel SIRT1 Activator E6155 Improves Insulin Sensitivity in Type 2 Diabetic KKAy Mice. Biochem. Biophys. Res. Commun. 2018, 498, 633–639. [Google Scholar] [CrossRef]
- North, B.J.; Verdin, E. Interphase Nucleo-Cytoplasmic Shuttling and Localization of SIRT2 during Mitosis. PLoS ONE 2007, 2, e784. [Google Scholar] [CrossRef]
- Jing, E.; Gesta, S.; Kahn, C.R. SIRT2 Regulates Adipocyte Differentiation through FoxO1 Acetylation/Deacetylation. Cell Metab. 2007, 6, 105–114. [Google Scholar] [CrossRef]
- Dryden, S.C.; Nahhas, F.A.; Nowak, J.E.; Goustin, A.-S.; Tainsky, M.A. Role for Human SIRT2 NAD-Dependent Deacetylase Activity in Control of Mitotic Exit in the Cell Cycle. Mol. Cell. Biol. 2003, 23, 3173–3185. [Google Scholar] [CrossRef]
- Funato, K.; Hayashi, T.; Echizen, K.; Negishi, L.; Shimizu, N.; Koyama-Nasu, R.; Nasu-Nishimura, Y.; Morishita, Y.; Tabar, V.; Todo, T.; et al. SIRT2-mediated Inactivation of P73 Is Required for Glioblastoma Tumorigenicity. EMBO Rep. 2018, 19, e45587. [Google Scholar] [CrossRef]
- Hong, J.Y.; Fernandez, I.; Anmangandla, A.; Lu, X.; Bai, J.J.; Lin, H. Pharmacological Advantage of SIRT2-Selective versus Pan-SIRT1–3 Inhibitors. ACS Chem. Biol. 2021, 16, 1266–1275. [Google Scholar] [CrossRef]
- Rotili, D.; Tarantino, D.; Nebbioso, A.; Paolini, C.; Huidobro, C.; Lara, E.; Mellini, P.; Lenoci, A.; Pezzi, R.; Botta, G.; et al. Discovery of Salermide-Related Sirtuin Inhibitors: Binding Mode Studies and Antiproliferative Effects in Cancer Cells Including Cancer Stem Cells. J. Med. Chem. 2012, 55, 10937–10947. [Google Scholar] [CrossRef]
- Liu, P.Y.; Xu, N.; Malyukova, A.; Scarlett, C.J.; Sun, Y.T.; Zhang, X.D.; Ling, D.; Su, S.-P.; Nelson, C.; Chang, D.K.; et al. The Histone Deacetylase SIRT2 Stabilizes Myc Oncoproteins. Cell Death Differ. 2013, 20, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Kudo, N.; Ito, A.; Arata, M.; Nakata, A.; Yoshida, M. Identification of a Novel Small Molecule That Inhibits Deacetylase but Not Defatty-Acylase Reaction Catalysed by SIRT2. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170070. [Google Scholar] [CrossRef]
- Farooqi, A.S.; Hong, J.Y.; Cao, J.; Lu, X.; Price, I.R.; Zhao, Q.; Kosciuk, T.; Yang, M.; Bai, J.J.; Lin, H. Novel Lysine-Based Thioureas as Mechanism-Based Inhibitors of Sirtuin 2 (SIRT2) with Anticancer Activity in a Colorectal Cancer Murine Model. J. Med. Chem. 2019, 62, 4131–4141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Au, Q.; Zhang, M.; Barber, J.R.; Ng, S.C.; Zhang, B. Identification of a Small Molecule SIRT2 Inhibitor with Selective Tumor Cytotoxicity. Biochem. Biophys. Res. Commun. 2009, 386, 729–733. [Google Scholar] [CrossRef] [PubMed]
- Spiegelman, N.A.; Price, I.R.; Jing, H.; Wang, M.; Yang, M.; Cao, J.; Hong, J.Y.; Zhang, X.; Aramsangtienchai, P.; Sadhukhan, S.; et al. Direct Comparison of SIRT2 Inhibitors: Potency, Specificity, Activity-Dependent Inhibition, and On-Target Anticancer Activities. ChemMedChem 2018, 13, 1890–1894. [Google Scholar] [CrossRef]
- Jing, H.; Hu, J.; He, B.; Negrón Abril, Y.L.; Stupinski, J.; Weiser, K.; Carbonaro, M.; Chiang, Y.-L.; Southard, T.; Giannakakou, P.; et al. A SIRT2-Selective Inhibitor Promotes c-Myc Oncoprotein Degradation and Exhibits Broad Anticancer Activity. Cancer Cell 2016, 29, 767–768. [Google Scholar] [CrossRef]
- Eren, G.; Bruno, A.; Guntekin-Ergun, S.; Cetin-Atalay, R.; Ozgencil, F.; Ozkan, Y.; Gozelle, M.; Kaya, S.G.; Costantino, G. Pharmacophore Modeling and Virtual Screening Studies to Identify Novel Selective SIRT2 Inhibitors. J. Mol. Graph. Model. 2019, 89, 60–73. [Google Scholar] [CrossRef]
- Seifert, T.; Malo, M.; Kokkola, T.; Engen, K.; Fridén-Saxin, M.; Wallén, E.A.A.; Lahtela-Kakkonen, M.; Jarho, E.M.; Luthman, K. Chroman-4-One- and Chromone-Based Sirtuin 2 Inhibitors with Antiproliferative Properties in Cancer Cells. J. Med. Chem. 2014, 57, 9870–9888. [Google Scholar] [CrossRef]
- Yang, L.-L.; Wang, H.-L.; Zhong, L.; Yuan, C.; Liu, S.-Y.; Yu, Z.-J.; Liu, S.; Yan, Y.-H.; Wu, C.; Wang, Y.; et al. X-Ray Crystal Structure Guided Discovery of New Selective, Substrate-Mimicking Sirtuin 2 Inhibitors That Exhibit Activities against Non-Small Cell Lung Cancer Cells. Eur. J. Med. Chem. 2018, 155, 806–823. [Google Scholar] [CrossRef]
- Moniot, S.; Forgione, M.; Lucidi, A.; Hailu, G.S.; Nebbioso, A.; Carafa, V.; Baratta, F.; Altucci, L.; Giacché, N.; Passeri, D.; et al. Development of 1,2,4-Oxadiazoles as Potent and Selective Inhibitors of the Human Deacetylase Sirtuin 2: Structure–Activity Relationship, X-Ray Crystal Structure, and Anticancer Activity. J. Med. Chem. 2017, 60, 2344–2360. [Google Scholar] [CrossRef]
- Kozako, T.; Mellini, P.; Ohsugi, T.; Aikawa, A.; Uchida, Y.-I.; Honda, S.-I.; Suzuki, T. Novel Small Molecule SIRT2 Inhibitors Induce Cell Death in Leukemic Cell Lines. BMC Cancer 2018, 18, 791. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.S.; Scian, M.; Sripathy, S.; Posakony, J.; Lao, U.; Loe, T.K.; Leko, V.; Thalhofer, A.; Schuler, A.D.; Bedalov, A.; et al. Development of Pyrazolone and Isoxazol-5-One Cambinol Analogues as Sirtuin Inhibitors. J. Med. Chem. 2014, 57, 3283–3294. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Sripathy, S.; Webster, A.; Park, A.; Lao, U.; Hsu, J.H.; Loe, T.; Bedalov, A.; Simon, J.A. Discovery of Selective SIRT2 Inhibitors as Therapeutic Agents in B-Cell Lymphoma and Other Malignancies. Molecules 2020, 25, 455. [Google Scholar] [CrossRef]
- Shah, A.A.; Ito, A.; Nakata, A.; Yoshida, M. Identification of a Selective SIRT2 Inhibitor and Its Anti-Breast Cancer Activity. Biol. Pharm. Bull. 2016, 39, 1739–1742. [Google Scholar] [CrossRef] [PubMed]
- Neugebauer, R.C.; Uchiechowska, U.; Meier, R.; Hruby, H.; Valkov, V.; Verdin, E.; Sippl, W.; Jung, M. Structure–Activity Studies on Splitomicin Derivatives as Sirtuin Inhibitors and Computational Prediction of Binding Mode. J. Med. Chem. 2008, 51, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Yeong, K.Y.; Khaw, K.Y.; Takahashi, Y.; Itoh, Y.; Murugaiyah, V.; Suzuki, T. Discovery of Gamma-Mangostin from Garcinia Mangostana as a Potent and Selective Natural SIRT2 Inhibitor. Bioorg. Chem. 2020, 94, 103403. [Google Scholar] [CrossRef]
- McCarthy, A.R.; Sachweh, M.C.C.; Higgins, M.; Campbell, J.; Drummond, C.J.; van Leeuwen, I.M.M.; Pirrie, L.; Ladds, M.J.G.W.; Westwood, N.J.; Laín, S. Tenovin-D3, a Novel Small-Molecule Inhibitor of Sirtuin SirT2, Increases P21 (CDKN1A) Expression in a P53-Independent Manner. Mol. Cancer Ther. 2013, 12, 352–360. [Google Scholar] [CrossRef]
- Yang, H.; Chen, Y.; Jiang, Y.; Wang, D.; Yan, J.; Zhou, Z. TP53 Mutation Influences the Efficacy of Treatment of Colorectal Cancer Cell Lines with a Combination of Sirtuin Inhibitors and Chemotherapeutic Agents. Exp. Ther. Med. 2020, 20, 1415–1422. [Google Scholar] [CrossRef]
- Outeiro, T.F.; Kontopoulos, E.; Altmann, S.M.; Kufareva, I.; Strathearn, K.E.; Amore, A.M.; Volk, C.B.; Maxwell, M.M.; Rochet, J.-C.; McLean, P.J.; et al. Sirtuin 2 Inhibitors Rescue Alpha-Synuclein-Mediated Toxicity in Models of Parkinson’s Disease. Science 2007, 317, 516–519. [Google Scholar] [CrossRef]
- Tatum, P.R.; Sawada, H.; Ota, Y.; Itoh, Y.; Zhan, P.; Ieda, N.; Nakagawa, H.; Miyata, N.; Suzuki, T. Identification of Novel SIRT2-Selective Inhibitors Using a Click Chemistry Approach. Bioorg. Med. Chem. Lett. 2014, 24, 1871–1874. [Google Scholar] [CrossRef]
- Taylor, D.M.; Balabadra, U.; Xiang, Z.; Woodman, B.; Meade, S.; Amore, A.; Maxwell, M.M.; Reeves, S.; Bates, G.P.; Luthi-Carter, R.; et al. A Brain-Permeable Small Molecule Reduces Neuronal Cholesterol by Inhibiting Activity of Sirtuin 2 Deacetylase. ACS Chem. Biol. 2011, 6, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Rumpf, T.; Schiedel, M.; Karaman, B.; Roessler, C.; North, B.J.; Lehotzky, A.; Oláh, J.; Ladwein, K.I.; Schmidtkunz, K.; Gajer, M.; et al. Selective Sirt2 Inhibition by Ligand-Induced Rearrangement of the Active Site. Nat. Commun. 2015, 6, 6263. [Google Scholar] [CrossRef] [PubMed]
- Lain, S.; Hollick, J.J.; Campbell, J.; Staples, O.D.; Higgins, M.; Aoubala, M.; McCarthy, A.; Appleyard, V.; Murray, K.E.; Baker, L.; et al. Discovery, in Vivo Activity, and Mechanism of Action of a Small-Molecule P53 Activator. Cancer Cell 2008, 13, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Harting, K.; Knöll, B. SIRT2-Mediated Protein Deacetylation: An Emerging Key Regulator in Brain Physiology and Pathology. Eur. J. Cell Biol. 2010, 89, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, Y.; Cao, W.; Wei, X.; Chen, J.; Ying, W. SIRT2 Plays Significant Roles in Lipopolysaccharides-Induced Neuroinflammation and Brain Injury in Mice. Neurochem. Res. 2016, 41, 2490–2500. [Google Scholar] [CrossRef]
- Spires-Jones, T.L.; Fox, L.M.; Rozkalne, A.; Pitstick, R.; Carlson, G.A.; Kazantsev, A.G. Inhibition of Sirtuin 2 with Sulfobenzoic Acid Derivative AK1 Is Non-Toxic and Potentially Neuroprotective in a Mouse Model of Frontotemporal Dementia. Front. Pharmacol. 2012, 3, 42. [Google Scholar] [CrossRef]
- Silva, D.F.; Esteves, A.R.; Oliveira, C.R.; Cardoso, S.M. Mitochondrial Metabolism Power SIRT2-Dependent Deficient Traffic Causing Alzheimer’s-Disease Related Pathology. Mol. Neurobiol. 2017, 54, 4021–4040. [Google Scholar] [CrossRef]
- Diaz-Perdigon, T.; Belloch, F.B.; Ricobaraza, A.; Elboray, E.E.; Suzuki, T.; Tordera, R.M.; Puerta, E. Early Sirtuin 2 Inhibition Prevents Age-Related Cognitive Decline in a Senescence-Accelerated Mouse Model. Neuropsychopharmacology 2020, 45, 347–357. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, J.Q.; Hong, T.T.; Sun, Y.H.; Huang, H.L.; Chen, F.; Chen, X.J.; Chen, H.Y.; Dong, S.S.; Cui, L.L.; et al. RTN4B-Mediated Suppression of Sirtuin 2 Activity Ameliorates β-Amyloid Pathology and Cognitive Impairment in Alzheimer’s Disease Mouse Model. Aging Cell 2020, 19, e13194. [Google Scholar] [CrossRef]
- Esteves, A.R.; Palma, A.M.; Gomes, R.; Santos, D.; Silva, D.F.; Cardoso, S.M. Acetylation as a Major Determinant to Microtubule-Dependent Autophagy: Relevance to Alzheimer’s and Parkinson Disease Pathology. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2008–2023. [Google Scholar] [CrossRef]
- Biella, G.; Fusco, F.; Nardo, E.; Bernocchi, O.; Colombo, A.; Lichtenthaler, S.F.; Forloni, G.; Albani, D. Sirtuin 2 Inhibition Improves Cognitive Performance and Acts on Amyloid-β Protein Precursor Processing in Two Alzheimer’s Disease Mouse Models. J. Alzheimers Dis. 2016, 53, 1193–1207. [Google Scholar] [CrossRef] [PubMed]
- Quinti, L.; Casale, M.; Moniot, S.; Pais, T.F.; van Kanegan, M.J.; Kaltenbach, L.S.; Pallos, J.; Lim, R.G.; Naidu, S.D.; Runne, H.; et al. SIRT2- and NRF2-Targeting Thiazole-Containing Compound with Therapeutic Activity in Huntington’s Disease Models. Cell Chem. Biol. 2016, 23, 849–861. [Google Scholar] [CrossRef] [PubMed]
- Luthi-Carter, R.; Taylor, D.M.; Pallos, J.; Lambert, E.; Amore, A.; Parker, A.; Moffitt, H.; Smith, D.L.; Runne, H.; Gokce, O.; et al. SIRT2 Inhibition Achieves Neuroprotection by Decreasing Sterol Biosynthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 7927–7932. [Google Scholar] [CrossRef] [PubMed]
- Chopra, V.; Quinti, L.; Kim, J.; Vollor, L.; Narayanan, K.L.; Edgerly, C.; Cipicchio, P.M.; Lauver, M.A.; Choi, S.H.; Silverman, R.B.; et al. The Sirtuin 2 Inhibitor AK-7 Is Neuroprotective in Huntington’s Disease Mouse Models. Cell Rep. 2012, 2, 1492–1497. [Google Scholar] [CrossRef]
- Ai, T.; Wilson, D.J.; More, S.S.; Xie, J.; Chen, L. 5-((3-Amidobenzyl)Oxy)Nicotinamides as Sirtuin 2 Inhibitors. J. Med. Chem. 2016, 59, 2928–2941. [Google Scholar] [CrossRef]
- Garske, A.L.; Smith, B.C.; Denu, J.M. Linking SIRT2 to Parkinson’s Disease. ACS Chem. Biol. 2007, 2, 529–532. [Google Scholar] [CrossRef]
- Harrison, I.F.; Smith, A.D.; Dexter, D.T. Pathological Histone Acetylation in Parkinson’s Disease: Neuroprotection and Inhibition of Microglial Activation through SIRT 2 Inhibition. Neurosci. Lett. 2018, 666, 48–57. [Google Scholar] [CrossRef]
- Wang, X.; Guan, Q.; Wang, M.; Yang, L.; Bai, J.; Yan, Z.; Zhang, Y.; Liu, Z. Aging-Related Rotenone-Induced Neurochemical and Behavioral Deficits: Role of SIRT2 and Redox Imbalance, and Neuroprotection by AK-7. Drug Des. Dev. Ther. 2015, 9, 2553–2563. [Google Scholar] [CrossRef]
- Guan, Q.; Wang, M.; Chen, H.; Yang, L.; Yan, Z.; Wang, X. Aging-Related 1-Methyl-4-Phenyl-1,2,3,6-Tetrahydropyridine-Induced Neurochemial and Behavioral Deficits and Redox Dysfunction: Improvement by AK-7. Exp. Gerontol. 2016, 82, 19–29. [Google Scholar] [CrossRef]
- di Fruscia, P.; Zacharioudakis, E.; Liu, C.; Moniot, S.; Laohasinnarong, S.; Khongkow, M.; Harrison, I.F.; Koltsida, K.; Reynolds, C.R.; Schmidtkunz, K.; et al. The Discovery of a Highly Selective 5,6,7,8-Tetrahydrobenzo [4,5]Thieno [2,3-d]Pyrimidin-4(3H)-One SIRT2 Inhibitor That Is Neuroprotective in an in Vitro Parkinson’s Disease Model. ChemMedChem 2015, 10, 69–82. [Google Scholar] [CrossRef]
- Chen, X.; Wales, P.; Quinti, L.; Zuo, F.; Moniot, S.; Herisson, F.; Rauf, N.A.; Wang, H.; Silverman, R.B.; Ayata, C.; et al. The Sirtuin-2 Inhibitor AK7 Is Neuroprotective in Models of Parkinson’s Disease but Not Amyotrophic Lateral Sclerosis and Cerebral Ischemia. PLoS ONE 2015, 10, e0116919. [Google Scholar] [CrossRef]
- Song, J.; Yang, B.; Jia, X.; Li, M.; Tan, W.; Ma, S.; Shi, X.; Feng, L. Distinctive Roles of Sirtuins on Diabetes, Protective or Detrimental? Front Endocrinol. 2018, 9, 724. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Dey, C.S. SIRT2 Negatively Regulates Insulin Resistance in C2C12 Skeletal Muscle Cells. Biochim. Biophys. Acta 2014, 1842, 1372–1378. [Google Scholar] [CrossRef] [PubMed]
- Onyango, P.; Celic, I.; McCaffery, J.M.; Boeke, J.D.; Feinberg, A.P. SIRT3, a Human SIR2 Homologue, Is an NAD-Dependent Deacetylase Localized to Mitochondria. Proc. Natl. Acad. Sci. USA 2002, 99, 13653–13658. [Google Scholar] [CrossRef] [PubMed]
- Ahn, B.-H.; Kim, H.-S.; Song, S.; Lee, I.H.; Liu, J.; Vassilopoulos, A.; Deng, C.-X.; Finkel, T. A Role for the Mitochondrial Deacetylase Sirt3 in Regulating Energy Homeostasis. Proc. Natl. Acad. Sci. USA 2008, 105, 14447–14452. [Google Scholar] [CrossRef] [PubMed]
- Bell, E.L.; Emerling, B.M.; Ricoult, S.J.H.; Guarente, L. SirT3 Suppresses Hypoxia Inducible Factor 1α and Tumor Growth by Inhibiting Mitochondrial ROS Production. Oncogene 2011, 30, 2986–2996. [Google Scholar] [CrossRef]
- Tao, R.; Coleman, M.C.; Pennington, J.D.; Ozden, O.; Park, S.-H.; Jiang, H.; Kim, H.-S.; Flynn, C.R.; Hill, S.; Hayes McDonald, W.; et al. Sirt3-Mediated Deacetylation of Evolutionarily Conserved Lysine 122 Regulates MnSOD Activity in Response to Stress. Mol. Cell 2010, 40, 893–904. [Google Scholar] [CrossRef]
- Chen, Y.; Fu, L.L.; Wen, X.; Wang, X.Y.; Liu, J.; Cheng, Y.; Huang, J. Sirtuin-3 (SIRT3), a Therapeutic Target with Oncogenic and Tumor-Suppressive Function in Cancer. Cell Death Dis. 2014, 5, e1047. [Google Scholar] [CrossRef]
- Alhazzazi, T.Y.; Kamarajan, P.; Xu, Y.; Ai, T.; Chen, L.; Verdin, E.; Kapila, Y.L. A Novel Sirtuin-3 Inhibitor, LC-0296, Inhibits Cell Survival and Proliferation, and Promotes Apoptosis of Head and Neck Cancer Cells. Anticancer Res. 2016, 36, 49–60. [Google Scholar]
- Ma, J.; Liu, B.; Yu, D.; Zuo, Y.; Cai, R.; Yang, J.; Cheng, J. SIRT3 Deacetylase Activity Confers Chemoresistance in AML via Regulation of Mitochondrial Oxidative Phosphorylation. Br. J. Haematol. 2019, 187, 49–64. [Google Scholar] [CrossRef]
- Li, M.; Chiang, Y.-L.; Lyssiotis, C.A.; Teater, M.R.; Hong, J.Y.; Shen, H.; Wang, L.; Hu, J.; Jing, H.; Chen, Z.; et al. Non-Oncogene Addiction to SIRT3 Plays a Critical Role in Lymphomagenesis. Cancer Cell 2019, 35, 916–931.e9. [Google Scholar] [CrossRef]
- Wang, X.; Zeng, Q.; Li, Z.; Yang, X.; Xia, W.; Chen, Z. Adjudin Synergizes with Paclitaxel and Inhibits Cell Growth and Metastasis by Regulating the Sirtuin 3-Forkhead Box O3a Axis in Human Small-Cell Lung Cancer. Thorac. Cancer 2019, 10, 642–658. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zou, L.; Shi, D.; Liu, J.; Zhang, J.; Zhao, R.; Wang, G.; Zhang, L.; Ouyang, L.; Liu, B. Structure-Guided Design of a Small-Molecule Activator of Sirtuin-3 That Modulates Autophagy in Triple Negative Breast Cancer. J. Med. Chem. 2021, 64, 14192–14216. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-J.; Liu, X.-Y.; Li, J.-H.; Guo, J.; Li, F.; Gui, Y.; Li, X.-H.; Yang, L.; Wu, C.-Y.; Yuan, Y.; et al. Gastrodin Attenuates Microglia Activation through Renin-Angiotensin System and Sirtuin3 Pathway. Neurochem. Int. 2018, 120, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-H.; Burgess, J.D.; Faroqi, A.H.; DeMeo, N.N.; Fiesel, F.C.; Springer, W.; Delenclos, M.; McLean, P.J. Alpha-Synuclein-Induced Mitochondrial Dysfunction Is Mediated via a Sirtuin 3-Dependent Pathway. Mol. Neurodegener. 2020, 15, 5. [Google Scholar] [CrossRef]
- Ramesh, S.; Govindarajulu, M.; Lynd, T.; Briggs, G.; Adamek, D.; Jones, E.; Heiner, J.; Majrashi, M.; Moore, T.; Amin, R.; et al. SIRT3 Activator Honokiol Attenuates β-Amyloid by Modulating Amyloidogenic Pathway. PLoS ONE 2018, 13, e0190350. [Google Scholar] [CrossRef]
- Lee, A.Y.; Christensen, S.M.; Duong, N.; Tran, Q.-A.; Xiong, H.M.; Huang, J.; James, S.; Vallabh, D.; Talbott, G.; Rose, M.; et al. Sirt3 Pharmacologically Promotes Insulin Sensitivity through PI3/AKT/MTOR and Their Downstream Pathway in Adipocytes. Int. J. Mol. Sci. 2022, 23, 3740. [Google Scholar] [CrossRef]
- Huang, J.-Y.; Hirschey, M.D.; Shimazu, T.; Ho, L.; Verdin, E. Mitochondrial Sirtuins. Biochim. Biophys. Acta 2010, 1804, 1645–1651. [Google Scholar] [CrossRef]
- Jeong, S.M.; Hwang, S.; Seong, R.H. SIRT4 Regulates Cancer Cell Survival and Growth after Stress. Biochem. Biophys. Res. Commun. 2016, 470, 251–256. [Google Scholar] [CrossRef]
- He, Q.; Chen, K.; Ye, R.; Dai, N.; Guo, P.; Wang, L. Associations of Sirtuins with Clinicopathological Variables and Prognosis in Human Ovarian Cancer. Oncol. Lett. 2020, 19, 3278–3288. [Google Scholar] [CrossRef] [PubMed]
- Igci, M.; Kalender, M.E.; Borazan, E.; Bozgeyik, I.; Bayraktar, R.; Bozgeyik, E.; Camci, C.; Arslan, A. High-Throughput Screening of Sirtuin Family of Genes in Breast Cancer. Gene 2016, 586, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, Y.; Gao, J.; Yuan, X. Tumor-Suppressive Function of SIRT4 in Neuroblastoma through Mitochondrial Damage. Cancer Manag. Res. 2018, 10, 5591–5603. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ding, X.; Wu, Q.; Qi, J.; Zhu, M.; Miao, C. Monomethyltransferase SET8 Facilitates Hepatocellular Carcinoma Growth by Enhancing Aerobic Glycolysis. Cell Death Dis. 2019, 10, 312. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Qin, Y.; Ji, S.; Xu, W.; Liu, W.; Sun, Q.; Zhang, Z.; Liu, M.; Ni, Q.; Yu, X.; et al. UHRF1 Promotes Aerobic Glycolysis and Proliferation via Suppression of SIRT4 in Pancreatic Cancer. Cancer Lett. 2019, 452, 226–236. [Google Scholar] [CrossRef]
- Shih, J.; Liu, L.; Mason, A.; Higashimori, H.; Donmez, G. Loss of SIRT4 Decreases GLT-1-Dependent Glutamate Uptake and Increases Sensitivity to Kainic Acid. J. Neurochem. 2014, 131, 573–581. [Google Scholar] [CrossRef]
- Buck, E.; Bayer, H.; Lindenberg, K.S.; Hanselmann, J.; Pasquarelli, N.; Ludolph, A.C.; Weydt, P.; Witting, A. Comparison of Sirtuin 3 Levels in ALS and Huntington’s Disease-Differential Effects in Human Tissue Samples vs. Transgenic Mouse Models. Front. Mol. Neurosci. 2017, 10, 156. [Google Scholar] [CrossRef] [PubMed]
- Haigis, M.C.; Mostoslavsky, R.; Haigis, K.M.; Fahie, K.; Christodoulou, D.C.; Murphy, A.J.; Valenzuela, D.M.; Yancopoulos, G.D.; Karow, M.; Blander, G.; et al. SIRT4 Inhibits Glutamate Dehydrogenase and Opposes the Effects of Calorie Restriction in Pancreatic Beta Cells. Cell 2006, 126, 941–954. [Google Scholar] [CrossRef]
- Hutton, J.C.; Sener, A.; Malaisse, W.J. Interaction of Branched Chain Amino Acids and Keto Acids upon Pancreatic Islet Metabolism and Insulin Secretion. J. Biol. Chem. 1980, 255, 7340–7346. [Google Scholar] [CrossRef]
- Huynh, F.K.; Hu, X.; Lin, Z.; Johnson, J.D.; Hirschey, M.D. Loss of Sirtuin 4 Leads to Elevated Glucose- and Leucine-Stimulated Insulin Levels and Accelerated Age-Induced Insulin Resistance in Multiple Murine Genetic Backgrounds. J. Inherit. Metab. Dis. 2018, 41, 59–72. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, H.; Zha, X. Overview of SIRT5 as a Potential Therapeutic Target: Structure, Function and Inhibitors. Eur. J. Med. Chem. 2022, 236, 114363. [Google Scholar] [CrossRef]
- Abril, Y.L.N.; Fernandez, I.R.; Hong, J.Y.; Chiang, Y.-L.; Kutateladze, D.A.; Zhao, Q.; Yang, M.; Hu, J.; Sadhukhan, S.; Li, B.; et al. Pharmacological and Genetic Perturbation Establish SIRT5 as a Promising Target in Breast Cancer. Oncogene 2021, 40, 1644–1658. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Franzini, A.; Pomicter, A.D.; Halverson, B.J.; Antelope, O.; Mason, C.C.; Ahmann, J.M.; Senina, A.V.; Vellore, N.A.; Jones, C.L.; et al. SIRT5 is a druggable metabolic vulnerability in acute myeloid leukemia. Blood Cancer Discov. 2021, 2, 266–287. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Shukla, S.K.; Vernucci, E.; He, C.; Wang, D.; King, R.J.; Jha, K.; Siddhanta, K.; Mullen, N.J.; Attri, K.S.; et al. Metabolic Rewiring by Loss of Sirt5 Promotes Kras-Induced Pancreatic Cancer Progression. Gastroenterology 2021, 161, 1584–1600. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Peritore, C.; Ginsberg, J.; Shih, J.; Arun, S.; Donmez, G. Protective Role of SIRT5 against Motor Deficit and Dopaminergic Degeneration in MPTP-Induced Mice Model of Parkinson’s Disease. Behav. Brain Res. 2015, 281, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wei, Y.; Li, J.; Bai, Y.; Yin, P.; Wang, S. SIRT5 Represses Neurotrophic Pathways and Aβ Production in Alzheimer’s Disease by Targeting Autophagy. ACS Chem. Neurosci. 2021, 12, 4428–4437. [Google Scholar] [CrossRef]
- Wang, G.; Meyer, J.G.; Cai, W.; Softic, S.; Li, M.E.; Verdin, E.; Newgard, C.; Schilling, B.; Kahn, C.R. Regulation of UCP1 and Mitochondrial Metabolism in Brown Adipose Tissue by Reversible Succinylation. Mol. Cell 2019, 74, 844–857.e7. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Fei, X. SIRT5 Regulates Pancreatic β-Cell Proliferation and Insulin Secretion in Type 2 Diabetes. Exp. Ther. Med. 2018, 16, 1417–1425. [Google Scholar] [CrossRef]
- You, W.; Rotili, D.; Li, T.-M.; Kambach, C.; Meleshin, M.; Schutkowski, M.; Chua, K.F.; Mai, A.; Steegborn, C. Structural Basis of Sirtuin 6 Activation by Synthetic Small Molecules. Angew. Chem. Int. Ed. Engl. 2017, 56, 1007–1011. [Google Scholar] [CrossRef]
- Iachettini, S.; Trisciuoglio, D.; Rotili, D.; Lucidi, A.; Salvati, E.; Zizza, P.; di Leo, L.; del Bufalo, D.; Ciriolo, M.R.; Leonetti, C.; et al. Pharmacological Activation of SIRT6 Triggers Lethal Autophagy in Human Cancer Cells. Cell Death Dis. 2018, 9, 996. [Google Scholar] [CrossRef]
- Huang, Z.; Zhao, J.; Deng, W.; Chen, Y.; Shang, J.; Song, K.; Zhang, L.; Wang, C.; Lu, S.; Yang, X.; et al. Identification of a Cellularly Active SIRT6 Allosteric Activator. Nat. Chem. Biol. 2018, 14, 1118–1126. [Google Scholar] [CrossRef]
- Shang, J.-L.; Ning, S.-B.; Chen, Y.-Y.; Chen, T.-X.; Zhang, J. MDL-800, an Allosteric Activator of SIRT6, Suppresses Proliferation and Enhances EGFR-TKIs Therapy in Non-Small Cell Lung Cancer. Acta Pharmacol. Sin. 2021, 42, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Zhu, Z.; Chen, Y.; Song, J.; Huang, Y.; Song, K.; Zhong, J.; Xu, X.; Wei, J.; Wang, C.; et al. Small-Molecule Activating SIRT6 Elicits Therapeutic Effects and Synergistically Promotes Anti-Tumor Activity of Vitamin D3 in Colorectal Cancer. Theranostics 2020, 10, 5845–5864. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sun, W.; Huang, S.; Zhang, H.; Lin, G.; Li, H.; Qiao, J.; Li, L.; Yang, S. Discovery of Potent Small-Molecule SIRT6 Activators: Structure-Activity Relationship and Anti-Pancreatic Ductal Adenocarcinoma Activity. J. Med. Chem. 2020, 63, 10474–10495. [Google Scholar] [CrossRef] [PubMed]
- Sociali, G.; Galeno, L.; Parenti, M.D.; Grozio, A.; Bauer, I.; Passalacqua, M.; Boero, S.; Donadini, A.; Millo, E.; Bellotti, M.; et al. Quinazolinedione SIRT6 Inhibitors Sensitize Cancer Cells to Chemotherapeutics. Eur. J. Med. Chem. 2015, 102, 530–539. [Google Scholar] [CrossRef]
- Damonte, P.; Sociali, G.; Parenti, M.D.; Soncini, D.; Bauer, I.; Boero, S.; Grozio, A.; von Holtey, M.; Piacente, F.; Becherini, P.; et al. SIRT6 Inhibitors with Salicylate-like Structure Show Immunosuppressive and Chemosensitizing Effects. Bioorg. Med. Chem. 2017, 25, 5849–5858. [Google Scholar] [CrossRef]
- Cagnetta, A.; Soncini, D.; Orecchioni, S.; Talarico, G.; Minetto, P.; Guolo, F.; Retali, V.; Colombo, N.; Carminati, E.; Clavio, M.; et al. Depletion of SIRT6 Enzymatic Activity Increases Acute Myeloid Leukemia Cells’ Vulnerability to DNA-Damaging Agents. Haematologica 2018, 103, 80–90. [Google Scholar] [CrossRef]
- Nicholatos, J.W.; Francisco, A.B.; Bender, C.A.; Yeh, T.; Lugay, F.J.; Salazar, J.E.; Glorioso, C.; Libert, S. Nicotine Promotes Neuron Survival and Partially Protects from Parkinson’s Disease by Suppressing SIRT6. Acta Neuropathol. Commun. 2018, 6, 120. [Google Scholar] [CrossRef]
- Ferrara, G.; Benzi, A.; Sturla, L.; Marubbi, D.; Frumento, D.; Spinelli, S.; Abbotto, E.; Ivaldi, F.; von Holtey, M.; Murone, M.; et al. Sirt6 Inhibition Delays the Onset of Experimental Autoimmune Encephalomyelitis by Reducing Dendritic Cell Migration. J Neuroinflamm. 2020, 17, 228. [Google Scholar] [CrossRef]
- Parenti, M.D.; Grozio, A.; Bauer, I.; Galeno, L.; Damonte, P.; Millo, E.; Sociali, G.; Franceschi, C.; Ballestrero, A.; Bruzzone, S.; et al. Discovery of Novel and Selective SIRT6 Inhibitors. J. Med. Chem. 2014, 57, 4796–4804. [Google Scholar] [CrossRef]
- Sociali, G.; Magnone, M.; Ravera, S.; Damonte, P.; Vigliarolo, T.; von Holtey, M.; Vellone, V.G.; Millo, E.; Caffa, I.; Cea, M.; et al. Pharmacological Sirt6 Inhibition Improves Glucose Tolerance in a Type 2 Diabetes Mouse Model. FASEB J. 2017, 31, 3138–3149. [Google Scholar] [CrossRef]
- Sun, W.; Chen, X.; Huang, S.; Li, W.; Tian, C.; Yang, S.; Li, L. Discovery of 5-(4-Methylpiperazin-1-Yl)-2-Nitroaniline Derivatives as a New Class of SIRT6 Inhibitors. Bioorg. Med. Chem. Lett. 2020, 30, 127215. [Google Scholar] [CrossRef] [PubMed]
- Grob, A.; Roussel, P.; Wright, J.E.; McStay, B.; Hernandez-Verdun, D.; Sirri, V. Involvement of SIRT7 in Resumption of RDNA Transcription at the Exit from Mitosis. J. Cell Sci. 2009, 122, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Ford, E.; Voit, R.; Liszt, G.; Magin, C.; Grummt, I.; Guarente, L. Mammalian Sir2 Homolog SIRT7 Is an Activator of RNA Polymerase I Transcription. Genes Dev. 2006, 20, 1075–1080. [Google Scholar] [CrossRef]
- Zhang, C.; Li, Y.; Liu, B.; Ning, C.; Li, Y.; Wang, Y.; Li, Z. Discovery of SIRT7 Inhibitor as New Therapeutic Options Against Liver Cancer. Front. Cell Dev. Biol. 2021, 9, 813233. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Kim, D.; Cho, S.J.; Jung, K.-Y.; Kim, J.-H.; Lee, J.M.; Jung, H.J.; Kim, K.R. Identification of a Novel SIRT7 Inhibitor as Anticancer Drug Candidate. Biochem. Biophys. Res. Commun. 2019, 508, 451–457. [Google Scholar] [CrossRef]
- Vazquez, B.N.; Thackray, J.K.; Serrano, L. Sirtuins and DNA Damage Repair: SIRT7 Comes to Play. Nucleus 2017, 8, 107–115. [Google Scholar] [CrossRef]
- Li, X.; Liu, J.; Lu, L.; Huang, T.; Hou, W.; Wang, F.; Yu, L.; Wu, F.; Qi, J.; Chen, X.; et al. Sirt7 Associates with ELK1 to Participate in Hyperglycemia Memory and Diabetic Nephropathy via Modulation of DAPK3 Expression and Endothelial Inflammation. Transl. Res. 2022, 247, 99–116. [Google Scholar] [CrossRef]
- Heltweg, B.; Gatbonton, T.; Schuler, A.D.; Posakony, J.; Li, H.; Goehle, S.; Kollipara, R.; Depinho, R.A.; Gu, Y.; Simon, J.A.; et al. Antitumor Activity of a Small-Molecule Inhibitor of Human Silent Information Regulator 2 Enzymes. Cancer Res. 2006, 66, 4368–4377. [Google Scholar] [CrossRef]
- Lu, B.; Zhang, D.; Wang, X.; Lin, D.; Chen, Y.; Xu, X. Targeting SIRT1 to Inhibit the Proliferation of Multiple Myeloma Cells. Oncol. Lett. 2021, 21, 306. [Google Scholar] [CrossRef]
- Portmann, S.; Fahrner, R.; Lechleiter, A.; Keogh, A.; Overney, S.; Laemmle, A.; Mikami, K.; Montani, M.; Tschan, M.P.; Candinas, D.; et al. Antitumor Effect of SIRT1 Inhibition in Human HCC Tumor Models in Vitro and in Vivo. Mol. Cancer Ther. 2013, 12, 499–508. [Google Scholar] [CrossRef]
- Marshall, G.M.; Liu, P.Y.; Gherardi, S.; Scarlett, C.J.; Bedalov, A.; Xu, N.; Iraci, N.; Valli, E.; Ling, D.; Thomas, W.; et al. SIRT1 Promotes N-Myc Oncogenesis through a Positive Feedback Loop Involving the Effects of MKP3 and ERK on N-Myc Protein Stability. PLoS Genet 2011, 7, e1002135. [Google Scholar] [CrossRef] [PubMed]
- Holloway, K.R.; Barbieri, A.; Malyarchuk, S.; Saxena, M.; Nedeljkovic-Kurepa, A.; Cameron Mehl, M.; Wang, A.; Gu, X.; Pruitt, K. SIRT1 Positively Regulates Breast Cancer Associated Human Aromatase (CYP19A1) Expression. Mol. Endocrinol. 2013, 27, 480–490. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, A.R.; Pirrie, L.; Hollick, J.J.; Ronseaux, S.; Campbell, J.; Higgins, M.; Staples, O.D.; Tran, F.; Slawin, A.M.Z.; Lain, S.; et al. Synthesis and Biological Characterisation of Sirtuin Inhibitors Based on the Tenovins. Bioorg. Med. Chem. 2012, 20, 1779–1793. [Google Scholar] [CrossRef]
- Ke, X.; Qin, Q.; Deng, T.; Liao, Y.; Gao, S.-J. Heterogeneous Responses of Gastric Cancer Cell Lines to Tenovin-6 and Synergistic Effect with Chloroquine. Cancers 2020, 12, 365. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.B.; Kim, Y.; Kim, D.; Cho, E.Y.; Han, J.; Kim, H.K.; Shim, Y.M.; Kim, D.-H. Metformin and Tenovin-6 Synergistically Induces Apoptosis through LKB1-Independent SIRT1 down-Regulation in Non-Small Cell Lung Cancer Cells. J. Cell Mol. Med. 2019, 23, 2872–2889. [Google Scholar] [CrossRef]
- Yuan, H.; Wang, Z.; Li, L.; Zhang, H.; Modi, H.; Horne, D.; Bhatia, R.; Chen, W. Activation of Stress Response Gene SIRT1 by BCR-ABL Promotes Leukemogenesis. Blood 2012, 119, 1904–1914. [Google Scholar] [CrossRef]
- Schnekenburger, M.; Goffin, E.; Lee, J.-Y.; Jang, J.Y.; Mazumder, A.; Ji, S.; Rogister, B.; Bouider, N.; Lefranc, F.; Miklos, W.; et al. Discovery and Characterization of R/S-N-3-Cyanophenyl-N’-(6-Tert-Butoxycarbonylamino-3,4-Dihydro-2,2-Dimethyl-2H-1-Benzopyran-4-Yl)Urea, a New Histone Deacetylase Class III Inhibitor Exerting Antiproliferative Activity against Cancer Cell Lines. J. Med. Chem. 2017, 60, 4714–4733. [Google Scholar] [CrossRef]
- Laaroussi, H.; Ding, Y.; Teng, Y.; Deschamps, P.; Vidal, M.; Yu, P.; Broussy, S. Synthesis of Indole Inhibitors of Silent Information Regulator 1 (SIRT1), and Their Evaluation as Cytotoxic Agents. Eur. J. Med. Chem. 2020, 202, 112561. [Google Scholar] [CrossRef]
- Grozinger, C.M.; Chao, E.D.; Blackwell, H.E.; Moazed, D.; Schreiber, S.L. Identification of a Class of Small Molecule Inhibitors of the Sirtuin Family of NAD-Dependent Deacetylases by Phenotypic Screening. J. Biol. Chem. 2001, 276, 38837–38843. [Google Scholar] [CrossRef]
- Ota, H.; Tokunaga, E.; Chang, K.; Hikasa, M.; Iijima, K.; Eto, M.; Kozaki, K.; Akishita, M.; Ouchi, Y.; Kaneki, M. Sirt1 Inhibitor, Sirtinol, Induces Senescence-like Growth Arrest with Attenuated Ras-MAPK Signaling in Human Cancer Cells. Oncogene 2006, 25, 176–185. [Google Scholar] [CrossRef]
- Zhou, W.; Ni, T.K.; Wronski, A.; Glass, B.; Skibinski, A.; Beck, A.; Kuperwasser, C. The SIRT2 Deacetylase Stabilizes Slug to Control Malignancy of Basal-like Breast Cancer. Cell Rep. 2016, 17, 1302–1317. [Google Scholar] [CrossRef]
- Kojima, K.; Ohhashi, R.; Fujita, Y.; Hamada, N.; Akao, Y.; Nozawa, Y.; Deguchi, T.; Ito, M. A Role for SIRT1 in Cell Growth and Chemoresistance in Prostate Cancer PC3 and DU145 Cells. Biochem. Biophys. Res. Commun. 2008, 373, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Lara, E.; Mai, A.; Calvanese, V.; Altucci, L.; Lopez-Nieva, P.; Martinez-Chantar, M.L.; Varela-Rey, M.; Rotili, D.; Nebbioso, A.; Ropero, S.; et al. Salermide, a Sirtuin Inhibitor with a Strong Cancer-Specific Proapoptotic Effect. Oncogene 2009, 28, 781–791. [Google Scholar] [CrossRef]
- Liu, G.; Su, L.; Hao, X.; Zhong, N.; Zhong, D.; Singhal, S.; Liu, X. Salermide Up-Regulates Death Receptor 5 Expression through the ATF4-ATF3-CHOP Axis and Leads to Apoptosis in Human Cancer Cells. J. Cell Mol. Med. 2012, 16, 1618–1628. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.J.; Lee, Y.T.; Yeong, K.Y.; Petersen, S.H.; Kono, K.; Tan, S.C.; Oon, C.E. Anticancer Activities of a Benzimidazole Compound through Sirtuin Inhibition in Colorectal Cancer. Future Med. Chem. 2018, 10, 2039–2057. [Google Scholar] [CrossRef] [PubMed]
- Spiegelman, N.A.; Hong, J.Y.; Hu, J.; Jing, H.; Wang, M.; Price, I.R.; Cao, J.; Yang, M.; Zhang, X.; Lin, H. A Small-Molecule SIRT2 Inhibitor That Promotes K-Ras4a Lysine Fatty-Acylation. ChemMedChem 2019, 14, 744–748. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, G.T.T.; Gertz, M.; Steegborn, C. Crystal Structures of Sirt3 Complexes with 4’-Bromo-Resveratrol Reveal Binding Sites and Inhibition Mechanism. Chem. Biol. 2013, 20, 1375–1385. [Google Scholar] [CrossRef]
- George, J.; Nihal, M.; Singh, C.K.; Ahmad, N. 4’-Bromo-Resveratrol, a Dual Sirtuin-1 and Sirtuin-3 Inhibitor, Inhibits Melanoma Cell Growth through Mitochondrial Metabolic Reprogramming. Mol. Carcinog. 2019, 58, 1876–1885. [Google Scholar] [CrossRef]
- Hui, Q.; Li, X.; Fan, W.; Gao, C.; Zhang, L.; Qin, H.; Wei, L.; Zhang, L. Discovery of 2-(4-Acrylamidophenyl)-Quinoline-4-Carboxylic Acid Derivatives as Potent SIRT3 Inhibitors. Front. Chem. 2022, 10, 880067. [Google Scholar] [CrossRef]
- Audrito, V.; Vaisitti, T.; Rossi, D.; Gottardi, D.; D’Arena, G.; Laurenti, L.; Gaidano, G.; Malavasi, F.; Deaglio, S. Nicotinamide Blocks Proliferation and Induces Apoptosis of Chronic Lymphocytic Leukemia Cells through Activation of the P53/MiR-34a/SIRT1 Tumor Suppressor Network. Cancer Res. 2011, 71, 4473–4483. [Google Scholar] [CrossRef]
- Alhazzazi, T.Y.; Kamarajan, P.; Joo, N.; Huang, J.-Y.; Verdin, E.; D’Silva, N.J.; Kapila, Y.L. Sirtuin-3 (SIRT3), a Novel Potential Therapeutic Target for Oral Cancer. Cancer 2011, 117, 1670–1678. [Google Scholar] [CrossRef] [PubMed]
- Contoyiannis, P.A.; Draginis, E.; Adamopoulos, D.A.; Triantafyllou, G. Letter: Levodopa in Coma Due to Fulminant Hepatitis. Br. Med. J. 1975, 1, 272. [Google Scholar] [CrossRef] [PubMed]
- Jung-Hynes, B.; Nihal, M.; Zhong, W.; Ahmad, N. Role of Sirtuin Histone Deacetylase SIRT1 in Prostate Cancer. A Target for Prostate Cancer Management via Its Inhibition? J. Biol. Chem. 2009, 284, 3823–3832. [Google Scholar] [CrossRef]
- Liu, D.; Pitta, M.; Jiang, H.; Lee, J.-H.; Zhang, G.; Chen, X.; Kawamoto, E.M.; Mattson, M.P. Nicotinamide Forestalls Pathology and Cognitive Decline in Alzheimer Mice: Evidence for Improved Neuronal Bioenergetics and Autophagy Procession. Neurobiol. Aging 2013, 34, 1564–1580. [Google Scholar] [CrossRef] [PubMed]
- Green, K.N.; Steffan, J.S.; Martinez-Coria, H.; Sun, X.; Schreiber, S.S.; Thompson, L.M.; LaFerla, F.M. Nicotinamide Restores Cognition in Alzheimer’s Disease Transgenic Mice via a Mechanism Involving Sirtuin Inhibition and Selective Reduction of Thr231-Phosphotau. J. Neurosci. 2008, 28, 11500–11510. [Google Scholar] [CrossRef] [PubMed]
- Hathorn, T.; Snyder-Keller, A.; Messer, A. Nicotinamide Improves Motor Deficits and Upregulates PGC-1α and BDNF Gene Expression in a Mouse Model of Huntington’s Disease. Neurobiol. Dis. 2011, 41, 43–50. [Google Scholar] [CrossRef]
- Lee, S.-J.; Choi, S.-E.; Jung, I.-R.; Lee, K.-W.; Kang, Y. Protective Effect of Nicotinamide on High Glucose/Palmitate-Induced Glucolipotoxicity to INS-1 Beta Cells Is Attributed to Its Inhibitory Activity to Sirtuins. Arch. Biochem. Biophys. 2013, 535, 187–196. [Google Scholar] [CrossRef]
- PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/ (accessed on 14 April 2022).
- Document Search-Web of Science Core Collection. Available online: https://www.webofscience.com/wos/woscc/basic-search (accessed on 14 April 2022).
- Huhtiniemi, T.; Suuronen, T.; Rinne, V.M.; Wittekindt, C.; Lahtela-Kakkonen, M.; Jarho, E.; Wallén, E.A.A.; Salminen, A.; Poso, A.; Leppänen, J. Oxadiazole-Carbonylaminothioureas as SIRT1 and SIRT2 Inhibitors. J. Med. Chem. 2008, 51, 4377–4380. [Google Scholar] [CrossRef]
- Huhtiniemi, T.; Wittekindt, C.; Laitinen, T.; Leppänen, J.; Salminen, A.; Poso, A.; Lahtela-Kakkonen, M. Comparative and Pharmacophore Model for Deacetylase SIRT1. J. Comput. Aided Mol. Des. 2006, 20, 589–599. [Google Scholar] [CrossRef]
- Sakkiah, S.; Krishnamoorthy, N.; Gajendrarao, P.; Thangapandian, S.; Lee, Y.; Kim, S.; Suh, J.K.; Kim, H.H.; Lee, K.W. Pharmacophore Mapping and Virtual Screening for SIRT1 Activators. Bull. Korean Chem. Soc. 2009, 30, 1152–1156. [Google Scholar]
- Maybridge Library | Thermo Fisher Scientific-IT. Available online: https://www.thermofisher.com/it/en/home/global/forms/lab-solutions/maybridge-library.html (accessed on 13 May 2022).
- Sakkiah, S.; Arooj, M.; Lee, K.W.; Torres, J.Z. Theoretical Approaches to Identify the Potent Scaffold for Human Sirtuin1 Activator: Bayesian Modeling and Density Functional Theory. Med. Chem. Res. 2014, 23, 3998–4010. [Google Scholar] [CrossRef]
- Vyas, V.K.; Goel, A.; Ghate, M.; Patel, P. Ligand and Structure-Based Approaches for the Identification of SIRT1 Activators. Chem. Biol. Interact. 2015, 228, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Shoichet, B.K. ZINC-A Free Database of Commercially Available Compounds for Virtual Screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Azminah, A.; Erlina, L.; Radji, M.; Mun’im, A.; Syahdi, R.R.; Yanuar, A. In Silico and in Vitro Identification of Candidate SIRT1 Activators from Indonesian Medicinal Plants Compounds Database. Comput. Biol. Chem. 2019, 83, 107096. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Case, A.W.; Riera, T.V.; Considine, T.; Lee, J.E.; Hamuro, Y.; Zhao, H.; Jiang, Y.; Sweitzer, S.M.; Pietrak, B.; et al. Crystallographic Structure of a Small Molecule SIRT1 Activator-Enzyme Complex. Nat. Commun. 2015, 6, 7645. [Google Scholar] [CrossRef]
- Basis Data Tanaman Obat Indonesia. Available online: http://herbaldb.farmasi.ui.ac.id/v3/ (accessed on 13 May 2022).
- Zhao, X.; Allison, D.; Condon, B.; Zhang, F.; Gheyi, T.; Zhang, A.; Ashok, S.; Russell, M.; MacEwan, I.; Qian, Y.; et al. The 2.5 Å Crystal Structure of the SIRT1 Catalytic Domain Bound to Nicotinamide Adenine Dinucleotide (NAD+) and an Indole (EX527 Analogue) Reveals a Novel Mechanism of Histone Deacetylase Inhibition. J. Med. Chem. 2013, 56, 963–969. [Google Scholar] [CrossRef] [PubMed]
- Asinex.Com-Home. Available online: https://www.asinex.com/ (accessed on 13 May 2022).
- Pulla, V.K.; Alvala, M.; Sriram, D.S.; Viswanadha, S.; Sriram, D.; Yogeeswari, P. Structure-Based Drug Design of Small Molecule SIRT1 Modulators to Treat Cancer and Metabolic Disorders. J. Mol. Graph. Model. 2014, 52, 46–56. [Google Scholar] [CrossRef]
- Autiero, I.; Costantini, S.; Colonna, G. Human Sirt-1: Molecular Modeling and Structure-Function Relationships of an Unordered Protein. PLoS ONE 2009, 4, e7350. [Google Scholar] [CrossRef]
- Padmanabhan, B.; Ramu, M.; Mathur, S.; Unni, S.; Thiyagarajan, S. Identification of New Inhibitors for Human SIRT1: An in-Silico Approach. Med. Chem. 2016, 12, 347–361. [Google Scholar] [CrossRef]
- Davenport, A.M.; Huber, F.M.; Hoelz, A. Structural and Functional Analysis of Human SIRT1. J. Mol. Biol. 2014, 426, 526–541. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A Comprehensive Resource for in Silico Drug Discovery and Exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
- Princeton BioMolecular Research, Inc. Available online: https://princetonbio.com/ (accessed on 13 May 2022).
- Ertan-Bolelli, T.; Bolelli, K. In Silico Design of Novel Sirtuin 1 Enzyme Activators for the Treatment of Age-Related Diseases and Life Span. Curr. Comput. Aided Drug. Des. 2021, 17, 412–420. [Google Scholar] [CrossRef]
- Wang, F.; Liu, D.; Wang, H.; Luo, C.; Zheng, M.; Liu, H.; Zhu, W.; Luo, X.; Zhang, J.; Jiang, H. Computational Screening for Active Compounds Targeting Protein Sequences: Methodology and Experimental Validation. J. Chem. Inf. Model. 2011, 51, 2821–2828. [Google Scholar] [CrossRef] [PubMed]
- Vapnik, V. The Nature of Statistical Learning Theory; Springer: New York, NY, USA, 1999. [Google Scholar]
- Zheng, M.; Liu, Z.; Xue, C.; Zhu, W.; Chen, K.; Luo, X.; Jiang, H. Mutagenic Probability Estimation of Chemical Compounds by a Novel Molecular Electrophilicity Vector and Support Vector Machine. Bioinformatics 2006, 22, 2099–2106. [Google Scholar] [CrossRef] [PubMed]
- Gertrudes, J.C.; Maltarollo, V.G.; Silva, R.A.; Oliveira, P.R.; Honório, K.M.; da Silva, A.B.F. Machine Learning Techniques and Drug Design. Curr. Med. Chem. 2012, 19, 4289–4297. [Google Scholar] [CrossRef] [PubMed]
- Specs-Compound Management Services and Research Compounds for the Life Science Industry. Available online: https://specs.net/ (accessed on 13 May 2022).
- Sun, Y.; Zhou, H.; Zhu, H.; Leung, S. Ligand-Based Virtual Screening and Inductive Learning for Identification of SIRT1 Inhibitors in Natural Products. Sci. Rep. 2016, 6, 19312. [Google Scholar] [CrossRef]
- Sanderson, K. Databases Aim to Bridge the East-West Divide of Drug Discovery. Nat. Med. 2011, 17, 1531. [Google Scholar] [CrossRef]
- Xue, R.; Fang, Z.; Zhang, M.; Yi, Z.; Wen, C.; Shi, T. TCMID: Traditional Chinese Medicine Integrative Database for Herb Molecular Mechanism Analysis. Nucleic Acids Res. 2013, 41, D1089–D1095. [Google Scholar] [CrossRef]
- Chiba, S.; Ohue, M.; Gryniukova, A.; Borysko, P.; Zozulya, S.; Yasuo, N.; Yoshino, R.; Ikeda, K.; Shin, W.H.; Kihara, D.; et al. A Prospective Compound Screening Contest Identified Broader Inhibitors for Sirtuin 1. Sci. Rep. 2019, 9, 19585. [Google Scholar] [CrossRef]
- Uciechowska, U.; Schemies, J.; Neugebauer, R.C.; Huda, E.-M.; Schmitt, M.L.; Meier, R.; Verdin, E.; Jung, M.; Sippl, W. Thiobarbiturates as Sirtuin Inhibitors: Virtual Screening, Free-Energy Calculations, and Biological Testing. ChemMedChem 2008, 3, 1965–1976. [Google Scholar] [CrossRef]
- ChemBridge | Home. Available online: https://www.chembridge.com/ (accessed on 13 May 2022).
- Uciechowska, U.; Schemies, J.; Scharfe, M.; Lawson, M.; Wichapong, K.; Jung, M.; Sippl, W. Binding Free Energy Calculations and Biological Testing of Novel Thiobarbiturates as Inhibitors of the Human NAD+ Dependent Histone Deacetylase Sirt2. MedChemComm 2012, 3, 167–173. [Google Scholar] [CrossRef]
- Sivaraman, P.; Mattegunta, S.; Subbaraju, G.V.; Satyanarayana, C.; Padmanabhan, B. Design of a Novel Nucleoside Analog as Potent Inhibitor of the NAD+ Dependent Deacetylase, SIRT2. Syst. Synth. Biol. 2010, 4, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Home-NCI Wiki. Available online: https://wiki.nci.nih.gov/dashboard.action (accessed on 13 May 2022).
- Sacconnay, L.; Ryckewaert, L.; Randazzo, G.M.; Petit, C.; Passos, C.D.S.; Jachno, J.; Michailovienė, V.; Zubrienė, A.; Matulis, D.; Carrupt, P.-A.; et al. 5-Benzylidene-Hydantoin Is a New Scaffold for SIRT Inhibition: From Virtual Screening to Activity Assays. Eur. J. Pharm. Sci. 2016, 85, 59–67. [Google Scholar] [CrossRef]
- Moniot, S.; Schutkowski, M.; Steegborn, C. Crystal Structure Analysis of Human Sirt2 and Its ADP-Ribose Complex. J. Struct. Biol. 2013, 182, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Tervo, A.J.; Kyrylenko, S.; Niskanen, P.; Salminen, A.; Leppänen, J.; Nyrönen, T.H.; Järvinen, T.; Poso, A. An In Silico Approach to Discovering Novel Inhibitors of Human Sirtuin Type 2. J. Med. Chem. 2004, 47, 6292–6298. [Google Scholar] [CrossRef] [PubMed]
- Tervo, A.J.; Suuronen, T.; Kyrylenko, S.; Kuusisto, E.; Kiviranta, P.H.; Salminen, A.; Leppanen, J.; Poso, A. Discovering Inhibitors of Human Sirtuin Type 2: Novel Structural Scaffolds. J. Med. Chem. 2006, 49, 7239–7241. [Google Scholar] [CrossRef] [PubMed]
- Sakkiah, S.; Thangapandian, S.; Park, C.; Son, M.; Lee, K.W. Molecular Docking and Dynamics Simulation, Receptor-Based Hypothesis: Application to Identify Novel Sirtuin 2 Inhibitors. Chem. Biol. Drug Des. 2012, 80, 315–327. [Google Scholar] [CrossRef]
- CHEMDIV INC-FULLY INTEGRATED TARGET-TO-CLINIC CONTRACT RESEARCH ORGANIZATION (CRO). Available online: https://www.chemdiv.com/ (accessed on 13 May 2022).
- Sakkiah, S.; Baek, A.; Lee, K.W. Pharmacophore Modeling and Molecular Dynamics Simulation to Identify the Critical Chemical Features against Human Sirtuin 2 Inhibitors. J. Mol. Struct. 2012, 1011, 66–75. [Google Scholar] [CrossRef]
- Schiedel, M.; Rumpf, T.; Karaman, B.; Lehotzky, A.; Oláh, J.; Gerhardt, S.; Ovádi, J.; Sippl, W.; Einsle, O.; Jung, M. Aminothiazoles as Potent and Selective Sirt2 Inhibitors: A Structure–Activity Relationship Study. J. Med. Chem. 2016, 59, 1599–1612. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Kessler, A.; Kouznetsova, V.L.; Tsigelny, I.F. Targeting Epigenetic Regulators Using Machine Learning: Potential Sirtuin 2 Inhibitors. J. Comput. Biophys. Chem. 2021, 20, 841–851. [Google Scholar] [CrossRef]
- Khanfar, M.A.; Alqtaishat, S. Discovery of Potent Natural-Product-Derived SIRT2 Inhibitors Using Structure- Based Exploration of SIRT2 Pharmacophoric Space Coupled With QSAR Analyses. Anticancer Agents Med. Chem. 2021, 21, 2278–2286. [Google Scholar] [CrossRef] [PubMed]
- Triballeau, N.; Acher, F.; Brabet, I.; Pin, J.P.; Bertrand, H.O. Virtual Screening Workflow Development Guided by the “Receiver Operating Characteristic” Curve Approach. Application to High-Throughput Docking on Metabotropic Glutamate Receptor Subtype 4. J. Med. Chem. 2005, 48, 2534–2547. [Google Scholar] [CrossRef] [PubMed]
- Rogers, D.; Hopfinger, A.J. Application of Genetic Function Approximation to Quantitative Structure-Activity Relationships and Quantitative Structure-Property Relationships. J. Chem. Inf. Comput. Sci. 1994, 34, 854–866. [Google Scholar] [CrossRef]
- Taha, M.O.; Bustanji, Y.; Al-Ghussein, M.A.S.; Mohammad, M.; Zalloum, H.; Al-Masri, I.M.; Atallah, N. Pharmacophore Modeling, Quantitative Structure–Activity Relationship Analysis, and in Silico Screening Reveal Potent Glycogen Synthase Kinase-3β Inhibitory Activities for Cimetidine, Hydroxychloroquine, and Gemifloxacin. J. Med. Chem. 2008, 51, 2062–2077. [Google Scholar] [CrossRef] [PubMed]
- Khanfar, M.A.; Banat, F.; Alabed, S.; Alqtaishat, S. Discovery of Potent NEK2 Inhibitors as Potential Anticancer Agents Using Structure-Based Exploration of NEK2 Pharmacophoric Space Coupled with QSAR Analyses. Mol. Divers. 2017, 21, 187–200. [Google Scholar] [CrossRef]
- Khanfar, M.A.; Alqtaishat, S. Discovery of Potent IRAK-4 Inhibitors as Potential Anti-Inflammatory and Anticancer Agents Using Structure-Based Exploration of IRAK-4 Pharmacophoric Space Coupled with QSAR Analyses. Comput. Biol. Chem. 2019, 79, 147–154. [Google Scholar] [CrossRef]
- MEGx Purified Natural Product Screening Compounds-AnalytiCon Discovery. Available online: https://ac-discovery.com/purified-natural-product-screening-compounds/ (accessed on 13 May 2022).
- Kokkonen, P.; Kokkola, T.; Suuronen, T.; Poso, A.; Jarho, E.; Lahtela-Kakkonen, M. Virtual Screening Approach of Sirtuin Inhibitors Results in Two New Scaffolds. Eur. J. Pharm. Sci. 2015, 76, 27–32. [Google Scholar] [CrossRef]
- Salo, H.S.; Laitinen, T.; Poso, A.; Jarho, E.; Lahtela-Kakkonen, M. Identification of Novel SIRT3 Inhibitor Scaffolds by Virtual Screening. Bioorg. Med. Chem. Lett. 2013, 23, 2990–2995. [Google Scholar] [CrossRef]
- Choubey, S.K.; Prabhu, D.; Nachiappan, M.; Biswal, J.; Jeyakanthan, J. Molecular Modeling, Dynamics Studies and Density Functional Theory Approaches to Identify Potential Inhibitors of SIRT4 Protein from Homo Sapiens: A Novel Target for the Treatment of Type 2 Diabetes. J. Biomol. Struct. Dyn. 2017, 35, 3316–3329. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ji, S.; Yu, Z.-J.; Wang, H.-L.; Cheng, X.; Li, W.-J.; Jing, L.; Yu, Y.; Chen, Q.; Yang, L.-L.; et al. Structure-Based Discovery of New Selective Small-Molecule Sirtuin 5 Inhibitors. Chem. Biol. Drug Des. 2018, 91, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Tenhunen, J.; Kučera, T.; Huovinen, M.; Küblbeck, J.; Bisenieks, E.; Vigante, B.; Ogle, Z.; Duburs, G.; Doležal, M.; Moaddel, R.; et al. Screening of SIRT6 Inhibitors and Activators: A Novel Activator Has an Impact on Breast Cancer Cells. Biomed. Pharmacother. 2021, 138, 111452. [Google Scholar] [CrossRef] [PubMed]
- Schlicker, C.; Boanca, G.; Lakshminarasimhan, M.; Steegborn, C. Structure-Based Development of Novel Sirtuin Inhibitors. Aging 2011, 3, 852–872. [Google Scholar] [CrossRef]
- Karaman, B.; Alhalabi, Z.; Swyter, S.; Mihigo, S.; Andrae-Marobela, K.; Jung, M.; Sippl, W.; Ntie-Kang, F. Identification of Bichalcones as Sirtuin Inhibitors by Virtual Screening and In Vitro Testing. Molecules 2018, 23, 416. [Google Scholar] [CrossRef]
- Sinha, S.; Patel, S.; Athar, M.; Vora, J.; Chhabria, M.T.; Jha, P.C.; Shrivastava, N. Structure-Based Identification of Novel Sirtuin Inhibitors against Triple Negative Breast Cancer: An in Silico and in Vitro Study. Int. J. Biol. Macromol. 2019, 140, 454–468. [Google Scholar] [CrossRef]
- SARI, S.; AVCI, A.; KOÇAK ASLAN, E. In Silico Repurposing of Drugs for Pan-HDAC and Pan-SIRT Inhibitors: Consensus Structure-Based Virtual Screening and Pharmacophore Modeling Investigations. Turk. J. Pharm. Sci. 2021, 18, 730–737. [Google Scholar] [CrossRef]
- Kadam, R.U.; Tavares, J.; Kiran, V.M.; Cordeiro, A.; Ouaissi, A.; Roy, N. Structure Function Analysis of Leishmania Sirtuin: An Ensemble of In Silico and Biochemical Studies. Chem. Biol. Drug Des. 2008, 71, 501–506. [Google Scholar] [CrossRef]
- Soares, M.B.P.; Silva, C.V.; Bastos, T.M.; Guimarães, E.T.; Figueira, C.P.; Smirlis, D.; Azevedo, W.F. Anti-Trypanosoma Cruzi Activity of Nicotinamide. Acta Trop. 2012, 122, 224–229. [Google Scholar] [CrossRef]
- Sacconnay, L.; Angleviel, M.; Randazzo, G.M.; Marçal Ferreira Queiroz, M.; Ferreira Queiroz, E.; Wolfender, J.-L.; Carrupt, P.-A.; Nurisso, A. Computational Studies on Sirtuins from Trypanosoma Cruzi: Structures, Conformations and Interactions with Phytochemicals. PLoS Negl. Trop. Dis. 2014, 8, e2689. [Google Scholar] [CrossRef]
- Singh, R.; Singh, S.; Pandey, P.N. In Silico Analysis of Sirt2 from Schistosoma Mansoni: Structures, Conformations, and Interactions with Inhibitors. J. Biomol. Struct. Dyn. 2016, 34, 1042–1051. [Google Scholar] [CrossRef] [PubMed]
- Huhtiniemi, T.; Salo, H.S.; Suuronen, T.; Poso, A.; Salminen, A.; Leppänen, J.; Jarho, E.; Lahtela-Kakkonen, M. Structure-Based Design of Pseudopeptidic Inhibitors for SIRT1 and SIRT2. J. Med. Chem. 2011, 54, 6456–6468. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Wei, W.; Jiang, Y.; Peng, H.; Cai, J.; Mao, C.; Dai, H.; Choy, W.; Bemis, J.E.; Jirousek, M.R.; et al. Crystal Structures of Human SIRT3 Displaying Substrate-Induced Conformational Changes. J. Biol. Chem. 2009, 284, 24394–24405. [Google Scholar] [CrossRef]
- Halgren, T. New Method for Fast and Accurate Binding-Site Identification and Analysis. Chem. Biol. Drug Des. 2007, 69, 146–148. [Google Scholar] [CrossRef] [PubMed]
- Szczepankiewicz, B.G.; Dai, H.; Koppetsch, K.J.; Qian, D.; Jiang, F.; Mao, C.; Perni, R.B. Synthesis of Carba-NAD and the Structures of Its Ternary Complexes with SIRT3 and SIRT5. J. Org. Chem. 2012, 77, 7319–7329. [Google Scholar] [CrossRef]
- QM-Polarized Ligand Docking Protocol. Available online: http://gohom.win/ManualHom/Schrodinger/Schrodinger_2015-2_docs/qpld/qpld_user_manual.pdf (accessed on 29 July 2022).
- Zhou, Y.; Zhang, H.; He, B.; Du, J.; Lin, H.; Cerione, R.A.; Hao, Q. The Bicyclic Intermediate Structure Provides Insights into the Desuccinylation Mechanism of Human Sirtuin 5 (SIRT5). J. Biol. Chem. 2012, 287, 28307–28314. [Google Scholar] [CrossRef]
- Wang, H.-L.; Liu, S.; Yu, Z.-J.; Wu, C.; Cheng, L.; Wang, Y.; Chen, K.; Zhou, S.; Chen, Q.; Yu, Y.; et al. Interactions between Sirtuins and Fluorogenic Small-Molecule Substrates Offer Insights into Inhibitor Design. RSC Adv. 2017, 7, 36214–36222. [Google Scholar] [CrossRef]
- Pan, P.W.; Feldman, J.L.; Devries, M.K.; Dong, A.; Edwards, A.M.; Denu, J.M. Structure and Biochemical Functions of SIRT6. J. Biol. Chem. 2011, 286, 14575–14587. [Google Scholar] [CrossRef]
- del Rio, A.; Barbosa, A.J.M.; Caporuscio, F.; Mangiatordi, G.F. CoCoCo: A Free Suite of Multiconformational Chemical Databases for High-Throughput Virtual Screening Purposes. Mol. Biosyst. 2010, 6, 2122. [Google Scholar] [CrossRef]
- Home-Enamine. Available online: https://enamine.net/ (accessed on 13 May 2022).
- Mai, A.; Valente, S.; Meade, S.; Carafa, V.; Tardugno, M.; Nebbioso, A.; Galmozzi, A.; Mitro, N.; de Fabiani, E.; Altucci, L.; et al. Study of 1,4-Dihydropyridine Structural Scaffold: Discovery of Novel Sirtuin Activators and Inhibitors. J. Med. Chem. 2009, 52, 5496–5504. [Google Scholar] [CrossRef]
- Valente, S.; Mellini, P.; Spallotta, F.; Carafa, V.; Nebbioso, A.; Polletta, L.; Carnevale, I.; Saladini, S.; Trisciuoglio, D.; Gabellini, C.; et al. 1,4-Dihydropyridines Active on the SIRT1/AMPK Pathway Ameliorate Skin Repair and Mitochondrial Function and Exhibit Inhibition of Proliferation in Cancer Cells. J. Med. Chem. 2016, 59, 1471–1491. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Lu, S.; Huang, Z.; Liu, X.; Mou, L.; Luo, Y.; Zhao, Y.; Liu, Y.; Chen, Z.; Hou, T.; et al. Allosite: A Method for Predicting Allosteric Sites. Bioinformatics 2013, 29, 2357–2359. [Google Scholar] [CrossRef] [PubMed]
- Priyanka, A.; Solanki, V.; Parkesh, R.; Thakur, K.G. Crystal Structure of the N-Terminal Domain of Human SIRT7 Reveals a Three-Helical Domain Architecture. Proteins Struct. Funct. Bioinform. 2016, 84, 1558–1563. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://Zhanggroup.Org/I-TASSER/ (accessed on 29 July 2022).
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER Server: New Development for Protein Structure and Function Predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, A.; Min, J.; Antoshenko, T.; Wang, C.L.; Allali-Hassani, A.; Dong, A.; Loppnau, P.; Vedadi, M.; Bochkarev, A.; Sternglanz, R.; et al. Structural Basis of Inhibition of the Human NAD+-Dependent Deacetylase SIRT5 by Suramin. Structure 2007, 15, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Amengual, J.E.; Clark-Garvey, S.; Kalac, M.; Scotto, L.; Marchi, E.; Neylon, E.; Johannet, P.; Wei, Y.; Zain, J.; O’Connor, O.A. Sirtuin and Pan-Class I/II Deacetylase (DAC) Inhibition Is Synergistic in Preclinical Models and Clinical Studies of Lymphoma. Blood 2013, 122, 2104–2113. [Google Scholar] [CrossRef]
- Park, M.A.; Mitchell, C.; Zhang, G.; Yacoub, A.; Allegood, J.; Häussinger, D.; Reinehr, R.; Larner, A.; Spiegel, S.; Fisher, P.B.; et al. Vorinostat and Sorafenib Increase CD95 Activation in Gastrointestinal Tumor Cells through a Ca2+ De Novo Ceramide-PP2A-Reactive Oxygen Species–Dependent Signaling Pathway. Cancer Res. 2010, 70, 6313–6324. [Google Scholar] [CrossRef]
- Ntie-Kang, F.; Amoa Onguéné, P.; Fotso, G.W.; Andrae-Marobela, K.; Bezabih, M.; Ndom, J.C.; Ngadjui, B.T.; Ogundaini, A.O.; Abegaz, B.M.; Meva’a, L.M. Virtualizing the P-ANAPL Library: A Step towards Drug Discovery from African Medicinal Plants. PLoS ONE 2014, 9, e90655. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.-B.; Jing, H.; Aramsangtienchai, P.; He, B.; Khan, S.; Hu, J.; Lin, H.; Hao, Q. Efficient Demyristoylase Activity of SIRT2 Revealed by Kinetic and Structural Studies. Sci. Rep. 2015, 5, 8529. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, K.; Goto, Y.; Nishimasu, H.; Morimoto, J.; Ishitani, R.; Dohmae, N.; Takeda, N.; Nagai, R.; Komuro, I.; Suga, H.; et al. Structural Basis for Potent Inhibition of SIRT2 Deacetylase by a Macrocyclic Peptide Inducing Dynamic Structural Change. Structure 2014, 22, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Rumpf, T.; Gerhardt, S.; Einsle, O.; Jung, M. Seeding for Sirtuins: Microseed Matrix Seeding to Obtain Crystals of Human Sirt3 and Sirt2 Suitable for Soaking. Acta Crystallogr. F Struct. Biol. Commun. 2015, 71, 1498–1510. [Google Scholar] [CrossRef]
- Gertz, M.; Fischer, F.; Nguyen, G.T.T.; Lakshminarasimhan, M.; Schutkowski, M.; Weyand, M.; Steegborn, C. Ex-527 Inhibits Sirtuins by Exploiting Their Unique NAD-Dependent Deacetylation Mechanism. Proc. Natl. Acad. Sci. USA 2013, 110, E2772–E2781. [Google Scholar] [CrossRef]
- You, W.; Steegborn, C. Structural Basis of Sirtuin 6 Inhibition by the Hydroxamate Trichostatin A: Implications for Protein Deacylase Drug Development. J. Med. Chem. 2018, 61, 10922–10928. [Google Scholar] [CrossRef]
- McGann, M. FRED and HYBRID Docking Performance on Standardized Datasets. J. Comput. Aided Mol. Des. 2012, 26, 897–906. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef]
- RCSB PDB-2B4Y: Crystal Structure of Human Sirtuin Homolog 5. Available online: https://www.rcsb.org/structure/2B4Y (accessed on 13 April 2022).
- Gaspar, L.; Coron, R.P.; KongThoo Lin, P.; Costa, D.M.; Perez-Cabezas, B.; Tavares, J.; Roura-Ferrer, M.; Ramos, I.; Ronin, C.; Major, L.L.; et al. Inhibitors of Trypanosoma Cruzi Sir2 Related Protein 1 as Potential Drugs against Chagas Disease. PLoS Negl. Trop. Dis. 2018, 12, e0006180. [Google Scholar] [CrossRef]
- Ghazy, E.; Abdelsalam, M.; Robaa, D.; Pierce, R.J.; Sippl, W. Histone Deacetylase (HDAC) Inhibitors for the Treatment of Schistosomiasis. Pharmaceuticals 2022, 15, 80. [Google Scholar] [CrossRef]
- Fioravanti, R.; Mautone, N.; Rovere, A.; Rotili, D.; Mai, A. Targeting Histone Acetylation/Deacetylation in Parasites: An Update (2017–2020). Curr. Opin. Chem. Biol. 2020, 57, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Ouaissi, M.; Ouaissi, A. Histone Deacetylase Enzymes as Potential Drug Targets in Cancer and Parasitic Diseases. J. Biomed. Biotechnol. 2006, 2006, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lancelot, J.; Cabezas-Cruz, A.; Caby, S.; Marek, M.; Schultz, J.; Romier, C.; Sippl, W.; Jung, M.; Pierce, R.J. Schistosome Sirtuins as Drug Targets. Future Med. Chem. 2015, 7, 765–782. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W. Sirtuins as Emerging Anti-Parasitic Targets. Eur. J. Med. Chem. 2013, 59, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Religa, A.A.; Waters, A.P. Sirtuins of Parasitic Protozoa: In Search of Function(s). Mol. Biochem. Parasitol. 2012, 185, 71–88. [Google Scholar] [CrossRef]
- Ihlenfeldt, W.-D.; Voigt, J.H.; Bienfait, B.; Oellien, F.; Nicklaus, M.C. Enhanced CACTVS Browser of the Open NCI Database. J. Chem. Inf. Comput. Sci. 2002, 42, 46–57. [Google Scholar] [CrossRef]
- Kadam, R.U.; Kiran, V.M.; Roy, N. Comparative Protein Modeling and Surface Analysis of Leishmania Sirtuin: A Potential Target for Antileishmanial Drug Discovery. Bioorg. Med. Chem. Lett. 2006, 16, 6013–6018. [Google Scholar] [CrossRef]
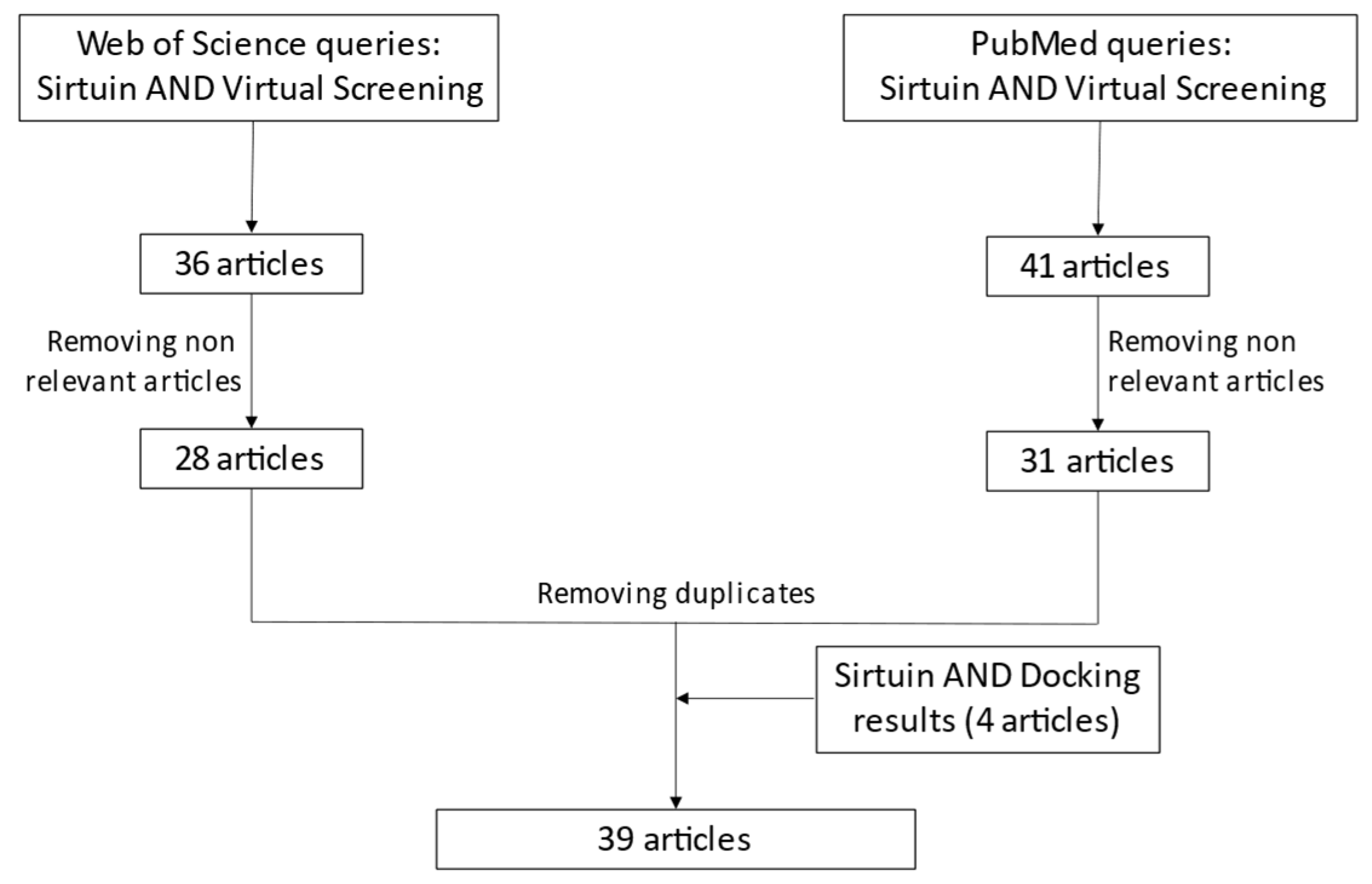
| Sirtuin | Enzymatic Activity | Cellular Localization |
|---|---|---|
| SIRT-1 | Deacetylase [7] | Nucleus, Cytoplasm [13,14] |
| SIRT-2 | Deacetylase, Deacylase [13,15,16] | Nucleus, Cytoplasm [13,15] |
| SIRT-3 | Deacetylase [17,18] | Mitochondria [17,18] |
| SIRT-4 | Mono-ADP-ribosyltransferase, Lipoamidase [19,20] | Mitochondria [19] |
| SIRT-5 | Deacylase, Desuccinylase, Demalonylase [21,22] | Mitochondria [23] |
| SIRT-6 | Deacetylase, Deacylase, Mono-ADP-ribosyl transferase [24,25,26,27] | Nucleus [13,14] |
| SIRT-7 | Deacetylase [19] | Nucleus (nucleolus) [13,14] |
| Sirtuin | Cancer | Neurodegenerative Diseases (AD, HD, PD, ALS, MS) | Type 2 Diabetes (T2D) |
|---|---|---|---|
| SIRT-1 | Activation or inhibition (depending on cancer type) | AD: activation HD: inhibition PD: activation ALS: inhibition MS: inhibition | Activation |
| SIRT-2 | Inhibition | AD: inhibition HD: inhibition PD: inhibition ALS: - MS: - | Inhibition |
| SIRT-3 | Activation or inhibition (depending on cancer type) | AD: activation HD: - PD: activation ALS: - MS: activation | Activation |
| SIRT-4 | - | - | - |
| SIRT-5 | Activation or inhibition (depending on cancer type) | - | - |
| SIRT-6 | Activation or inhibition (depending on cancer type) | AD: - HD: - PD: inhibition ALS: - MS: inhibition | Inhibition |
| SIRT-7 | Inhibition (depending on cancer type) | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbotto, E.; Scarano, N.; Piacente, F.; Millo, E.; Cichero, E.; Bruzzone, S. Virtual Screening in the Identification of Sirtuins’ Activity Modulators. Molecules 2022, 27, 5641. https://doi.org/10.3390/molecules27175641
Abbotto E, Scarano N, Piacente F, Millo E, Cichero E, Bruzzone S. Virtual Screening in the Identification of Sirtuins’ Activity Modulators. Molecules. 2022; 27(17):5641. https://doi.org/10.3390/molecules27175641
Chicago/Turabian StyleAbbotto, Elena, Naomi Scarano, Francesco Piacente, Enrico Millo, Elena Cichero, and Santina Bruzzone. 2022. "Virtual Screening in the Identification of Sirtuins’ Activity Modulators" Molecules 27, no. 17: 5641. https://doi.org/10.3390/molecules27175641
APA StyleAbbotto, E., Scarano, N., Piacente, F., Millo, E., Cichero, E., & Bruzzone, S. (2022). Virtual Screening in the Identification of Sirtuins’ Activity Modulators. Molecules, 27(17), 5641. https://doi.org/10.3390/molecules27175641










