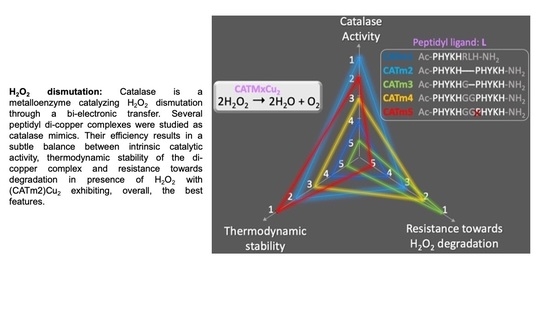
Improvement of Peptidyl Copper Complexes Mimicking Catalase: A Subtle Balance between Thermodynamic Stability and Resistance towards H2O2 Degradation
Abstract
1. Introduction
with an overall reaction: 2H2O2 → O2 + 2H2O
2. Results and Discussion
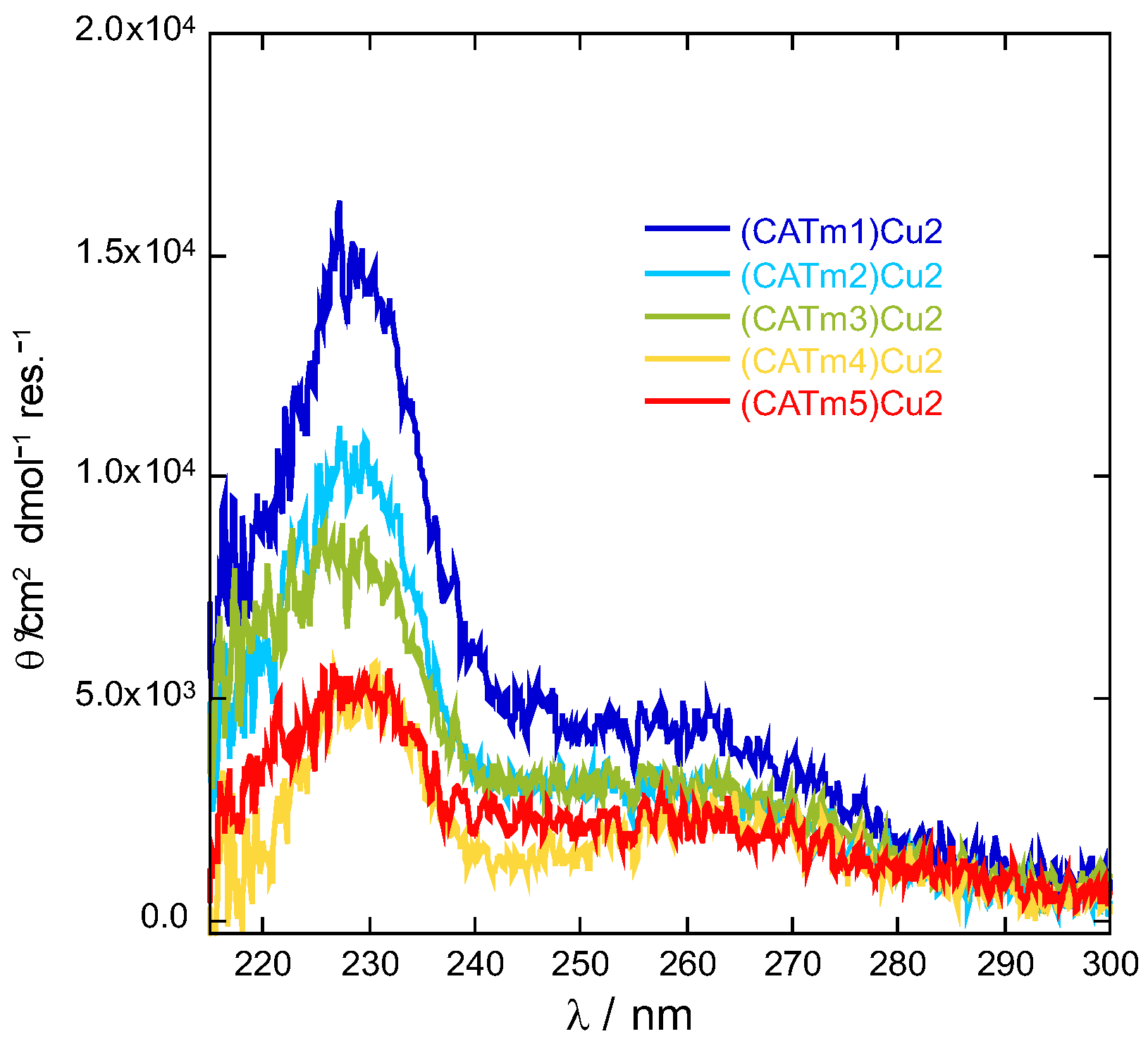
2.1. Design and Synthesis
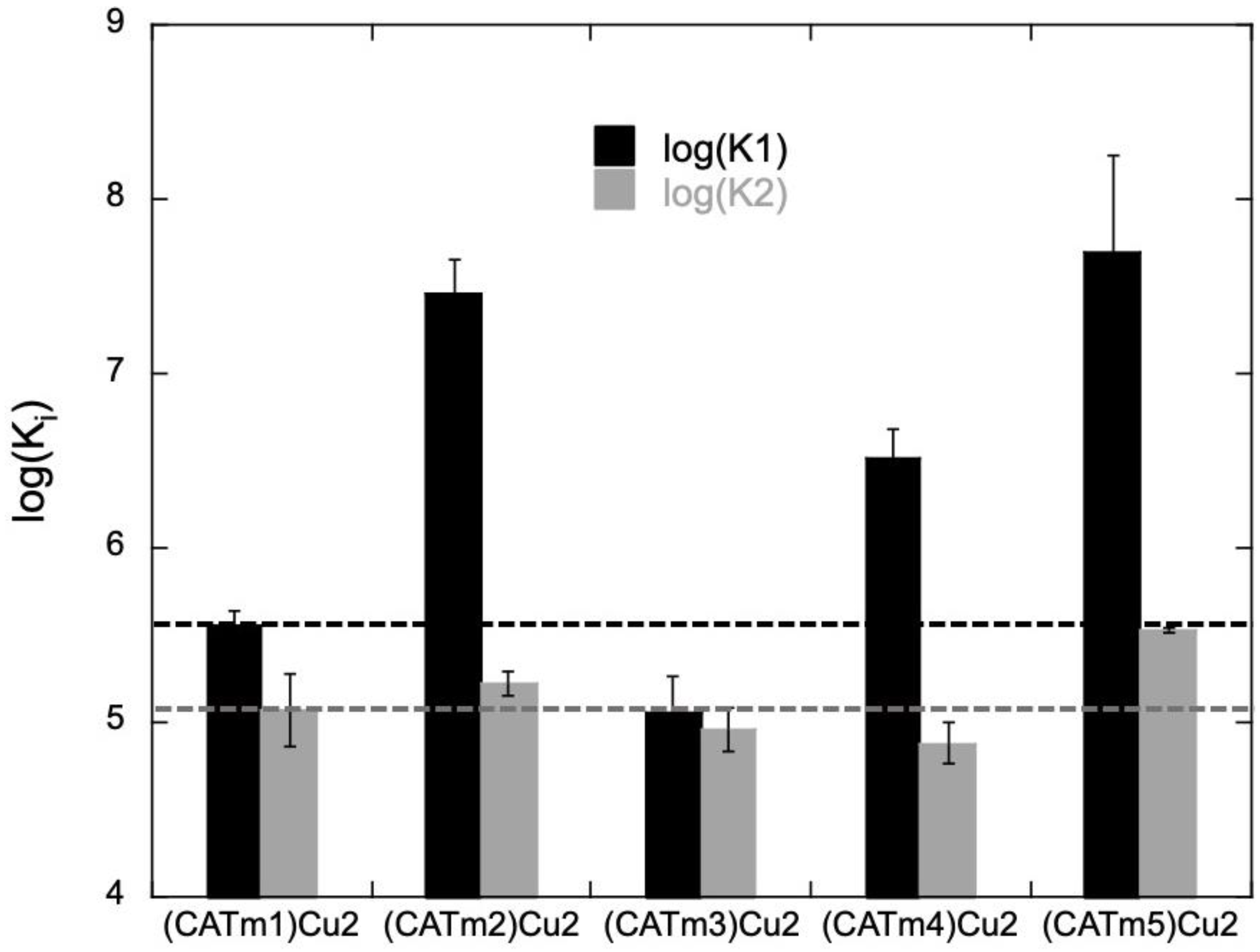
2.2. Complexes’ Thermodynamic Stability
2.3. Kinetic Study of Complex Degradation in the Presence of H2O2
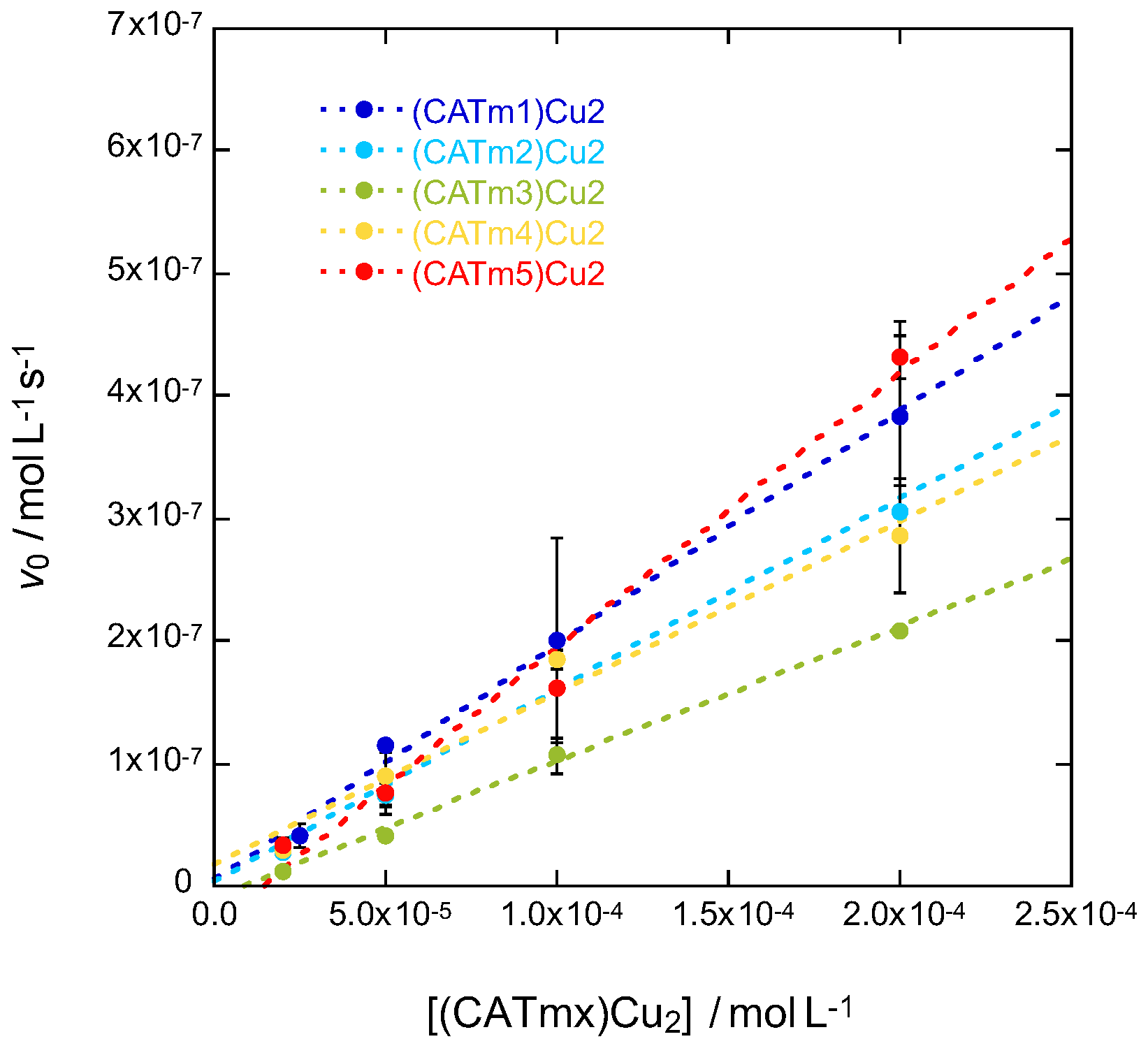
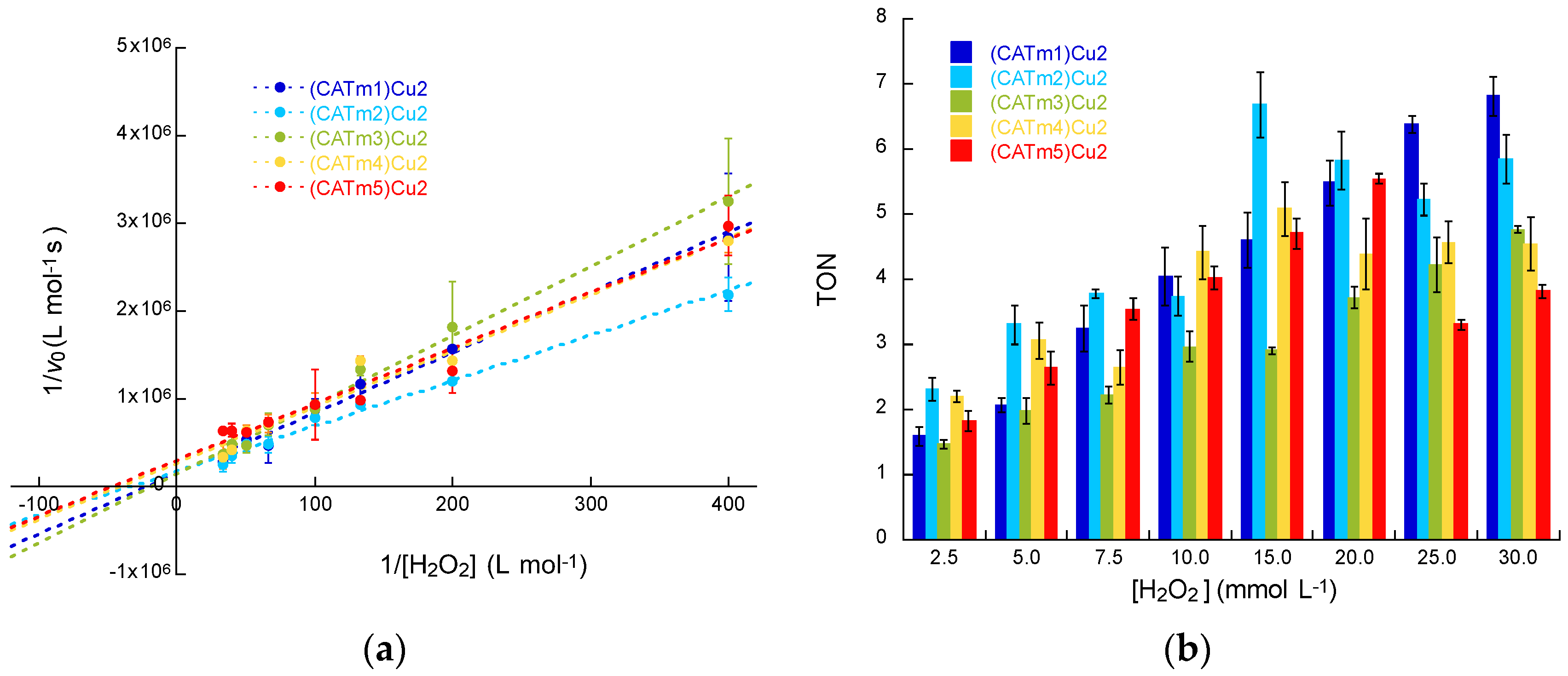
2.4. Catalytic Activity
3. Conclusions
4. Experimental Section
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Halliwell, B.; Gutteridge, J.M.C. Oxygen Toxicity, Oxygen Radicals, Transition Metals and Disease. Biochem. J. 1984, 219, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Auten, R.L.; Davis, J.M. Oxygen Toxicity and Reactive Oxygen Species: The Devil Is in the Details. Pediatr. Res. 2009, 66, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P.; Holmgren, A.; Larsson, N.-G.; Halliwell, B.; Chang, C.J.; Kalyanaraman, B.; Rhee, S.G.; Thornalley, P.J.; Partridge, L.; Gems, D.; et al. Unraveling the Biological Roles of Reactive Oxygen Species. Cell Metab. 2011, 13, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative Stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef]
- Sies, H.; Jones, D.P. Reactive Oxygen Species (ROS) as Pleiotropic Physiological Signalling Agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef]
- McCord, J.M.; Edeas, M.A. SOD, Oxidative Stress and Human Pathologies: A Brief History and a Future Vision. Biomed. Pharmacoter. 2005, 59, 139–142. [Google Scholar] [CrossRef]
- Sheng, Y.; Abreu, I.A.; Cabelli, D.E.; Maroney, M.J.; Miller, A.-F.; Teixeira, M.; Valentine, J.S. Superoxide Dismutases and Superoxide Reductases. Chem. Rev. 2014, 114, 3854–3918. [Google Scholar] [CrossRef]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide Dismutases: Dual Roles in Controlling ROS Damage and Regulating ROS Signaling. J. Cell. Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef]
- Veal, E.A.; Day, A.M.; Morgan, B.A. Hydrogen Peroxide Sensing and Signaling. Mol. Cell 2007, 26, 1–14. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Biological Production, Detection, and Fate of Hydrogen Peroxide. Antioxid. Redox Signal. 2018, 29, 541–551. [Google Scholar] [CrossRef]
- Gardner, P.R.; Raineri, I.; Epstein, L.B.; White, C.W. Superoxide Radical and Iron Modulate Aconitase Activity in Mammalian Cells *. J. Biol. Chem. 1995, 270, 13399–13405. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine, 5th ed.; Oxford University Press: New York, NY, USA, 2015; ISBN 978-0-19-871748-5. [Google Scholar]
- Salvemini, D.; Muscoli, C.; Riley, D.P.; Cuzzocrea, S. Superoxide Dismutase Mimetics. Pulm. Pharmacol. Ther. 2002, 15, 439–447. [Google Scholar] [CrossRef]
- Batinic-Haberle, I.; Tovmasyan, A.; Roberts, E.R.H.; Vujaskovic, Z.; Leong, K.W.; Spasojevic, I. SOD Therapeutics: Latest Insights into Their Structure-Activity Relationships and Impact on the Cellular Redox-Based Signaling Pathways. Antioxid Redox Signal. 2014, 20, 2372–2415. [Google Scholar] [CrossRef]
- Vincent, A.; Thauvin, M.; Quévrain, E.; Mathieu, E.; Layani, S.; Seksik, P.; Batinic-Haberle, I.; Vriz, S.; Policar, C.; Delsuc, N. Evaluation of the Compounds Commonly Known as Superoxide Dismutase and Catalase Mimics in Cellular Models. J. Inorg. Biochem. 2021, 219, 111431. [Google Scholar] [CrossRef]
- Policar, C.; Bouvet, J.; Bertrand, H.C.; Delsuc, N. SOD Mimics: From the Tool Box of the Chemists to Cellular Studies. Curr. Opin. Chem. Biol. 2022, 67, 102109. [Google Scholar] [CrossRef]
- Signorella, S.; Hureau, C. Bioinspired Functional Mimics of the Manganese Catalases. Coord. Chem. Rev. 2012, 256, 1229–1245. [Google Scholar] [CrossRef]
- Tovmasyan, A.; Maia, C.G.C.; Weitner, T.; Carballal, S.; Sampaio, R.S.; Lieb, D.; Ghazaryan, R.; Ivanovic-Burmazovic, I.; Ferrer-Sueta, G.; Radi, R.; et al. A Comprehensive Evaluation of Catalase-like Activity of Different Classes of Redox-Active Therapeutics. Free Radic. Biol. Med. 2015, 86, 308–321. [Google Scholar] [CrossRef]
- Coulibaly, K.; Thauvin, M.; Melenbacher, A.; Testard, C.; Trigoni, E.; Vincent, A.; Stillman, M.J.; Vriz, S.; Policar, C.; Delsuc, N. A Di-Copper Peptidyl Complex Mimics the Activity of Catalase, a Key Antioxidant Metalloenzyme. Inorg. Chem. 2021, 60, 9309–9319. [Google Scholar] [CrossRef]
- Kirkman, H.N.; Gaetani, G.F. Mammalian Catalase: A Venerable Enzyme with New Mysteries. Trends Biochem. Sci. 2007, 32, 44–50. [Google Scholar] [CrossRef]
- Whittaker, J.W. Non-Heme Manganese Catalase—The ‘Other’ Catalase. Arch. Biochem. Biophys. 2012, 525, 111–120. [Google Scholar] [CrossRef]
- Barynin, V.V.; Whittaker, M.M.; Antonyuk, S.V.; Lamzin, V.S.; Harrison, P.M.; Artymiuk, P.J.; Whittaker, J.W. Crystal Structure of Manganese Catalase from Lactobacillus Plantarum. Structure 2001, 9, 725–738. [Google Scholar] [CrossRef]
- Day, B.J. Catalase and Glutathione Peroxidase Mimics. Biochem. Pharmacol. 2009, 77, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Haukka, M.; McKenzie, C.J.; Nordlander, E. High Turnover Catalase Activity of a Mixed-Valence MnIIMnIII Complex with Terminal Carboxylate Donors. Eur. J. Inorg. Chem. 2018, 2015, 3485–3492. [Google Scholar] [CrossRef]
- Mu, J.; Zhang, L.; Zhao, M.; Wang, Y. Co3O4 Nanoparticles as an Efficient Catalase Mimic: Properties, Mechanism and Its Electrocatalytic Sensing Application for Hydrogen Peroxide. J. Mol. Cat. A Chem. 2013, 378, 30–37. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, Y.; Lan, L.; Zhang, C.; Pan, M.; Wang, Y.; Han, B.; Wang, Z.; Jiao, J.; Chen, Q. A Novel Catalase Mimicking Nanocomposite of Mn(II)-Poly-L-Histidine-Carboxylated Multi Walled Carbon Nanotubes and the Application to Hydrogen Peroxide Sensing. Anal. Biochem. 2019, 567, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Martell, A.E.; Reibenspies, J.H. Novel Dicopper(II) Catalase-like Model Complexes: Synthesis, Crystal Structure, Properties and Kinetic Studies. Inorg. Chim. Acta 2003, 346, 32–42. [Google Scholar] [CrossRef]
- Kaizer, J.; Csonka, R.; Speier, G.; Giorgi, M.; Réglier, M. Synthesis, Structure and Catalase-like Activity of New Dicopper(II) Complexes with Phenylglyoxylate and Benzoate Ligands. J. Mol. Cat. A Chem. 2005, 236, 12–17. [Google Scholar] [CrossRef]
- Kaizer, J.; Csay, T.; Speier, G.; Réglier, M.; Giorgi, M. Synthesis, Structure and Catalase-like Activity of Cu(N-Baa)2(Phen) (Phen = 1,10-Phenanthroline, N-BaaH = N-Benzoylanthranilic Acid). Inorg. Chem. Commun. 2006, 9, 1037–1039. [Google Scholar] [CrossRef]
- Ramadan, A.E.-M.M. Syntheses and Characterization of New Tetraazamacrocyclic Copper(II) Complexes as a Dual Functional Mimic Enzyme (Catalase and Superoxide Dismutase). J. Coord. Chem. 2012, 65, 1417–1433. [Google Scholar] [CrossRef]
- Pires, B.M.; Silva, D.M.; Visentin, L.C.; Rodrigues, B.L.; Carvalho, N.M.F.; Faria, R.B. Synthesis and Characterization of Cobalt(III), Nickel(II) and Copper(II) Mononuclear Complexes with the Ligand 1,3-Bis[(2-Aminoethyl)Amino]-2-Propanol and Their Catalase-Like Activity. PLoS ONE 2015, 10, e0137926. [Google Scholar] [CrossRef][Green Version]
- Richezzi, M.; Ferreyra, J.; Puzzolo, J.; Milesi, L.; Palopoli, C.M.; Moreno, D.M.; Hureau, C.; Signorella, S.R. Versatile Activity of a Copper(II) Complex Bearing a N4-Tetradentate Schiff Base Ligand with Reduced Oxygen Species. Eur. J. Inorg. Chem. 2022, 2022, e202101042. [Google Scholar] [CrossRef]
- Krieger, F.; Möglich, A.; Kiefhaber, T. Effect of Proline and Glycine Residues on Dynamics and Barriers of Loop Formation in Polypeptide Chains. J. Am. Chem. Soc. 2005, 127, 3346–3352. [Google Scholar] [CrossRef]
- Hureau, C.; Faller, P. Aβ-Mediated ROS Production by Cu Ions: Structural Insights, Mechanisms and Relevance to Alzheimer’s Disease. Biochimie 2009, 91, 1212–1217. [Google Scholar] [CrossRef]
- Pires dos Santos, M.L.; Faljoni-Alário, A.; Mangrich, A.S.; Costa Ferreira, A.M. da Antioxidant and Pro-Oxidant Properties of Some Di-Schiff Base Copper(II) Complexes. J. Inorg. Biochem. 1998, 71, 71–78. [Google Scholar] [CrossRef]
- Tang, Q.; Wu, J.-Q.; Li, H.-Y.; Feng, Y.-F.; Zhang, Z.; Liang, Y.-N. Dinuclear Cu(II) Complexes Based on p-Xylylene-Bridged Bis(1,4,7-Triazacyclononane) Ligands: Synthesis, Characterization, DNA Cleavage Abilities and Evaluation of Superoxide Dismutase- and Catalase-like Activities. Appl. Organomet. Chem. 2018, 32, e4297. [Google Scholar] [CrossRef]
- Guerreiro, J.F.; Gomes, M.A.G.B.; Pagliari, F.; Jansen, J.; Marafioti, M.G.; Nistico, C.; Hanley, R.; Costa, R.O.; Ferreira, S.S.; Mendes, F.; et al. Iron and Copper Complexes with Antioxidant Activity as Inhibitors of the Metastatic Potential of Glioma Cells. RSC Adv. 2020, 10, 12699–12710. [Google Scholar] [CrossRef]
- Shank, M.; Barynin, V.; Dismukes, G.C. Protein Coordination to Manganese Determines the High Catalytic Rate of Dimanganese Catalases. Comparison to Functional Catalase Mimics. Biochemistry 2002, 33, 15433–15436. [Google Scholar] [CrossRef]

| Name | Peptide Sequence |
|---|---|
| CATm1 | Ac(PHYKH)RLH-NH2 |
| CATm2 | Ac(PHYKH)(PHYKH)-NH2 |
| CATm3 | Ac(PHYKH)G(PHYKH)-NH2 |
| CATm4 | Ac(PHYKH)GG(PHYKH)-NH2 |
| CATm5 | Ac(PHYKH)GGHYKH-NH2 |
| kobs (s−1) | |
|---|---|
| (CATm1)Cu2 a | 1.90 × 10−3 |
| (CATm2)Cu2 | 1.56 × 10−3 |
| (CATm3)Cu2 | 1.10 × 10−3 |
| (CATm4)Cu2 | 1.41 × 10−3 |
| (CATm5)Cu2 | 2.24 × 10−3 |
| KM (M) | kcat (s−1) | kcat/KM (M−1·s−1) | References | |
|---|---|---|---|---|
| (CATm1)Cu2 [a] | 4.8 × 10−2 | 1.4 × 10−1 | 2.9 | This work, [19] |
| (CATm2)Cu2 [a] | 2.9 × 10−2 | 1.1 × 10−1 | 3.9 | This work |
| (CATm3)Cu2 [a] | 5.2 × 10−2 | 1.3 × 10−1 | 2.5 | This work |
| (CATm4)Cu2 [a] | 2.4 × 10−2 | 0.8 × 10−1 | 3.1 | This work |
| (CATm5)Cu2 [a] | 2.1 × 10−2 | 0.7 × 10−1 | 3.2 | This work |
| Cu(N-baa)2(phen) [b] | 5.2 × 10−2 | 6.6 × 10−2 | 1.3 | [29] |
| [Cu(HL1)]2+ [c] | 1.7 × 101 | 1.5 × 10−3 | 8.9 × 10−5 | [31] |
| Cu2(pxdiprbtacn)Cl4 [d] | 1.5 | 1.24 | 0.8 | [36] |
| CuL2 [e] | 4.2 × 10−2 | 3.6 × 10−1 | 8.25 | [37] |
| [Cu(apzpn)]2+ [f] | 1.10 | [35] | ||
| [Cu(py2pn)]2+ [g] | 0.8 × 10−4 | [32] | ||
| Catalase | 8.3 × 10−2 | 2.6 × 105 | 3.1 × 106 | [38] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Hadj Hammouda, Y.; Coulibaly, K.; Bathily, A.; Teoh Sook Han, M.; Policar, C.; Delsuc, N. Improvement of Peptidyl Copper Complexes Mimicking Catalase: A Subtle Balance between Thermodynamic Stability and Resistance towards H2O2 Degradation. Molecules 2022, 27, 5476. https://doi.org/10.3390/molecules27175476
Ben Hadj Hammouda Y, Coulibaly K, Bathily A, Teoh Sook Han M, Policar C, Delsuc N. Improvement of Peptidyl Copper Complexes Mimicking Catalase: A Subtle Balance between Thermodynamic Stability and Resistance towards H2O2 Degradation. Molecules. 2022; 27(17):5476. https://doi.org/10.3390/molecules27175476
Chicago/Turabian StyleBen Hadj Hammouda, Yaqine, Koudedja Coulibaly, Alimatou Bathily, Magdalene Teoh Sook Han, Clotilde Policar, and Nicolas Delsuc. 2022. "Improvement of Peptidyl Copper Complexes Mimicking Catalase: A Subtle Balance between Thermodynamic Stability and Resistance towards H2O2 Degradation" Molecules 27, no. 17: 5476. https://doi.org/10.3390/molecules27175476
APA StyleBen Hadj Hammouda, Y., Coulibaly, K., Bathily, A., Teoh Sook Han, M., Policar, C., & Delsuc, N. (2022). Improvement of Peptidyl Copper Complexes Mimicking Catalase: A Subtle Balance between Thermodynamic Stability and Resistance towards H2O2 Degradation. Molecules, 27(17), 5476. https://doi.org/10.3390/molecules27175476