Coordinative Changes in Metabolites in Grape Cells Exposed to Endophytic Fungi and Their Extracts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of In Vitro Grape Callus
2.2. Preparation of Endophytic Fungi (EF) and the Corresponding Fungal Extracts (CFE)
2.3. Treatment of Grape Cells with Living Fungi and Fungal Extracts
2.4. Metabolites Profiling
2.5. Statistical Data Analysis
3. Results
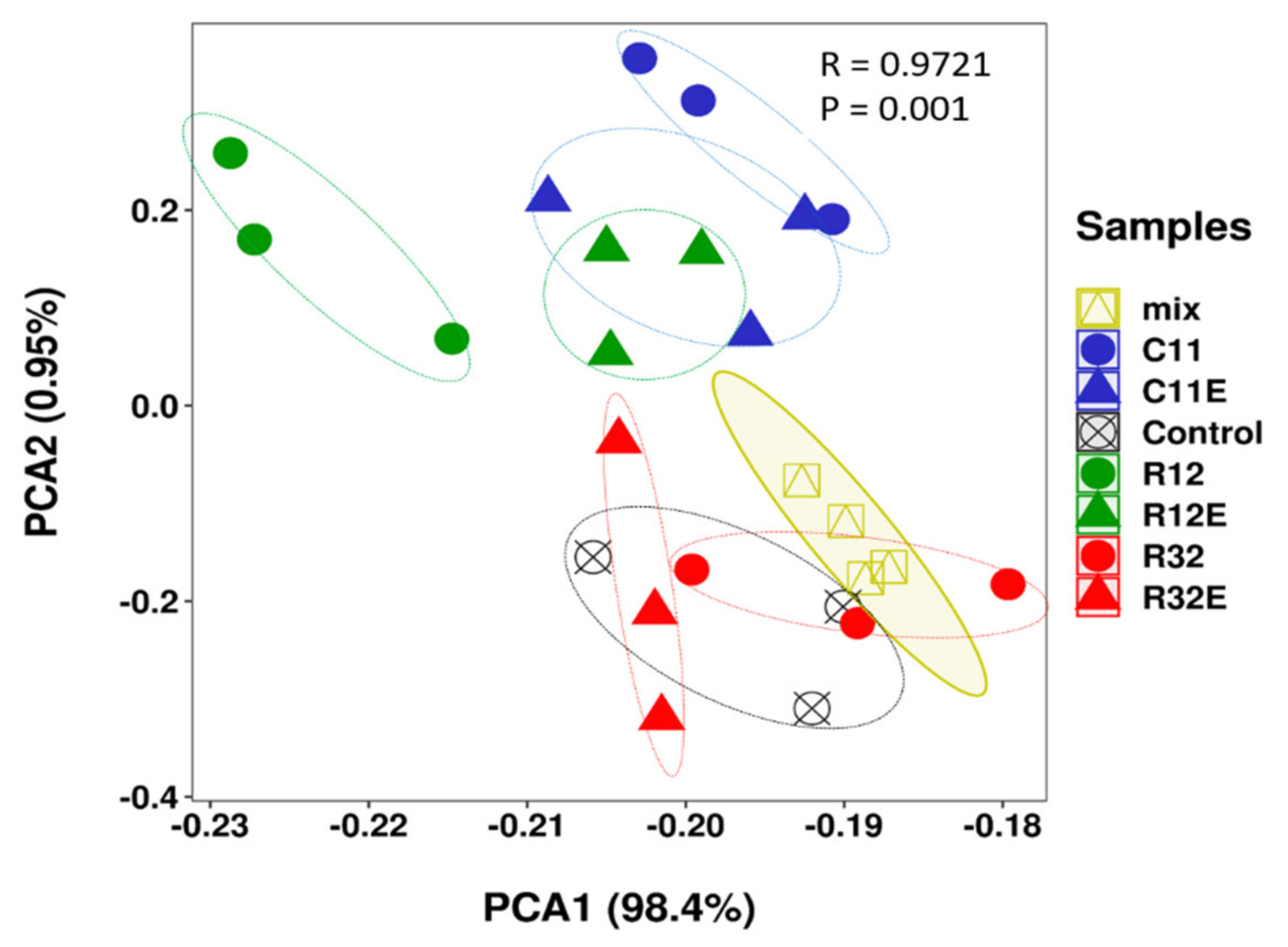
3.1. Grape Cells Exposed to Different EF and Different EF Derived Extracts Differentially Modified the Metabolite Profiles
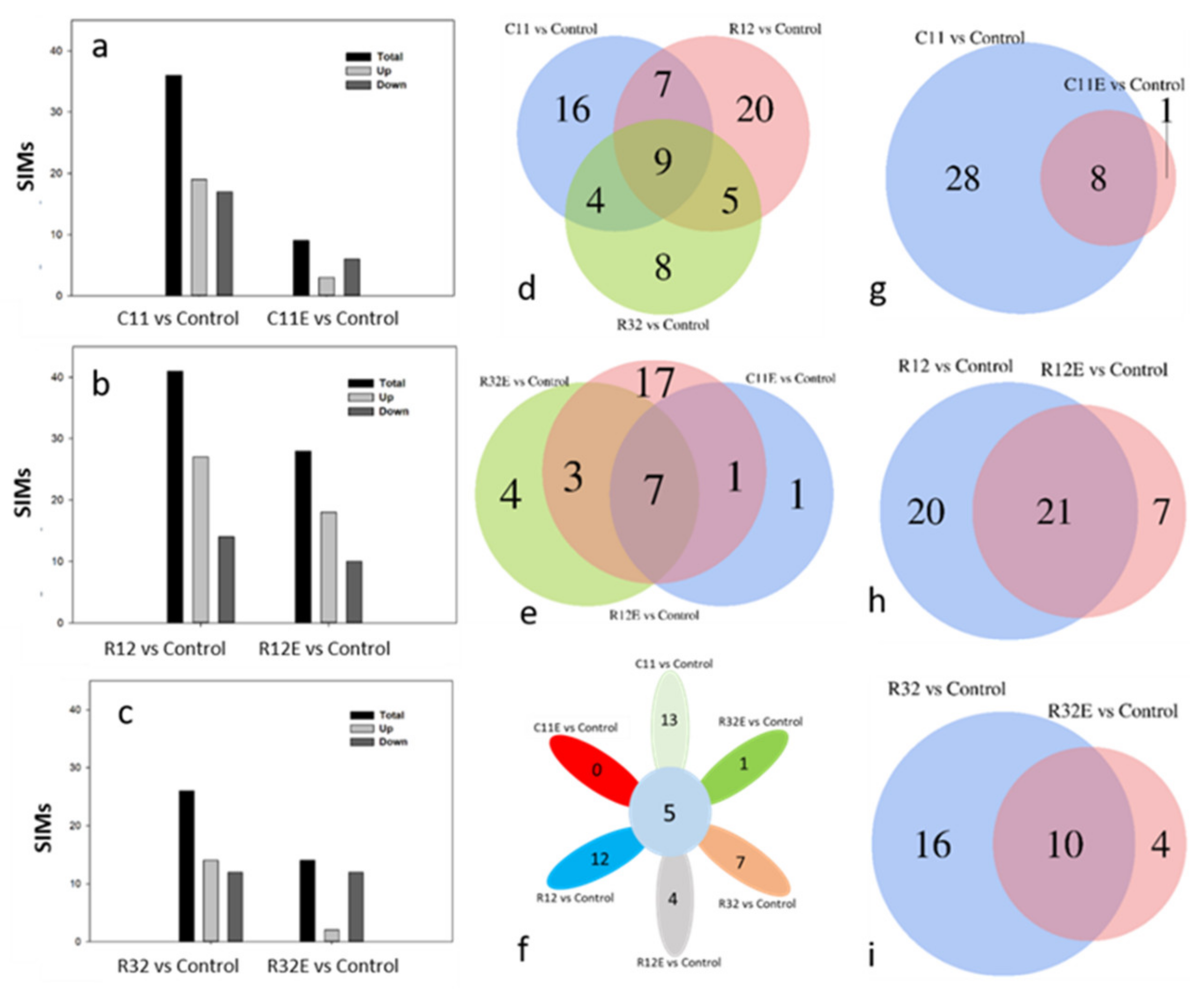
3.2. Proportions of SIMs Specific to a Certain Endophytic Fungal Strain Were Co-Initiated in EF- and CFE-Exposed Grape Cells
3.3. Grape Cells Exposed to EF and CFEs Selectively Influenced Different Classes of Metabolites
3.4. Coordinative and Differential Responses in Metabolites between EF- and CFE-Exposed Grape Cells
4. Discussion
4.1. Grape Cells Exposed to Different Fungi and Different Fungal Derived Extracts Differentially Modified the Metabolite Profiles
4.2. The Use of Different Forms of Regents Expanded the Functions of Endophytes in Crop Biochemical Quality Regulations
4.3. Majority of Metabolites Coordinately Responded in EF- and CFE-Exposed Grape Cells
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| EF | Endophytic Fungi |
| CFE | Corresponding Fungal Extracts |
| SIM | Significantly Influenced Metabolite |
| DRM | Differentially Responding Metabolite |
| CRM | Coordinately Responding Metabolite |
| RI | Response Index |
| PDA | Potato Dextrose Agar |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttila, A.M.; Compant, S.; Campisano, A.; Doring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Perez, L.I.; Gundel, P.E.; Marrero, H.J.; Arzac, A.G.; Omacini, M. Symbiosis with systemic fungal endophytes promotes host es cape from vector-borne disease. Oecologia 2017, 184, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Busby, P.E.; Ridout, M.; Newcombe, G. Fungal endophytes: Modifiers of plant disease. Plant Mol. Biol. 2016, 90, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Li, X.; Gao, Y.; Liu, H.; Gao, Y.B.; Heijden, M.G.A.V.D.; Ren, A.Z. Plant endophytes and arbuscular mycorrhizal fungi alter plant competition. Funct. Ecol. 2017, 32, 11681179. [Google Scholar] [CrossRef]
- Pacifico, D.; Squartini, A.; Crucitti, D.; Barizza, E.; Schiavo, F.L.; Muresu, R.; Carimi, F.; Zottini, M. The Role of the endophytic microbiome in the grapevine response to environmental triggers. Front. Plant Sci. 2019, 10, 1256. [Google Scholar] [CrossRef]
- Fernandez, O.; Vandesteene, L.; Feil, R.; Baillieul, F.; Lunn, J.E.; Clément, C. Trehalose metabolism is activated upon chilling in grapevine and might participate inBurkholderia phytofirmansinduced chilling tolerance. Planta 2012, 236, 355–369. [Google Scholar] [CrossRef]
- Ignacio, P.; Thomas, L.; Macarena, G.; Poupin María, J. Burkholderia phytofirmans PsJN induces long-term metabolic and transcriptional changes involved in arabidopsis thaliana salt tolerance. Front. Plant Sci. 2015, 6, 466. [Google Scholar]
- Musetti, R.; Vecchione, A.; Stringher, L.; Borselli, S.; Zulini, L.; Marzani, C.; Ambrosio, M.D.; Sanità, L.; Pertot, I. Inhibition of Sporulation and Ultrastructural Al terations of Grapevine Downy Mildew by the Endophytic Fungus Alterna alternata. Phytopathology 2006, 96, 689–698. [Google Scholar] [CrossRef]
- Yang, M.Z.; Ma, M.D.; Yuan, M.Q.; Huang, Z.Y.; Yang, W.X.; Zhang, H.B.; Huang, L.H.; Ren, A.Y.; Shan, H. Fungal Endophytes as a Metabolic Fine-Tuning Regulator for Wine Grape. PLoS ONE 2016, 11, e0163186. [Google Scholar] [CrossRef]
- Huang, L.H.; Yuan, M.Q.; Ao, X.J.; Ren, A.Y.; Zhang, H.B.; Yang, M.Z. Endophytic fungi specifically introduce novel metabolites into grape flesh cells in vitro. PLoS ONE 2018, 13, e0196996. [Google Scholar] [CrossRef]
- Cordovez, V.; Dini-Andreote, F.; Carrion, V.; Raaijimakers, J.M. Ecology and Evolution of Plant Microbiomes. Annu. Rev. Microbiol. 2019, 73, 69–88. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.Z.; Huang, L.H.; Ao, X.J.; Ren, A.Y.; Yuan, M.Q.; Zhang, H.B. Endophytic Fungal strains Specifically Modified the Bio chemical Status of Grape Cells. J. Plant Biol. 2018, 61, 210–216. [Google Scholar] [CrossRef]
- Huang, L.H.; Ao, X.J.; Shan, H.; Li, H.X.; Yang, W.X.; Zhang, H.B.; Yang, M.Z. “In vitro” specific interactions revealed the infective characteristics of fungal endophytes to grapevine. Vitis 2017, 56, 71–77. [Google Scholar]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Ludwig-Müller, J. Plants and endophytes: Equal partners in secondary metabolite production? Biotechnol. Lett. 2015, 37, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Chen, J.C.; Qu, J.Z.; Liu, F.; Zhou, M.; Ma, Y.M.; Xiang, S.Y.; Pan, X.X.; Zhang, H.B.; Yang, M.Z. Exposure to endophytic fungi quantitatively and compositionally alters anthocyanins in grape cells. Plant Physiol. Biochem. 2020, 149, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Wu, L.; Wan, H.; Yang, K.; Si, J.; Qin, L. Fungal elicitors stimulate biomass and active ingredients accumulation in Den drobium catenatum plantlets. Biologia 2018, 73, 917–926. [Google Scholar] [CrossRef]
- Senda, K.; Doke, N.; Kawakita, K. Effect of mastoparan on phospholipase A2 activity in potato tubers treated with fun gal elicitor. Plant Cell Physiol. 1998, 39, 1080–1098. [Google Scholar] [CrossRef]
- Zhang, R.; Li, P.; Xu, L.; Chen, Y.; Sui, P.; Zhou, L.; Li, J. Enhancement of diosgenin production in dioscorea zingiberensis cell culture by oligosaccharide elicitor from its endophytic fungus Fusarium oxysporum Dzf17. Nat. Prod. Commun. 2009, 4, 1459–1462. [Google Scholar] [CrossRef]
- Buchanan, B.B.; Gruissem, W.; Jones, R.L. Biochemistry and Molecular Biology of Plants, 2nd ed.; Wiley: Hoboken, NJ, USA, 2015. [Google Scholar]
- Wu, Y.; Duan, S.; Zhao, L.; Gao, Z.; Luo, M.; Song, S.; Xu, W.; Zhang, C.; Ma, C.; Wang, S. Aroma characteri-zation based on aromatic series analysis in table grapes. Sci. Rep. 2016, 6, 31116. [Google Scholar] [CrossRef]
- Knapp, D.G.; Lazar, A.; Molnar, A.; Vajna, B.; Karacsony, Z.; Vaczy, K.Z.; Kovacs, G.M. Above-ground parts of white grapevine Vitis vinifera cv. Furmint share core members of the fungal microbiome. Environ. Microbiol. Rep. 2021, 13, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.C.; Yu, M.; Liu, F.; Qu, J.Z.; Pan, X.X.; Zhang, H.B.; Yang, M.Z. Diversity distributions and the anthocyanin associations of fungal endophytes in different colored grapevine Leaves. J. Plant Biol. 2020, 62, 107–117. [Google Scholar] [CrossRef]
- Deyett, E.; Rolshausen, P.E. Endophytic microbial assemblage in grapevine. FEMS Microbiol. Ecol. 2020, 96, fiaa053. [Google Scholar] [CrossRef] [PubMed]
- Varanda, C.M.R.; Oliveira, M.; Materatski, P.; Landum, M.; Félix, M.D.R. Fungal endophytic communities associated to the phyllosphere of grapevine cultivars under different types of management. Fungal Biol. 2016, 120, 1525–1536. [Google Scholar] [CrossRef] [PubMed]
- Harb, J.; Alseekh, S.; Tohge, T.; Fernie, A.R. Profiling of primary metabolites and flavonols in leaves of two table grape vari eties collected from semiarid and temperate regions. Phytochemistry 2015, 117, 444–455. [Google Scholar] [CrossRef] [PubMed]
- Victoriamoreno-Arribas, M.; Carmenpolo, M. Wine Chemistry and Biochemistry; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]


| Metabolites Class (SIMs/TDM) | Metabolites | Significance Compared to the Control | |||||
|---|---|---|---|---|---|---|---|
| C11 | C1E | R12 | R12E | R32 | R32E | ||
| Alkaloids (1/12) | Caffeine | −1 | −1 | − * | −1 | − *** | − * |
| Amino Acids and Derivatives (16/61) | 5-Aminovaleric acid | − * | −1 | −1 | 0 | −1 | 0 |
| Trans-4-Hydroxy-L-proline | − * | −1 | −1 | −1 | −1 | 0 | |
| L-Tyramine | − * | − * | − * | − * | − * | − *** | |
| 1,2-N-Methylpipecolic acid | − * | −1 | −1 | −1 | −1 | − * | |
| N-Acetyl-L-leucine | +* | +1 | 0 | −1 | +1 | +1 | |
| N-Acetylaspartate | +* | 0 | 0 | +1 | 0 | +1 | |
| N-Acetyl-L-glutamic acid | +* | 0 | +1 | 0 | −1 | 0 | |
| N-Acetylmethionine | − * | − * | − * | − * | − * | − ** | |
| N-α-Acetyl-L-arginine | +* | 0 | 0 | 0 | 0 | 1 | |
| N-Acetyl-L-tyrosine | +* | +1 | +1 | 0 | +1 | +1 | |
| Lysine butyrate | − * | − * | −1 | −1 | 0 | 0 | |
| N-(3-Indolylacetyl)-L-alanine | +* | +1 | +* | +1 | +1 | +1 | |
| L-Homocystine | −1 | 0 | − * | −1 | 0 | −1 | |
| Leucylphenylalanine | −1 * | 0 | − * | −1 | 0 | 0 | |
| L-Glutamic acid O-glycoside | −1 | 0 | −1 | −1 * | −1 * | −1 | |
| L-Glutaminyl-L-valyl-L-valyl-L-cysteine | +** | N/A | +** | +** | +** | N/A | |
| Lipids (16/56) | γ-Linolenic acid | −1 | −1 | −1 | − * | −1 | −1 |
| 11-Octadecanoic acid(Vaccenic acid) | +* | −1 | +1 | 0 | +1 | +1 | |
| 13-HOTrE(r) | −1 | −1 | +* | 0 | 0 | −1 | |
| 1-α-Linolenoyl-glycerol | −1 | −1 | − * | −1 | −1 | −1 | |
| LysoPC 18:1 | +1 | +1 | +* | +1 | +1 | +1 | |
| LysoPC 18:1(2n isomer) | +1 | +1 | +* | +1 | +1 | +1 | |
| LysoPC 18:0 | 0 | 0 | +* | +* | 0 | 0 | |
| LysoPC 18:0(2n isomer) | 0 | +1 | +1 | +* | 0 | 0 | |
| LysoPE 16:0 | +1 | +1 | +* | +* | −1 | +1 | |
| LysoPE 16:0(2n isomer) | 0 | +1 | +* | +* | 0 | +1 | |
| LysoPE 18:3 | −1 | −1 | +1 | +1 | − ** | −1 | |
| LysoPE 18:2 | 0 | +1 | +* | +* | −1 | +1 | |
| LysoPE 18:2(2n isomer) | +1 | +1 | +* | +* | 0 | 1 | |
| LysoPE 18:1 | −1 | +1 | +* | +* | − ** | +1 | |
| LysoPE 18:1(2n isomer) | +1 | +1 | +* | +* | +1 | +1 | |
| Choline alfoscerate | −1 | +1 | +1 | 0 | −1 | − * | |
| Nucleotides and Derivatives (3/40) | 5-Methylcytosine | − * | 0 | −1 | −1 | −1 | −1 |
| Xanthine | − * | −1 | −1 | −1 | −1 | −1 | |
| 9-(β-D-Arabinofuranosyl) hypoxanthine | +1 | 0 | +* | −1 | 0 | −1 | |
| Saccharides and Alcohols (3/24) | D-Glucoronic acid | − * | 0 | −1 | − * | −1 | −1 |
| D-(+)-Melezitose | +1 | 0 | +* | +1 | −1 | +1 | |
| D(+)-Melezitose O-rhamnoside | +* | +1 | +* | +1 | +1 | +1 | |
| Vitamins (2/11) | Nicotinamide | −1 | +1 | +1 | 0 | − * | −1 |
| Pyridoxine | +1 | 0 | +* | +1 | +1 | +1 | |
| Organic acids (7/30) | 2-Furanoic acid | +1 | +1 | 0 | −1 | − * | 0 |
| 3-Hydroxy-3-methyl butyric acid | +1 | 0 | +1 | 0 | +* | 0 | |
| 6-Aminocaproic acid | − * | −1 | −1 | −1 | −1 | 0 | |
| 3-Hydroxyanthranilic acid | − ** | 0 | +1 | +1 | −1 | +1 | |
| Diethyl phosphate | +1 | 0 | − * | −1 | 0 | −1 | |
| 3,4-Dihydroxybenzeneacetic acid | +* | +1 | +* | +1 | +1 | +1 | |
| Trans-4-Hydroxycinnamic acid methyl ester | −1 | −1 | − * | −1 | −1 | −1 | |
| Phenolic Acids (11/42) | Methyl ferulate | +* | 0 | +1 | +* | +* | +* |
| 3-Hydroxy-5-Methylphenol-1-oxy-β-D-Glucose | 0 | +1 | +1 | +* | 0 | −1 | |
| Isosalicylic acid O-glycoside | +1 | 0 | +1 | +1 | +* | +1 | |
| Feruloylmalic acid | +* | −1 | +* | +* | +* | +* | |
| 3-Hydroxy-4-isopropylbenzylalcohol 3-glucoside | 0 | +* | +* | +* | +1 | +1 | |
| 1′-O-Vanilloyl-β-D-glucoside | −1 | +1 | +1 | +* | 0 | −1 | |
| Feruloyl glucose | −1 | +1 | +1 | +1 | +* | +1 | |
| Syringic acid O-glucoside | +1 | −1 | − * | +1 | +* | −1 | |
| Trihydroxycinnamoylquinic acid | +* | −1 | − ** | −1 | +* | − ** | |
| Syringin | +1 | 0 | +* | +* | +* | 0 | |
| p-Coumaroylcaffeoyltartaric acid | +* | +1 | +1 | 0 | +* | +1 | |
| Others (14/28) | Indole | − * | −1 | 0 | −1 | −1 | 0 |
| Piceid | +* | +1 | +1 * | +* | +* | +1 | |
| ε-Viniferin | +* | − ** | +* | − ** | 0 | − ** | |
| Resveratrol-O-diglucoside | −1 | +1 | +1 | 0 | − * | −1 | |
| Camaldulenic acid | − *** | +1 * | − *** | − *** | − *** | − *** | |
| 2-Hydroxyoleanolic acid | − * | +* | − * | − * | − * | − * | |
| 6-Hydroxyrumicin-8-O-D-glucopyranoside | +* | −1 | 0 | +1 | +* | 0 | |
| Skimmin | +* | +1 | +1 | +1 | +* | +1 | |
| 2-O-Galloyl-β-D-glucose | +1 | −1 | − * | +* | +* | − * | |
| Eriodictyol 7-O-glucoside | +1 | +1 | +* | +* | +1 | −1 | |
| Hesperetin 7-O-neohesperidoside (Neohesperidin) | − *** | − * | − ** | − *** | −1 | − *** | |
| Isorhamnetin-3-O-β-D-glucoside | 0 | +1 | +* | +1 | +1 | 0 | |
| Octadecenoic amide | +1 | −1 | +* | 0 | +1 | −1 | |
| Propyl 2-(trimethylammonio)ethyl phosphate | +* | +1 | +* | +1 | +1 | +1 | |
| α-Viniferin | − *** | −1 * | +1 * | − *** | − * | − * | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, J.-Z.; Liu, F.; Pan, X.-X.; Liao, C.-M.; Li, T.; Zhang, H.-B.; Yang, M.-Z. Coordinative Changes in Metabolites in Grape Cells Exposed to Endophytic Fungi and Their Extracts. Molecules 2022, 27, 5566. https://doi.org/10.3390/molecules27175566
Qu J-Z, Liu F, Pan X-X, Liao C-M, Li T, Zhang H-B, Yang M-Z. Coordinative Changes in Metabolites in Grape Cells Exposed to Endophytic Fungi and Their Extracts. Molecules. 2022; 27(17):5566. https://doi.org/10.3390/molecules27175566
Chicago/Turabian StyleQu, Jin-Zhuo, Fang Liu, Xiao-Xia Pan, Chang-Mei Liao, Tong Li, Han-Bo Zhang, and Ming-Zhi Yang. 2022. "Coordinative Changes in Metabolites in Grape Cells Exposed to Endophytic Fungi and Their Extracts" Molecules 27, no. 17: 5566. https://doi.org/10.3390/molecules27175566
APA StyleQu, J.-Z., Liu, F., Pan, X.-X., Liao, C.-M., Li, T., Zhang, H.-B., & Yang, M.-Z. (2022). Coordinative Changes in Metabolites in Grape Cells Exposed to Endophytic Fungi and Their Extracts. Molecules, 27(17), 5566. https://doi.org/10.3390/molecules27175566






