Abstract
Rhamnose-associated molecules are attracting attention because they are present in bacteria but not mammals, making them potentially useful as antibacterial agents. Additionally, they are also valuable for tumor immunotherapy. Thus, studies on the functions and biosynthetic pathways of rhamnose-containing compounds are in progress. In this paper, studies on the biosynthetic pathways of three rhamnose donors, i.e., deoxythymidinediphosphate-L-rhamnose (dTDP-Rha), uridine diphosphate-rhamnose (UDP-Rha), and guanosine diphosphate rhamnose (GDP-Rha), are firstly reviewed, together with the functions and crystal structures of those associated enzymes. Among them, dTDP-Rha is the most common rhamnose donor, and four enzymes, including glucose-1-phosphate thymidylyltransferase RmlA, dTDP-Glc-4,6-dehydratase RmlB, dTDP-4-keto-6-deoxy-Glc-3,5-epimerase RmlC, and dTDP-4-keto-Rha reductase RmlD, are involved in its biosynthesis. Secondly, several known rhamnosyltransferases from Geobacillus stearothermophilus, Saccharopolyspora spinosa, Mycobacterium tuberculosis, Pseudomonas aeruginosa, and Streptococcus pneumoniae are discussed. In these studies, however, the functions of rhamnosyltransferases were verified by employing gene knockout and radiolabeled substrates, which were almost impossible to obtain and characterize the products of enzymatic reactions. Finally, the application of rhamnose-containing compounds in disease treatments is briefly described.
1. Introduction
Glycans are important components of various glycoconjugates, such as glycoproteins, glycolipids, and proteoglycans, and play pivotal roles in many biological processes, including intracellular trafficking, cell adhesion and development, cancer progression, host–pathogen interactions, and immune responses [1]. For a detailed structure–activity relationship analysis of functional glycans, it is necessary to obtain molecules in structurally homogeneous forms, which is not easy to achieve via the isolation of natural products from biological sources. Therefore, the total synthesis of polysaccharides and their oligomeric analogs has become a hot research topic. Rhamnose (Rha)-containing compounds (RCCs) are especially interesting due to their potential applications, including antibacterial vaccines and killing tumors [2,3]. Additionally, Rha is a common component of various bacterial polysaccharides, such as lipopolysaccharides (LPSs) [4], extracellular polysaccharides (EPSs) [5], capsular polysaccharides (CPSs) [6], and cell wall polysaccharides [7]. In addition to bacteria, Rha is also found in viruses [8], fungi [9], plants [10], and lower animals [11]. Interestingly, Rha has not been found in humans or other mammals. In recent years, more evidence has emerged about its essential roles in many pathogenic bacteria, making it a potentially attractive therapeutic target. Furthermore, RCCs are also candidates for vaccines, antitumor drugs, and antibacterial drugs [3,12]. Thus, there is a keen desire to obtain and characterize RCCs. However, due to the complexity of the target molecules and the difficulty in constructing certain glycosidic linkages, such as β-linked Rha, via chemical glycosylation, enzymatic synthesis is particularly attractive [13]. For the enzymatic synthesis of RCCs or their conjugates, rhamnosyl donors are the key substrates [14,15], which are utilized by rhamnosyltransferases (Rha-Ts) and attached to sugar acceptors [16]. Therefore, the catalysis of Rha-Ts and the preparation of Rha donors and acceptors in vitro are hot topics [13,17,18]. In this paper, the biosynthetic pathways of Rha donors are reviewed, and the development of Rha-Ts and their medical perspectives are also explored. Such knowledge expands our understandings of the biosynthetic pathways of RCCs and could facilitate their enzymatic synthesis.
2. Biosynthetic Pathways of Donors of RCCs
Three sugar nucleotides, including deoxythymidinediphosphate-L-rhamnose (dTDP-Rha), guanosine diphosphate rhamnose (GDP-Rha), and uridine diphosphate-rhamnose (UDP-Rha), can serve as Rha donors in reactions catalyzed by Rha-Ts. dTDP-Rha and GDP-Rha are present in bacteria and fungi, whereas UDP-Rha is only found in plants. There are probably other Rha donors involving in Rha biosynthetic pathways in Mycoplasma [19]. The biosynthetic pathways of these three Rha donors and structural, mechanistic, and biochemical aspects of the key enzymes involved are reviewed below.
2.1. Biosynthetic Pathways of dTDP-Rha
The dTDP-Rha is one of the most important sugar precursors. Four enzymes, glucose-1-phosphate thymidylyltransferase (RmlA), dTDP-D-glucose 4,6-dehydratase (RmlB), dTDP-4-keto-6-deoxy-D-glucose3,5-epimerase (RmlC), and dTDP-4-keto-L-Rha reductase (RmlD), are responsible for the formation of dTDP-Rha (Figure 1) [20]. The rmlA, rmlB, rmlC, and rmlD genes are usually located in biosynthetic gene clusters of polysaccharides in conserved gene orders with few exceptions [20]. Below, we discuss what is known about the steps involved in the biosynthesis of dTDP-Rha, as well as the functions, physicochemical properties, and crystal structures of RmlA, RmlB, RmlC, and RmlD [21].
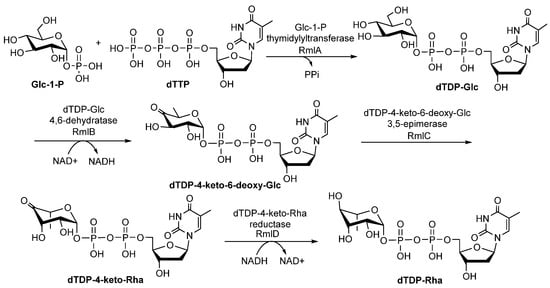
Figure 1.
The biosynthetic pathway of dTDP-Rha from Glc-1-P in bacteria [22,23].
RmlA is a nucleotidyltransferase that catalyzes the first reaction to form dTDP-glucose (dTDP-Glc) by transferring a deoxythymidine triphosphate (dTTP) to glucose-1-phosphate (Glc-1-P) via a single sequential displacement mechanism [24]. Based on the reverse reaction, RmlA is also known to be a pyrophosphorylase [24]. RmlA has attracted more attention because it displays unusual promiscuity toward both sugar-1-phosphates and nucleotide triphosphate substrates, which could be harnessed in glycorandomization [25,26]. The inherent sugar-1-phosphate and/or nucleotide triphosphate (NTP) promiscuity of RmlA was further expanded by mutation studies. For example, L89T [27], E162D, Y177F [27], T201A, and W224H [28] mutants increased its sugar-1-phosphate tolerance and conversion [27], whereas Q24S [29] and Q83D/S [30] mutants altered the preference for the NTP of wildtype RmlA (also called the inherent NTP purine/pyrimidine bias). RmlA and its variants can utilize 57 sugar-1-phosphates ranging from all epimers [26], substituted compounds (amino [30], N-acetyl [31], methyl [32], azido [32], thiol [32], and alkyl [33]), deoxy sugars of D-glucose [32], two anomers of L-fucose [34], to pentofuranosyl-1-phosphate [34]. In addition to sugar-1-phosphate substrates, Moretti et al. reported that RmlA could recognize all eight natural NTPs as substrates despite its reduced activity toward purine NTPs [35]. Furthermore, Cps2L (a RmlA homolog) can act on deoxythymidine 5-tetraphosphate (p4dT) and Glc-1-P to form dTDP-Glc and triphosphate (PPPi) [32]. To date, 154 (d) nucleotide diphosphate (NDP)-sugars have been produced by RmlA and its variants (Figure 2 and Supplementary information Table S1) [36,37,38,39,40,41,42].
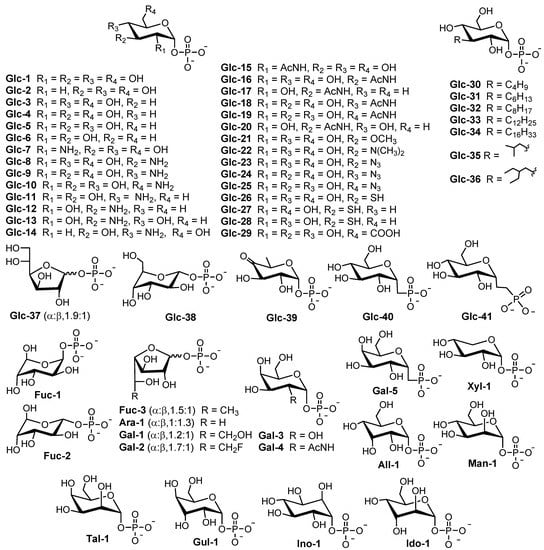
Figure 2.
Sugar-1-phosphates recognized by RmlA [22].
The activity of RmlA is inhibited by dTDP-Glc, inorganic pyrophosphate (PPi) and dTDP-Rha [43], and dTDP-Rha is both a competitive and a noncompetitive inhibitor [43]. Although the mechanism of noncompetitive inhibition of RmlA by dTDP-Rha proposed by Mmot et al. remains unclear and needs to be further studied [43], the mechanism of competitive inhibition is well understood [44], which involved: (1) dTDP-Rha occupies the same site as dTDP-Glc; E161 of RmlA interacts with O2′ and O3′ of Rha through a bidentate hydrogen, similar to the dTDP-Glc complex (Figure 3D); (2) The two phosphates of dTDP-Rha move into the active site and form strong salt bridges with R194, which is absent in the dTDP-Glc complex; (3) A hydrogen bond between ribose O3 of dTDP-Rha and the side chain of D110 is likely replaced by the α-phosphate, resulting in the decomposition of dTDP-Rha. Thus, it was concluded that targeting these sites could provide a potential basis for inhibitor design. In addition, the R15 loop probably affects catalytic activity because it is different in the active site of the dTDP-Rha complex [44]. Crystal structures of RmlA from Pseudomonas aeruginosa [44], Escherichia coli [45], and Salmonella typhimurium [45] showed that RmlA is a homotetramer (Figure 3A).
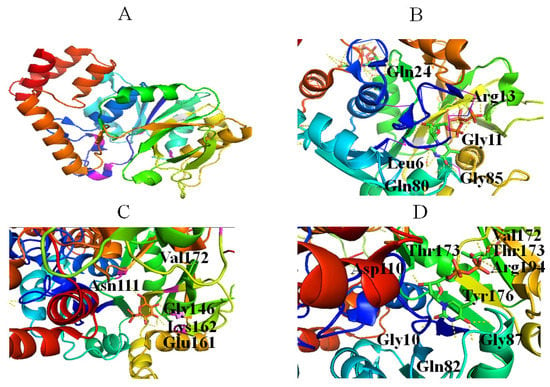
Figure 3.
Stereo views of RmlA (Protein Data Bank (PDB) entry 4HO3) (A), dTTP (PDB entry 4HO3) (B), Glc-1-P (PDB entry 1G23) (C), and dTDP-Rha (PDB entry 1G3L) (D) bound to RmlA. Hydrogen bonds are shown as red lines. Helices, sheets, and loops of RmlA are colored blue, purple, and beige, respectively. C, N, O, and P elements of ligands are shown in green, blue, red, and brown, respectively. RmlA is shown in cartoon representation, and ligands are shown as sticks [22].
The active center of RmlA lies in a deep pocket formed by core and sugar-binding domains [45]. G11, Q80, and G85 form hydrogen bonds with the thymine: N3 and O4 of thymine engage in hydrogen bonds with Q80; O4 of thymine also forms hydrogen bonds with the N atom of G85; O2 of the thymine base engages in hydrogen bonds with G11 (Figure 3B) [45]. Neither methyl group of the pyrimidine ring nor the 2-OH of ribose interacts with RmlA, which explains why RmlA can accept UTP and dTTP as substrates.
The 3-hydroxyl group of ribose contacts Q24 (Figure 3B) [45], and glucose residue interacts with RmlA via hydrogen bonds. Specifically, O2 and O3 of glucose form hydrogen bonds with E161; O2, O3, and O4 of glucose form hydrogen bonds with G146 and L172; O6 of glucose forms hydrogen bonds with N111 (Figure 3C). In addition, Q26, G11, S13, and two water molecules bind magnesium. Therefore, the crystal structures of RmlA helped to reveal the reaction mechanism and provide a basis for active site engineering of RmlA [35].
The second step in the dTDP-Rha biosynthetic pathway is the dehydration of dTDP-Glc to form dTDP-4-keto-6-deoxy-D-glucose (dT4k6dG), which is catalyzed by RmlB. Four steps have been proposed during the reaction: (1) NAD+ extracts a hydride from C4 of the glucose ring; (2) Glu135 removes a C5 proton; (3) elimination of a water molecule between C5 and C6 generates 4-keto-5,6-glucosene as an intermediate; and (4) a hydride is transferred from NADH to C6 of the glucose ring [46]. The substrate tolerance of RmlB is more limited compared with that of RmlA, probably because it catalyzes the committal step in the dTDP-Rha biosynthetic pathway [47]. The crystal structure of RmlB from Salmonella enterica serovar Typhimurium showed that it functions as a homodimer (Figure 4A). RmlB has two domains: a larger N-terminal domain consisting of seven β-strands and ten α-helices to bind the nucleotide cofactor NAD+; and a smaller C-terminal domain composed of four β-strands and six α-helices to bind dTDP-Glc [46]. The two domains create a deep cavity in the enzyme to form the active site (Figure 4A) [46]. The key residues interacting with NAD+ include (1) a hydrogen bond (Asp62) and a hydrophobic crevice consisting of Ile21, Ala57, Ile59, Val77, Ala81, and Leu107 binding to the adenine portion of NAD+, and (2) Asp37, Tyr161, and Lys171 forming hydrogen bonds with the ribose sugar (Figure 4B) [46]. In addition, Thr133, Asp134, Glu135, Asn196, Arg231, and Asn266 make contacts with dTDP-Glc (Figure 4C) [46]. Specifically, Thr133, Glu135, and Asp134 bind to the 4, 6-hydroxyl groups of the glucose ring (Figure 4C), while Asn196 and Arg231 interact with the phosphoryl oxygen atom (Figure 4C), and Asn266 hydrogen binds to the 3-hydroxyl group of the ribose sugar (Figure 4D) [46]. Notably, Asn266 may also control the selectivity for the deoxy-nucleotide sugar substrate in the binding site [46].
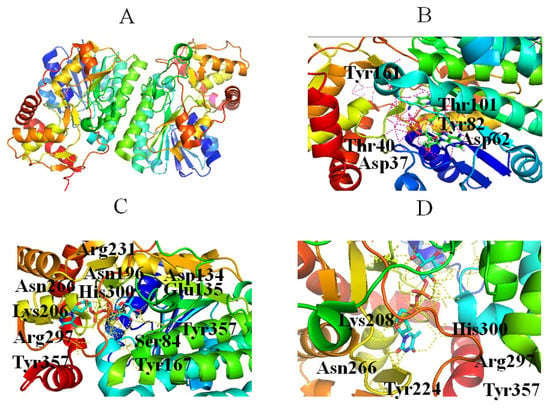
Figure 4.
Stereo view of RmlB (PDB entry 1KEP) (A), NAD+ (PDB entry 1KEP) (B), and dTDP-Glc (PDBentry 1KEU) (C) bound to RmlB, and the position of the Asn266 side chain as the sugar moves to stack against Tyr224 (D). Hydrogen bonds are shown as red lines. Helices, sheets, and loops of RmlB are colored blue, purple, and beige, respectively. C, N, O, and P elements of ligands are green, blue, red, and brown, respectively. RmlB is shown in cartoon representation, and ligands are shown as sticks [22].
RmlC catalyzes the third step in the dTDP-Rha biosynthetic pathway, in which the C3 and C5 positions of dT4k6dG are epimerized to generate dTDP-4-keto-Rha [48]. The catalytic mechanism of this catalytic reaction is proposed as follows: (1) a proton is abstracted from C5 of glucose of dT4k6dG accompanied by epimerization, then proton donation to C5, resulting in a mono-epimerized intermediate; (2) a proton from C3 of glucose is abstracted accompanied by epimerization, followed by proton donation to C3; (3) a ring flip occurs [49]. These reactions need strict stereo control and a cofactor is not required [50]. RmlC and/or RmlC co-complex structures have been obtained with dTDP-phenol, dTDP, dTDP-Glc and dTDP-D-xylose [49,51]. RmlC functions as a homodimer (Figure 5A). The monomer consists of 11 β-strands and seven α-helices that can be divided into three parts, including an N-terminal portion, a core active site, and a C-terminal portion. A His-Asp dyad (Figure 5B) in the active site is crucial in the RmlC catalytic mechanism because a conserved His65 residue from the His-Asp dyad extracts C5 and C3 protons (Figure 5B). Moreover, Tyr134 is essential for epimerization and for proton incorporation at C5. However, a water molecule may replace Tyr134 to facilitate C3 proton incorporation (Figure 5B) [49].
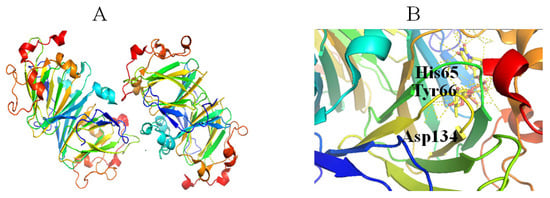
Figure 5.
Stereo view of the RmlC (PDB entry 2IXL) (A) and His-Asp dyad of RmlC (PDB entry 2IXT) (B). Helix, sheet, and loop of RmlC are shown as blue, purple, and beige, respectively. C, N, O, and P elements of ligands are green, blue, red, and brown, respectively. RmlC is shown in cartoon representation, and ligands are shown as sticks [22].
RmlD catalyzes the last step in the dTDP-Rha biosynthetic pathway, in which the C4 keto group of dTDP-4-keto-Rha is reduced to a hydroxyl group to produce dTDP-Rha (Figure 1) [52,53]. During the reaction, proton transferred from the nicotinamide ring of the cofactor to the C4 keto group requires the assistance of Mg2+ [52]. RmlD is a homodimer, and the monomer consists of two domains: an N-terminal domain that binds NAD(H), and a C-terminal domain that binds substrate [52]. Various residues are involved in interactions with NAD(P)H, including (1) a ribose moiety located in the space formed by Ala62, Ala63, Gly7 and Gly10, in which the 2′- and 3′-hydroxyl groups of the ribose ring and Lys132 from the conserved YXXXK motif engage in two hydrogen bonds (Figure 6A); (2) the adenine ring of the cofactor located in a pocket formed by Val31, Asp39, Phe40, Ala62, Ala63, Leu80, and Phe40, in which Asp39 interacts with adenine via hydrogen bonds (Figure 6A); (3) Gln11 and Thr 65 interact with diphosphate (Figure 6A) [52]. Three glutamic acids (Glu15, Glu190, and Glu292) of two monomers bind to Mg2+ [52], and dTDP-Rha binds in a pocket of RmlD built from the hydrophobic parts of the side chains of Thr65, Tyr106, Tyr128, and Val67, together with the nicotinamide ring of the cofactor [52]. Additionally, Thr104, 105, Trp153, the carboxamide group of the cofactor, and a water molecule interact with L-Rha (Figure 6B) [52].
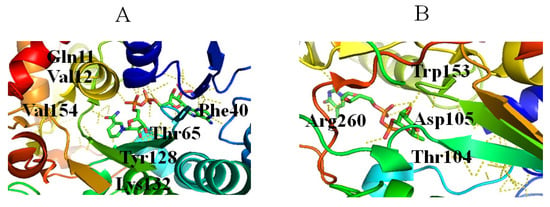
Figure 6.
Stereo view of RmlD bound to NADH (PDB entry 1KC3) (A) and dTDP-Rha (PDB entry 1KC3) (B). Hydrogen bonds are shown as red lines. Helices, sheets, and loops of RmlD are colored blue, purple, and beige, respectively. C, N, O, and P elements of ligands are green, blue, red, and brown, respectively. RmlD is shown in cartoon representation, and ligands are shown as sticks [22].
2.2. Biosynthetic Pathway of GDP-Rha
D-Rha is a rare 6-deoxy monosaccharide found in the LPS of pathogenic bacteria [54]. GDP-Rha is the precursor for the biosynthesis of D-Rha-containing compounds, and it is synthesized in two steps: (1) GDP-mannose-4,6-dehydratase (GMD) catalyzes the conversion of GDP-D-mannose (GDP-Man) to GDP-4-keto-6-deoxy-D-Man; (2) GDP-6-deoxy-D-lyxo-hexos-4-ulose-4-reductase (RMD) catalyzes the production of GDP-Rha (Figure 7). Both GMD and RMD are members of the short-chain dehydrogenase/reductase (SDR) family. GMD is homologous to RmlB, while RMD is homologous to RmlD. However, GMD and RMD cannot catalyze the conversion of dT4k6dG to dTDP-Rha, indicating that enzymes involved in the GDP-D-Rha biosynthesis pathway possess strict substrate specificity. The functions of GMD and RMD from Aneurinibacillus thermoaerophilus strain L420-91 (T) [55] and Pseudomonas aeruginosa [56] have been confirmed in vitro.
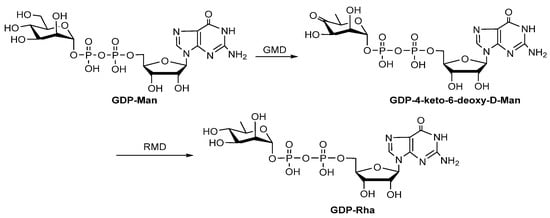
Figure 7.
Biosynthesis pathway of GDP-Rha [22].
GMD is present in bacteria [57], plants [58], and animals [59], and its production serves as a branch point for several different deoxyhexoses, such as GDP-Rha, GDP-L-fucose, GDP-6-deoxy-D-talose, and the GDP-dideoxy amino sugars [56]. GMD functions as a homodimer [60] or a homotetramer [56] in cells. In particular, PBCV-1 GMD behaves as a bifunctional enzyme, displaying not only dehydratase activity but also a strong NAD(P)H-dependent reductase activity toward GDP-4-keto-6-deoxy-D-Man (the dehydration product), leading to the formation of GDP-Rha [61]. The crystal structures of GMD from E. coli [62], Arabidopsis thaliana [58], P. aeruginosa [56], and Paramecium bursaria Chlorella virus 1 (PBCV-1) [63] have been reported. The GMD monomer folds into two domains: a N-terminal cofactor-binding domain and a C-terminal substrate-binding domain. Residues of GMDs that contact the GDP moiety are highly conserved, including Val190, Asn179, Lys193, Arg218, Arg279, and Glu282. However, the hexose moiety has not been successfully crystallized. The crystal structure of RMD from Aneurinibacillus thermoaerophilus was reported in 2008, but the quality of the crystal structure was not good [64].
2.3. Biosynthesis Pathway of UDP-Rha
UDP-Rha is found in fungi and plants, and its biosynthesis pathway involves dehydration, epimerization, and reduction, similar to dTDP-Rha (Figure 8) [9]. Three isoenzymes (UDP-Rha synthases, RHMs) RHM1, RHM2/RHM4 and RHM3 convert UDP-Glc to UDP-Rha via UDP-4-keto-6-deoxy-glucose (U4k6dG) as an intermediate [65]. All the RHMs function as the activities of UDP-D-Glc 4,6-dehydratase, UDP-4-keto-6-deoxy-D-Glc 3,5-epimerase, and UDP-4-keto-L-Rha4-keto-reductase, respectively [66]. Other enzymes, such as a bifunctional enzyme named nucleotide-Rha synthase/epimerase-reductase (NRS/ER), can also act on the intermediate U4k6dG to form UDP-Rha [56,65]. Notably, it is not known what substrates are utilized by Rha-Ts as donors in plants because there are two rhamnose donors in plants (UDP-Rha and dTDP-Rha).
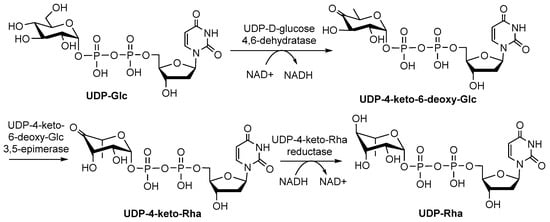
Figure 8.
Biosynthesis pathway of UDP-Rha [22].
3. Rha-Ts Generating RCCs in Bacteria
Glycosyltransferases (GTs) are a large family of enzymes that catalyze the transfer of saccharide moieties from glycosyl donors to a broad range of acceptor substrates, including monosaccharides, oligosaccharides and polysaccharides, lipids, proteins, nucleic acids, and small organic molecules, to form complex carbohydrates and glycoconjugates that are essential to many fundamental biological processes [1]. There are three main methods for the classification of GTs: Firstly, based on the anomeric configuration of reactants and products, GTs are classified as inverting or retaining enzymes; Secondly, GT-A, GT-B, and GT-C topologies of GTs are divided based on Rossmann-fold domains and the locations of donors and acceptors; Thirdly, according to sequence similarity, GTs are divided into 114 different families, as listed in the carbohydrate-active enzymes (CAZy) database (http://www.cazy.org accessed on 14 March 2022). Rha-Ts are GTs that generate RCCs, which are universally present in bacteria [67,68]. However, biochemical knowledge on Rha-Ts is still limited.
3.1. Rha-Ts from Geobacillus Stearothermophilus
The S-layer protein of Geobacillus stearothermophilus NRS 2004/3a serves as a model for investigating O-glycosylation pathways in bacteria, and the glycans of this protein are 2-OMe-α-L-Rha-(1→3)-β-L-Rha-(1→2)-α-L-Rha-(1→[2)-α-L-Rha-(1→3)-β-L-Rha-(1→2)-α-L-Rha-(1→]n=13–18[2)-α-L-Rha-(1→]n=1–23)-α-L-Rha-(1→3)-β-D-Gal-(1→Protein (Figure 9) [69,70].
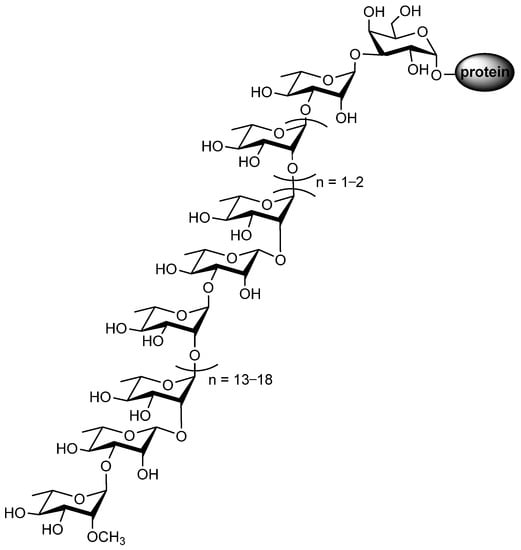
Figure 9.
The glycan structure of the Geobacillus stearothermophilus S-layer glycoprotein.
The polycistronic S-layer glycosylation (slg) gene cluster encodes four GTs, of which three Rha-Ts (WsaC, WsaD and WsaF) catalyze the biosynthesis of the glycan [70]. The biosynthesis pathway for this glycan is initiated by the transfer of a galactose residue to a membrane-associated lipid carrier, followed by two steps catalyzed by α-1,3-Rha-Ts (WsaC and WsaD) that add Rha to build up the [2)-α-L-Rha-(1→]n=1–23)-α-L-Rha-(1→3) linker, and two Rha-Ts (WsaE and WsaF) then extend the glycan chain by adding the repeating trisaccharide motif [→2)-α-L-Rha-(1→3)-β-L-Rha-(1→2)-α-L-Rha-(1→]n=13–18 through addition of Rha. The complete glycan chain is thereafter transported across the membrane by an ATP-binding cassette transporter (ABC transporter) and transferred to the S-layer protein by oligosaccharyltransferase WsaB [70]. The functions of WsaC−WsaF were proved by using the chemically synthesized β-D-Gal-(1→O)-octyl as substrate. Among them, both WsaC and WsaD are transmembrane proteins, and the activity of WsaC requires membranes, while WsaD can only recognize the natural substrate; WsaE is a multifunctional enzyme, and the N-terminal domain of WsaE possesses methylase activity, whereas the central and C-terminal domains of WsaE possess Rha-Ts activity, generating α-1,2 and α-1,3 linkages; WsaF is a β-1,2-Rha-T enzyme (Figure 10) [70].
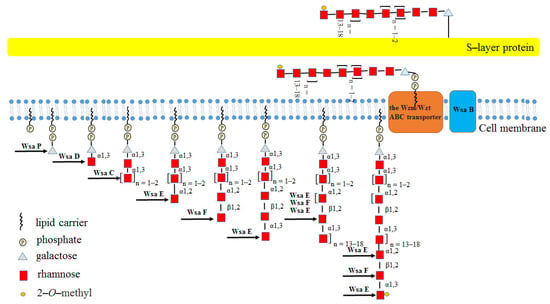
Figure 10.
Diagram of the biosynthesis pathway of the S-layer glycoprotein glycan of Geobacillus stearothermoph ilus NRS 2004/3a.
WsaF is a dimer formed by two monomers that consist of two GT-B-fold domains and a cleft between the two domains [69]. dTDP-Rha interacts with WsaF, and the dTDP-WsaF and dTDP-Rha-WsaF complex structures revealed that thymidine contacts with K302 and L303, while V282 and G283 interact with thymidine via van der Waals forces, pyrophosphate binds to G63, R249 and K302 through hydrogen bonds, Rha contacts N227, K225 and E333 directly, and Y329 engages in a stacking interaction with the hydrophobic face of Rha [69]. The crystal structure of the WsaF–acceptor complex has not been reported. However, the acceptor fragments of both α-L-Rha-(1-2)-α-L-Rha-(1-3)-α-L-Rha and α-L-Rha-(1-2)-α-L-Rha-(1-3)-α-L-Rha-(1-3)-α-D-Gal were modeled manually in the tunnel using PyMOL, suggesting that G63, I65, P54, S55, A140, Q170, D171, E173 and F176 would form van der Waals interactions with the acceptor fragments. This was confirmed by mutant studies [69].
3.2. Spinosyn SpnG Rha-T from Saccharopolyspora Spinosa
Spinosyn from S. spinosa is a type of macrocyclic lactone that has been used as an agricultural antibiotic [71,72]. The entire spinosyn biosynthetic gene cluster contains 19 genes, including 5 large genes (spnA, spnB, spnC, spnD, and spnE) encoding a type I polyketide synthase, 4 genes (spnF, spnJ, spnL, and spnM) encoding proteins involved in intramolecular C-C bond formation, 4 genes (spnG, spnI, spnK, and spnH) encoding proteins involved in Rha attachment and methylation, and 6 genes (spnP, spnO, spnN, spnQ, spnR, and spnS) encoding proteins involved in forosamine biosynthesis [72]. SpnG is known to be capable of transferring Rha from dTDP-Rha donors to spinosyn aglycone (AGL) and to display relaxed substrate specificity [73].
SpnG forms a C2-symmetric homodimer, and each monomer contains two domains connected by a long loop (residues 183–209) [71]. The C-terminal domain binds the donor substrate (dTDP-Rha) and the N-terminal domain binds the acceptor substrate [71,73]. Specifically, interactions between dTDP-Rha and SpnG include thymine contacting L279, V277, P257, and L279; α-phosphate forming hydrogen bonds with G296, T297, and T297; β-phosphate making hydrogen bonds with M227 and V228; the 3-OH group of deoxyribose directly forming a hydrogen bond with N202; and Rha contacting with D316, Q317, Y314, and W142 [71]. The crystal structure of a SpnG–acceptor complex has not been reported.
3.3. WbbL from Mycobacterium Tuberculosis
The cell wall of M. tuberculosis, essential for cell proliferation and growth, is composed of peptidoglycan, arabinogalactan, and mycolic acids [7]. The galactan of arabinogalactan combines with peptidoglycan via a disaccharide linker, α-L-Rha-(1→3)-α-D-GlcNAc-(1→P), to form the integrated mycobacterial cell wall [74]. The Rha-T enzyme WbbL forms the disaccharide linker by transferring Rha from dTDP-Rha to decaprenyldiphosphoryl-α-D-N-acetyl glucosamine (GlcNAc-PP-DP) [75]. The wbbL gene is essential for mycobacterial viability and is found in the genomes of all mycobacteria [76]; hence, it is an attractive target for antituberculosis therapeutics. Activity analysis of WbbL was performed using endogenous GlcNAc-PP-DP as a substrate, and a microtiter plate method was established [74]. The bioinformatics analysis of WbbL showed that it belongs to the GT2 family with a fold characteristic of the GT-A superfamily [74], members of which can utilize dTDP-β-Rha as a substrate and produce an α-Rha product. In addition, this protein has a N-terminal GT domain, no signal peptide or transmembrane helices, and it is located outside the membrane.
3.4. Rha-Ts from Pseudomonas Aeruginosa
Pseudomonas aeruginosa is a pathogen of plants and animals, and an opportunistic human pathogen that causes serious nosocomial infections [77]. LPSs are major virulence factors composed of three distinct regions, i.e., lipid A, core oligosaccharide (OS), and O polysaccharide (O antigen), which contain diverse repeating saccharide units. In this section, we focus on Rha found in OS and O antigens.
OS is divided into two types: one is capped (linked to O polysaccharides) with O antigen through an α-1,3-linked L-Rha, while the other is uncapped (devoid of O polysaccharides) and contains an α-1,6-linked L-Rha [78]. Gene knockout analysis showed that migA and wapR genes encode α-1,3 Rha-Ts and α-1,6 Rha-Ts, respectively, which are responsible for the biosynthesis of the α-1,3-linked L-Rha and α-1,6-linked L-Rha [78]. O antigen can also be divided into two types: heteropolymeric O antigen (formerly called B band), containing mannosuronic acid derivatives with N-acetyl-D-fucosamine (D-FucNAc), and an alternative LPS containing the common polysaccharide antigen (CPA; formerly called A band). There are repeating units of O-polysaccharides of A band, namely (→3-α-D-Rha-(1→2)-α-D-Rha (1→3)-α-D-Rha α-1→), containing a D-Rha moiety [79]. D-Rha-Ts (including WbpY, WbpX and WbpZ) catalyze the transfer of D-Rha to an acceptor [80,81].
Rhamnolipids are detergents composed of α-D-(α-D-hydroxyalkanoyloxy) alkanoic acids (HAA) derivatized with one or two Rha sugars (monorhamnolipids and dirhamnolipids; Figure 11), which are secreted by P. aeruginosa [82]. Rha-Ts I [83] and Rha-Ts II [84] generate rhamnolipids, and their mechanism has been determined: (1) Rha-Ts I are encoded by rhlA and rhlB genes, and gene knockout analysis of these genes indicated that RhlA forms HAA, while RhlB is a Rha-T enzyme [85], and the heterologous expression of RhlA and RhlB was achieved [86]; (2) gene knock-in assay proved that RhlC encodes Rha-Ts II, which transfers the second Rha to dirhamnolipids [84].
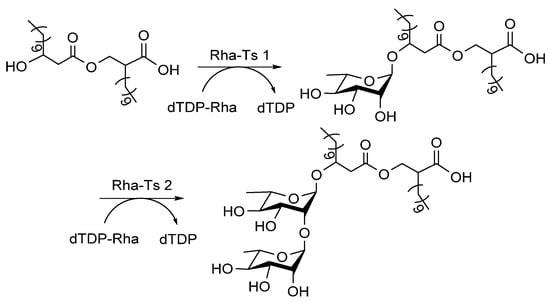
Figure 11.
Rha-Ts involved in the biosynthesis of rhamnolipids [22].
Additionally, a Rha-T EarP derived from P. aeruginosa has been reported that transfers Rha from dTDP-Rha to Arg32 of the translation elongation factor P (EF-P) [87,88]. This rhamnosylation of Arg32 by EarP can activate the functions of EF-P, which is important in the process of protein translation in ribosome. EarP is also discovered in other clinically relevant bacteria [89,90], indicating that this type of post-translational modification strategy is crucial for protein translation and bacteria pathogenicity [87,88].
3.5. Rha-Ts from Streptococcus Pneumoniae
Capsular polysaccharides (CPSs) are produced by almost all isolates of S. pneumoniae recovered from cases of invasive disease, and they are major virulence factors and immunogens [91]. Rha-containing CPS has been identified in at least 27 serotypes. Rha-containing CPS of S. pneumoniae is particularly attractive: (1) L-Rha may increase the immunogenicity of CPS based on the immune analysis of 23F CPS, showing that α-(1→2)-linked L-Rha is a dominant antigen [92]; (2) modified L-Rha may increase the stability of CPS based on the analysis of a 19F CPS analog in which a residue of carba-L-Rha was inserted into the natural trisaccharide, and this increased the stability of CPS [93]; (3) Rha-Ts are the most prevalent GT genes in S. pneumoniae cps loci [94]. Therefore, studies on Rha of S. pneumonia CPS may provide a new strategy for developing novel drugs to treat anti-pneumococcal infections. However, new serotypes should be identified, and attempts to determine the structures of CPSs and Rha-Ts have been reported [95,96].
3.6. Rha-Tss from Other Bacteria
Although numerous Rha-Ts have been predicted, the in vitro biochemical knowledge of these enzymes is limited. Gene mutants have confirmed the functions of some Rha-Ts, including RgpF [97], WbgA [97], AceR [98], AntB [99], and GacB [100]. Heterologous expression has also been used to confirm the functions of Rha-Ts, as exemplified by HlpA/RtfA [101]. Rha-Ts from Mycobacterium smegmatis [93], Streptococcus anginosus [102], serotype VIII capsular polysaccharide (CPS) of Group B Streptococci (GBS) [103], and Vibrio cholera [104] have also been reported.
This review mainly overviews the research advance of the Rha-Ts derived from bacterial; however, there are also other enzymes involved in RCC biosynthesis that will not be described in detail here. For example, several Rha-Ts from plants have been reported [105,106,107]. Additionally, in recent years, α-L-rhamnosidase has been found to synthesize RCCs by a reverse hydrolyzing mechanism, which has attracted extensive attention [108,109].
4. Application of RCC in the Research and Development of New Drugs
4.1. Rha Increases the Immunogenicity of Tumour-Associated Carbohydrate Antigen (TACA) Vaccines
TACAs are carbohydrates expressed at high levels on the surface of tumor cells [110,111], and anti-TACA vaccines have been well developed [112]. However, the immunogenicity of TACAs is very low [113]. Saccharide conjugating to proteins can increase its immunogenicity, and this approach was then widely applied in conjugation vaccinations [114]. Although some glycoconjugate TACA cancer vaccines have shown promising therapeutic potential, no vaccine has yet achieved a satisfactory survival rate in clinical trials [115,116]. Guo group developed both positive and negative immunotherapies with unnatural TACAs for testing against cancers [117,118]. However, the quality control of reactions was difficult, and unexpected immune responses to proteins and linkages limited their application.
To solve these problems, two strategies have been developed: using a low-molecular-weight peptide (such as YAF) in place of proteins to increase immunogenicity of TACAs, and antigens targeting antigen-presenting cells (APCs) [117,118]. Additionally, saccharide binding to Rha can improve immunogenicity, as demonstrated by Oyelaran et al. who reported that human serum contains high levels of anti-Rha antibody [119]. Zhang et al. reported that L-Rha conjugated with truncated MAGE-A3 enhanced the immunogenicity of melanoma-associated antigen A3, thereby stimulating antitumor immune responses [120]. A study by Sarkar et al. showed that L-Rha binding to carbohydrate antigens enhanced antigenicity in mice [121]. In 2013, this team also successfully formulated a MUC1 VNTR TACA conjugate into a liposome-based anticancer vaccine, and the immunogenicity of the vaccine was further augmented by incorporating surface-displayed L-Rha epitopes onto liposomes [122]. Li et al. reported a strategy targeting tumor cells using ligand-incorporated Rha-functionalized liposomes [123]. Additionally, L-Rha epitopes can also enhance cellular immunogenicity. Partha et al. reported that the Rha-decorated liposomal Pam3Cys-MUC1-Tn vaccine showed higher cellular immunogenicity [2]. In addition, the immunogenicity of Rha-decorated liposomal Pam3Cys-MUC1-Tn was further augmented in mice when received human anti-Rha antibodies prior to its vaccination [124]. Additionally, Rha and sTn antigen, co-conjugated to bovine serum albumin (BSA), significantly enhanced antigen uptake through the involvement of Rha-specific antibodies [125]. Together, these studies showed that TACA vaccines containing Rha can increase immunogenicity. Compared with Galα1-3Galβ1-4GlcNAc-R (α-Gal epitope), Rha not only increases the immunogenicity of TACAs, but also can be used directly in wild-type mice [126].
In addition to TACA vaccines, the strategies of enhancing the monoclonal antibodies’ (mAbs) efficacy were also developed by using high levels of anti-Rha antibody of the human serum [127,128]. MAbs are one of the most rapidly growing drug classes used for the clinical practice, such as cancer and infectious and autoimmune diseases. Complement-dependent cytotoxicity (CDC) and antibody-dependent cell-mediated cytotoxicity (ADCC) are effector functions for antibodies to deplete target cells [128]. Rituximab is one of the commercially available mAbs, which is site-specifically conjugated with the Rha hapten to generate rituximab–Rha conjugates, to recruit anti-Rha antibodies onto the cancer cell surface and further form an immune complex that leads to magnifying ADCC and CDC simultaneously [128]. Ou et al. reported an efficient chemoenzymatic synthesis of structurally well-defined conjugates of antibody–rhamnose clusters to recruit natural anti-rhamnose antibodies for the enhancement of the CDC effects [127]. In addition, Coen et al. reported on antibody-recruiting glycopolymers (ARGPs) that consist of polymeric copies of a rhamnose motif, which can bind anti-Rha antibody of the human serum, for the design of potent immunotherapeutics that mark target cells for destruction by the immune system through ADCC [129]. These studies developed general and cost-effective approaches to augment the mAb effector functions with the engagement of anti-Rha antibody of the human serum that may have broad applications.
4.2. Rha-Containing Tumor-Killing Agents
Many natural products are known to have human health benefits, such as saponins and tumor-killing agents. The relationships between biological activity and chemical structure of some tumor-killing agents indicate that Rha may play a crucial role in determining biological properties. For example, kaempferol-3-O-(3″,4″-di-O-acetyl-α-L-rhamnopyranoside; SL0101) from Forsteronia refracta can inhibit the activity of Ser/Thr protein kinases (RSKs) that are closely related to the proliferation and metastasis of many tumor cells [130]. During this process, acylation of the Rha moiety of SL0101 is required for high-affinity binding and selectivity [118]. In addition, the Rha moiety of solamargine and solasonine is a key factor in anticancer activity [131,132]. Lou group demonstrated why Rha plays an important role in the anticancer activity of solasodine-derived rhamnosides; they reported that Rha-binding lectins (RBLs) on the surface of tumor cells conjugated with Rha to mediate the transportation of rhamnosides [133].
Furthermore, due to specificity of the interactions between carbohydrates and cell receptors, a lectin-directed enzyme-activated prodrug therapy (LEAPT) strategy was developed [134]. Specifically, in the first phase of this strategy, a glycosylated enzyme is targeted to specific cell types or tissues; in the second phase, prodrugs capped with sugars are administered; the glycosylated enzyme is then able to activate the prodrugs at the site of interest by cleaving the prodrug linkage; the interaction of both prodrug and enzyme relies on their precise glycosylation, and Rha-doxorubicin and Rha-5-fluorouracil are effective examples [134]. Although the Rha of tumor-killing agents is a key factor in tumor killing, L-Rha cannot kill tumor cells directly because it does not affect energy metabolism [135].
4.3. Inhibitors of Rha Synthetases as Drug Targets
Many prevalent and opportunistic pathogens, including M. tuberculosis, P. aeruginosa, and S. pneumoniae, are particularly difficult to treat due to their intrinsic chemo-resistance and their ability to acquire further resistance mechanisms against antimicrobial agents. Rha biosynthesis pathways have been discovered in numerous bacteria and fungi, but they have not been discovered in humans, hence they might be potential therapeutic targets [136,137]. The first nanomolar inhibitors of RmlA from P. aeruginosa were thymine analogs, and some inhibitors also showed inhibitory activity against M. tuberculosis [138]. In addition, L-Rha-1-C-phosphonate is the best inhibitor of Cps2L, and a fluorine atom at C1 can increase inhibition by 25%, but two fluorine atoms at C1 had an adverse effect [139]. Furthermore, RmlC is the most promising therapeutic target because it possesses high substrate specificity and it does not require a cofactor [140].
5. Conclusions
RCCs are present in bacteria but not in humans and other mammals, making them valuable for tumor immunotherapy and treating antibacterial infections. To date, RCCs have been studied extensively, and produced a series of excellent results, i.e., the discovery of the biosynthetic pathways of three rhamnose donors, the discovery of Rha-Ts, and their application to the treatment of various diseases. In this review, the biosynthesis pathways and the properties of the related enzymes from three donor substrates, including dTDP-Rha, GDP-Rha, and UDP-Rha, were reviewed in detail, which is of great significance for the development of the strategies for the preparation of donor substrates of Rha-Ts in vitro. In addition, the functions and properties of Rha-Ts were also reviewed, which provides theoretical guidance for the development of Rha-Ts and the enzymatic synthesis of RCCs. It is important to note the complex and diverse structures of the receptor substrates of Rha-Ts, which need to be further studied. However, the research of the synthesis pathways of RCCs from different cells, the properties of related enzymes and their catalytic mechanisms is rather little; therefore, further studies on the biosynthesis and applications of RCCs are being carried out at present and subsequently via the latest biochemical technologies, such as molecular biology, structural biology, and computational biochemistry techniques.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27165315/s1, Table S1: Sugar-1-phosphate substrates of RmlA and their conversion into corresponding NDP-sugars.
Author Contributions
S.L. and L.W. made the bibliography, analyzed the literature, wrote the first drafts, and revised the final version of the manuscript. F.C., Y.L. and H.L. corrected the manuscript and wrote the final draft. G.G. and E.L. suggested the topic of the article, revised the manuscript, and adjusted it to be in a suitable form for publication. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of Henan Province (grant number 212300410203, 202300410279), the Key Science and Technology Innovation Demonstration Projects of Henan Province, (grant number 191110110600),the Scientific and Technological Research Project Foundation of Henan Provincial Scientific and Technological Department (grant number 202102310479, 222102110180), the Young Backbone Teachers Fundation of Huanghuai University, the Scientific Research Foundation for Advanced Talents of Huanghuai University, grant number 1000.12.01.1678.
Conflicts of Interest
The authors declare no conflict of interest.
References
- McArthur, J.B.; Chen, X. Glycosyltransferase engineering for carbohydrate synthesis. Biochem. Soc. Trans. 2016, 44, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, P.; Lee, K.; Sarkar, S.; Wall, K.A.; Sucheck, S.J. Synthesis of a Liposomal MUC1 Glycopeptide-Based Immunotherapeutic and Evaluation of the Effect of L-Rhamnose Targeting on Cellular Immune Responses. Bioconjug. Chem. 2016, 27, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.X.; Cheng, L.; Pan, M.; Qian, Q.; Zhu, Y.L.; Xu, L.Y.; Ding, Q. D Rhamnose beta-Hederin against human breast cancer by reducing tumor-derived exosomes. Oncol. Lett. 2018, 16, 5172–5178. [Google Scholar] [PubMed]
- Vanacore, A.; Vitiello, G.; Wanke, A.; Cavasso, D.; Clifton, L.A.; Mahdi, L.; Campanero-Rhodes, M.A.; Solis, D.; Wuhrer, M.; Nicolardi, S.; et al. Lipopolysaccharide O-antigen molecular and supramolecular modifications of plant root microbiota are pivotal for host recognition. Carbohydr. Polym. 2022, 277, 118839. [Google Scholar] [CrossRef] [PubMed]
- Concordio-Reis, P.; Alves, V.D.; Moppert, X.; Guezennec, J.; Freitas, F.; Reis, M.A.M. Characterization and Biotechnological Potential of Extracellular Polysaccharides Synthesized by Alteromonas Strains Isolated from French Polynesia Marine Environments. Mar. Drugs 2021, 19, 522. [Google Scholar] [CrossRef]
- Garcia-Vello, P.; Sharma, G.; Speciale, I.; Molinaro, A.; Im, S.H.; De Castro, C. Structural features and immunological perception of the cell surface glycans of Lactobacillus plantarum: A novel rhamnose-rich polysaccharide and teichoic acids. Carbohydr. Polym. 2020, 233, 115857. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zhou, P.; Qian, S.; Qin, X.; Fan, Z.; Fu, Q.; Zhan, Z.; Pei, H. Cloning, expression, identification and bioinformatics analysis of Rv3265c gene from Mycobacterium tuberculosis in Escherichia coli. Asian Pac. J. Trop. Med. 2011, 4, 266–270. [Google Scholar] [CrossRef]
- De Castro, C.; Molinaro, A.; Piacente, F.; Gurnon, J.R.; Sturiale, L.; Palmigiano, A.; Lanzetta, R.; Parrilli, M.; Garozzo, D.; Tonetti, M.G.; et al. Structure of N-linked oligosaccharides attached to chlorovirus PBCV-1 major capsid protein reveals unusual class of complex N-glycans. Proc. Natl. Acad. Sci. USA 2013, 110, 13956–13960. [Google Scholar] [CrossRef]
- Martinez, V.; Ingwers, M.; Smith, J.; Glushka, J.; Yang, T.; Bar-Peled, M. Biosynthesis of UDP-4-keto-6-deoxyglucose and UDP-rhamnose in pathogenic fungi Magnaporthe grisea and Botryotinia fuckeliana. J. Biol. Chem. 2012, 287, 879–892. [Google Scholar] [CrossRef]
- Yu, Y.; Wen, Q.; Song, A.; Liu, Y.; Wang, F.; Jiang, B. Isolation and immune activity of a new acidic Cordyceps militaris exopolysaccharide. Int. J. Biol. Macromol. 2022, 194, 706–714. [Google Scholar] [CrossRef]
- Allen, S.; Richardson, J.M.; Mehlert, A.; Ferguson, M.A. Structure of a complex phosphoglycan epitope from gp72 of Trypanosoma cruzi. J. Biol. Chem. 2013, 288, 11093–11105. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.K.; Vartak, A.; Sucheck, S.J.; Wall, K.A. Synthesis and Immunological Evaluation of a Single Molecular Construct MUC1 Vaccine Containing L-Rhamnose Repeating Units. Molecules 2020, 25, 3137. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, B.A.; Zorzoli, A.; Dorfmueller, H.C. NDP-rhamnose biosynthesis and rhamnosyltransferases: Building diverse glycoconjugates in nature. Biochem. J. 2021, 478, 685–701. [Google Scholar] [CrossRef]
- Oh, J.; Lee, S.G.; Kim, B.G.; Sohng, J.K.; Liou, K.; Lee, H.C. One-pot enzymatic production of dTDP-4-keto-6-deoxy-D-glucose from dTMP and glucose-1-phosphate. Biotechnol. Bioeng. 2003, 84, 452–458. [Google Scholar] [CrossRef]
- Glaser, L. The synthesis of thymidine-linked sugars. III. On the mechanism of thymidine diphosphate-L-rhamnose formation. Biochim. Biophys. Acta 1961, 51, 169–171. [Google Scholar] [CrossRef]
- Thibodeaux, C.J.; Melancon, C.R.; Liu, H.W. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew. Chem. Int. Ed. Engl. 2008, 47, 9814–9859. [Google Scholar] [CrossRef]
- Pal, D.; Mukhopadhyay, B. Chemical Synthesis of beta-L-Rhamnose Containing the Pentasaccharide Repeating Unit of the O-Specific Polysaccharide from a Halophilic Bacterium Halomonas ventosae RU5S2EL in the Form of Its 2-Aminoethyl Glycoside. J. Org. Chem. 2021, 86, 8683–8694. [Google Scholar] [CrossRef] [PubMed]
- Cloutier, M.; Prevost, M.J.; Lavoie, S.; Feroldi, T.; Piochon, M.; Groleau, M.C.; Legault, J.; Villaume, S.; Crouzet, J.; Dorey, S.; et al. Total synthesis, isolation, surfactant properties, and biological evaluation of ananatosides and related macrodilactone-containing rhamnolipids. Chem. Sci. 2021, 12, 7533–7546. [Google Scholar] [CrossRef]
- Jordan, D.S.; Daubenspeck, J.M.; Dybvig, K. Rhamnose biosynthesis in mycoplasmas requires precursor glycans larger than monosaccharide. Mol. Microbiol. 2013, 89, 918–928. [Google Scholar] [CrossRef]
- Madduri, K.; Waldron, C.; Merlo, D.J. Rhamnose biosynthesis pathway supplies precursors for primary and secondary metabolism in Saccharopolyspora spinosa. J. Bacteriol. 2001, 183, 5632–5638. [Google Scholar] [CrossRef]
- Yang, S.; An, X.; Gu, G.; Yan, Z.; Jiang, X.; Xu, L.; Xiao, M. Novel dTDP-L-Rhamnose Synthetic Enzymes (RmlABCD) From Saccharothrix syringae CGMCC 4.1716 for One-Pot Four-Enzyme Synthesis of dTDP-L-Rhamnose. Front. Microbiol. 2021, 12, 772839. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Studies and Application of Rhamnosyltransferase and Related Enzymes from Streptococcus pneumonia Serotype 23F. Ph.D. Thesis, Shandong University, Shandong, China, 2017. [Google Scholar]
- Li, S.; Wang, H.; Ma, J.; Gu, G.; Chen, Z.; Guo, Z. One-pot four-enzyme synthesis of thymidinediphosphate-L-rhamnose. Chem. Commun. 2016, 52, 13995–13998. [Google Scholar] [CrossRef] [PubMed]
- Pazur, J.H.; Anderson, J.S. Thymidine triphosphate: Alpha-D-galactose-L-phosphate thymj-L-phosphate thymidylyltransferase from streptococcus faecalis grown on d-galactose. J. Biol. Chem. 1963, 238, 3155–3160. [Google Scholar] [CrossRef]
- Barton, W.A.; Lesniak, J.; Biggins, J.B.; Jeffrey, P.D.; Jiang, J.; Rajashankar, K.R.; Thorson, J.S.; Nikolov, D.B. Structure, mechanism and engineering of a nucleotidylyltransferase as a first step toward glycorandomization. Nat. Struct. Biol. 2001, 8, 545–551. [Google Scholar] [CrossRef]
- Jiang, J.; Biggins, J.B.; Thorson, J.S. A General Enzymatic Method for the Synthesis of Natural and “Unnatural” UDP- and TDP-Nucleotide Sugars. J. Am. Chem. Soc. 2000, 122, 6803–6804. [Google Scholar] [CrossRef]
- Barton, W.A.; Biggins, J.B.; Jiang, J.; Thorson, J.S.; Nikolov, D.B. Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering. Proc. Natl. Acad. Sci. USA 2002, 99, 13397–13402. [Google Scholar] [CrossRef]
- Moretti, R.; Chang, A.; Peltier-Pain, P.; Bingman, C.A.; Phillips, G.J.; Thorson, J.S. Expanding the nucleotide and sugar 1-phosphate promiscuity of nucleotidyltransferase RmlA via directed evolution. J. Biol. Chem. 2011, 286, 13235–13243. [Google Scholar] [CrossRef]
- Jakeman, D.L.; Young, J.L.; Huestis, M.P.; Peltier, P.; Daniellou, R.; Nugier-Chauvin, C.; Ferrieres, V. Engineering ribonucleoside triphosphate specificity in a thymidylyltransferase. Biochemistry 2008, 47, 8719–8725. [Google Scholar] [CrossRef]
- Jiqing Jiang, J.B.B.A. Expanding the Pyrimidine Diphosphosugar Repertoire: The Chemoenzymatic Synthesis of Amino- and Acetamidoglucopyranosyl Derivatives. Angew. Chem. Int. Ed. Engl. 2001, 8, 1502–1505. [Google Scholar] [CrossRef]
- Timmons, S.C.; Mosher, R.H.; Knowles, S.A.; Jakeman, D.L. Exploiting nucleotidylyltransferases to prepare sugar nucleotides. Org. Lett. 2007, 9, 857–860. [Google Scholar] [CrossRef]
- Forget, S.M.; Smithen, D.A.; Jee, A.; Jakeman, D.L. Mechanistic evaluation of a nucleoside tetraphosphate with a thymidylyltransferase. Biochemistry 2015, 54, 1703–1707. [Google Scholar] [CrossRef] [PubMed]
- Huestis, M.P.; Aish, G.A.; Hui, J.P.; Soo, E.C.; Jakeman, D.L. Lipophilic sugar nucleotide synthesis by structure-based design of nucleotidylyltransferase substrates. Org. Biomol. Chem. 2008, 6, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Timmons, S.C.; Hui, J.P.; Pearson, J.L.; Peltier, P.; Daniellou, R.; Nugier-Chauvin, C.; Soo, E.C.; Syvitski, R.T.; Ferrieres, V.; Jakeman, D.L. Enzyme-catalyzed synthesis of furanosyl nucleotides. Org. Lett. 2008, 10, 161–163. [Google Scholar] [CrossRef] [PubMed]
- Moretti, R.; Thorson, J.S. Enhancing the latent nucleotide triphosphate flexibility of the glucose-1-phosphate thymidylyltransferase RmlA. J. Biol. Chem. 2007, 282, 16942–16947. [Google Scholar] [CrossRef]
- Yang, J.; Hoffmeister, D.; Liu, L.; Fu, X.; Thorson, J.S. Natural product glycorandomization. Bioorg. Med. Chem. 2004, 12, 1577–1584. [Google Scholar] [CrossRef]
- Zhang, C.; Moretti, R.; Jiang, J.; Thorson, J.S. The in vitro characterization of polyene glycosyltransferases AmphDI and NysDI. ChemBioChem 2008, 9, 2506–2514. [Google Scholar] [CrossRef]
- Jiang, J.; Albermann, C.; Thorson, J.S. Application of the nucleotidylyltransferase Ep toward the chemoenzymatic synthesis of dTDP-desosamine analogues. ChemBioChem 2003, 4, 443–446. [Google Scholar] [CrossRef]
- Albermann, C.; Soriano, A.; Jiang, J.; Vollmer, H.; Biggins, J.B.; Barton, W.A.; Lesniak, J.; Nikolov, D.B.; Thorson, J.S. Substrate specificity of NovM: Implications for novobiocin biosynthesis and glycorandomization. Org. Lett. 2003, 5, 933–936. [Google Scholar] [CrossRef]
- Williams, G.J.; Gantt, R.W.; Thorson, J.S. The impact of enzyme engineering upon natural product glycodiversification. Curr. Opin. Chem. Biol. 2008, 12, 556–564. [Google Scholar] [CrossRef]
- Thorson, J.S.; Barton, W.A.; Hoffmeister, D.; Albermann, C.; Nikolov, D.B. Structure-based enzyme engineering and its impact on in vitro glycorandomization. ChemBioBhem 2004, 5, 16–25. [Google Scholar] [CrossRef]
- Beaton, S.A.; Huestis, M.P.; Sadeghi-Khomami, A.; Thomas, N.R.; Jakeman, D.L. Enzyme-catalyzed synthesis of isosteric phosphono-analogues of sugar nucleotides. Chem. Commun. 2009, 8, 238–240. [Google Scholar] [CrossRef] [PubMed]
- Melo, A.; Glaser, L. The nucleotide specificity and feedback control of thymidine diphosphate D-glucose pyrophosphorylase. J. Biol. Chem. 1965, 240, 398–405. [Google Scholar] [CrossRef]
- Blankenfeldt, W.; Giraud, M.F.; Leonard, G.; Rahim, R.; Creuzenet, C.; Lam, J.S.; Naismith, J.H. The purification, crystallization and preliminary structural characterization of glucose-1-phosphate thymidylyltransferase (RmlA), the first enzyme of the dTDP-L-rhamnose synthesis pathway from Pseudomonas aeruginosa. Acta. Crystallogr. D. Biol. Crystallogr. 2000, 56 Pt 11, 1501–1504. [Google Scholar] [CrossRef] [PubMed]
- Sivaraman, J.; Sauve, V.; Matte, A.; Cygler, M. Crystal structure of Escherichia coli glucose-1-phosphate thymidylyltransferase (RffH) complexed with dTTP and Mg2+. J. Biol. Chem. 2002, 277, 44214–44219. [Google Scholar] [CrossRef]
- Allard, S.T.; Beis, K.; Giraud, M.F.; Hegeman, A.D.; Gross, J.W.; Wilmouth, R.C.; Whitfield, C.; Graninger, M.; Messner, P.; Allen, A.G.; et al. Toward a structural understanding of the dehydratase mechanism. Structure 2002, 10, 81–92. [Google Scholar] [CrossRef]
- James, D.B.; Yother, J. Genetic and biochemical characterizations of enzymes involved in Streptococcus pneumoniae serotype 2 capsule synthesis demonstrate that Cps2T (WchF) catalyzes the committed step by addition of beta1-4 rhamnose, the second sugar residue in the repeat unit. J. Bacteriol. 2012, 194, 6479–6489. [Google Scholar] [CrossRef]
- Dong, C.; Major, L.L.; Allen, A.; Blankenfeldt, W.; Maskell, D.; Naismith, J.H. High-resolution structures of RmlC from Streptococcus suis in complex with substrate analogs locate the active site of this class of enzyme. Structure 2003, 11, 715–723. [Google Scholar] [CrossRef]
- Dong, C.; Major, L.L.; Srikannathasan, V.; Errey, J.C.; Giraud, M.F.; Lam, J.S.; Graninger, M.; Messner, P.; McNeil, M.R.; Field, R.A.; et al. RmlC, a C3′ and C5′ carbohydrate epimerase, appears to operate via an intermediate with an unusual twist boat conformation. J. Mol. Biol. 2007, 365, 146–159. [Google Scholar] [CrossRef]
- Giraud, M.F.; Leonard, G.A.; Field, R.A.; Berlind, C.; Naismith, J.H. RmlC, the third enzyme of dTDP-L-rhamnose pathway, is a new class of epimerase. Nat. Struct. Biol. 2000, 7, 398–402. [Google Scholar]
- Giraud, M.F.; Gordon, F.M.; Whitfield, C.; Messner, P.; McMahon, S.A.; Naismith, J.H. Purification, crystallization and preliminary structural studies of dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase (RmlC), the third enzyme of the dTDP-L-rhamnose synthesis pathway, from Salmonella enterica serovar typhimurium. Acta Crystallogr. D Biol. Crystallogr. 1999, 55 Pt 3, 706–708. [Google Scholar] [CrossRef]
- Blankenfeldt, W.; Kerr, I.D.; Giraud, M.F.; McMiken, H.J.; Leonard, G.; Whitfield, C.; Messner, P.; Graninger, M.; Naismith, J.H. Variation on a theme of SDR. dTDP-6-deoxy-L- lyxo-4-hexulose reductase (RmlD) shows a new Mg2+-dependent dimerization mode. Structure 2002, 10, 773–786. [Google Scholar] [CrossRef]
- Graninger, M.; Nidetzky, B.; Heinrichs, D.E.; Whitfield, C.; Messner, P. Characterization of dTDP-4-dehydrorhamnose 3,5-epimerase and dTDP-4-dehydrorhamnose reductase, required for dTDP-L-rhamnose biosynthesis in Salmonella enterica serovar Typhimurium LT2. J. Biol. Chem. 1999, 274, 25069–25077. [Google Scholar] [CrossRef] [PubMed]
- Ovod, V.; Rudolph, K.; Knirel, Y.; Krohn, K. Immunochemical characterization of O polysaccharides composing the alpha-D-rhamnose backbone of lipopolysaccharide of Pseudomonas syringae and classification of bacteria into serogroups O1 and O2 with monoclonal antibodies. J. Bacteriol. 1996, 178, 6459–6465. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kneidinger, B.; Graninger, M.; Adam, G.; Puchberger, M.; Kosma, P.; Zayni, S.; Messner, P. Identification of two GDP-6-deoxy-D-lyxo-4-hexulose reductases synthesizing GDP-D-rhamnose in Aneurinibacillus thermoaerophilus L420-91T. J. Biol. Chem. 2001, 276, 5577–5583. [Google Scholar] [CrossRef] [PubMed]
- Webb, N.A.; Mulichak, A.M.; Lam, J.S.; Rocchetta, H.L.; Garavito, R.M. Crystal structure of a tetrameric GDP-D-mannose 4,6-dehydratase from a bacterial GDP-D-rhamnose biosynthetic pathway. Protein Sci. 2004, 13, 529–539. [Google Scholar] [CrossRef]
- Ginsburg, V. Formation of guanosine diphosphate L-fucose from guanosine diphosphate D-mannose. J. Biol. Chem. 1960, 235, 2196–2201. [Google Scholar] [CrossRef]
- Mulichak, A.M.; Bonin, C.P.; Reiter, W.D.; Garavito, R.M. Structure of the MUR1 GDP-mannose 4,6-dehydratase from Arabidopsis thaliana: Implications for ligand binding and specificity. Biochemistry 2002, 41, 15578–15589. [Google Scholar] [CrossRef]
- Reitman, M.L.; Trowbridge, I.S.; Kornfeld, S. Mouse lymphoma cell lines resistant to pea lectin are defective in fucose metabolism. J. Biol. Chem. 1980, 255, 9900–9906. [Google Scholar] [CrossRef]
- Bisso, A.; Sturla, L.; Zanardi, D.; De Flora, A.; Tonetti, M. Structural and enzymatic characterization of human recombinant GDP-D-mannose-4,6-dehydratase. FEBS. Lett. 1999, 456, 370–374. [Google Scholar] [CrossRef]
- Tonetti, M.; Zanardi, D.; Gurnon, J.R.; Fruscione, F.; Armirotti, A.; Damonte, G.; Sturla, L.; De Flora, A.; Van Etten, J.L. Paramecium bursaria Chlorella virus 1 encodes two enzymes involved in the biosynthesis of GDP-L-fucose and GDP-D-rhamnose. J. Biol. Chem. 2003, 278, 21559–21565. [Google Scholar] [CrossRef]
- Somoza, J.R.; Menon, S.; Schmidt, H.; Joseph-McCarthy, D.; Dessen, A.; Stahl, M.L.; Somers, W.S.; Sullivan, F.X. Structural and kinetic analysis of Escherichia coli GDP-mannose 4,6 dehydratase provides insights into the enzyme’s catalytic mechanism and regulation by GDP-fucose. Structure 2000, 8, 123–135. [Google Scholar] [CrossRef]
- Rosano, C.; Zuccotti, S.; Sturla, L.; Fruscione, F.; Tonetti, M.; Bolognesi, M. Quaternary assembly and crystal structure of GDP-D-mannose 4,6 dehydratase from Paramecium bursaria Chlorella virus. Biochem. Biophys. Res. Commun. 2006, 339, 191–195. [Google Scholar] [CrossRef] [PubMed]
- King, J.D.; Poon, K.K.; Webb, N.A.; Anderson, E.M.; McNally, D.J.; Brisson, J.R.; Messner, P.; Garavito, R.M.; Lam, J.S. The structural basis for catalytic function of GMD and RMD, two closely related enzymes from the GDP-D-rhamnose biosynthesis pathway. FEBS J. 2009, 276, 2686–2700. [Google Scholar] [CrossRef]
- Qian, L.; Han, X.; Zhang, L.; Liu, X. Structural and biochemical insights into nucleotide-rhamnose synthase/epimerase-reductase from Arabidopsis thaliana. Biochim. Biophys. Acta 2015, 10 Pt A, 1476–1486. [Google Scholar]
- Oka, T.; Nemoto, T.; Jigami, Y. Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-D-glucose to UDP-L-rhamnose conversion. J. Biol. Chem. 2007, 282, 5389–5403. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, J.J.; Kasimova, A.A.; Sviridova, A.N.; Shpirt, A.M.; Shneider, M.M.; Mikhaylova, Y.V.; Shelenkov, A.A.; Popova, A.V.; Perepelov, A.V.; Shashkov, A.S.; et al. Correlation of Acinetobacter baumannii K144 and K86 capsular polysaccharide structures with genes at the K locus reveals the involvement of a novel multifunctional rhamnosyltransferase for structural synthesis. Int. J. Biol. Macromol. 2021, 193 Pt B, 1294–1300. [Google Scholar] [CrossRef]
- Kenyon, J.J.; Arbatsky, N.P.; Sweeney, E.L.; Zhang, Y.; Senchenkova, S.N.; Popova, A.V.; Shneider, M.M.; Shashkov, A.S.; Liu, B.; Hall, R.M.; et al. Involvement of a multifunctional rhamnosyltransferase in the synthesis of three related Acinetobacter baumannii capsular polysaccharides, K55, K74 and K85. Int. J. Biol. Macromol. 2021, 166, 1230–1237. [Google Scholar] [CrossRef]
- Steiner, K.; Hagelueken, G.; Messner, P.; Schaffer, C.; Naismith, J.H. Structural basis of substrate binding in WsaF, a rhamnosyltransferase from Geobacillus stearothermophilus. J. Mol. Biol. 2010, 397, 436–447. [Google Scholar] [CrossRef]
- Steiner, K.; Novotny, R.; Werz, D.B.; Zarschler, K.; Seeberger, P.H.; Hofinger, A.; Kosma, P.; Schaffer, C.; Messner, P. Molecular basis of S-layer glycoprotein glycan biosynthesis in Geobacillus stearothermophilus. J. Biol. Chem. 2008, 283, 21120–21133. [Google Scholar] [CrossRef]
- Isiorho, E.A.; Liu, H.W.; Keatinge-Clay, A.T. Structural studies of the spinosyn rhamnosyltransferase, SpnG. Biochemistry. 2012, 51, 1213–1222. [Google Scholar] [CrossRef]
- Huang, K.X.; Xia, L.; Zhang, Y.; Ding, X.; Zahn, J.A. Recent advances in the biochemistry of spinosyns. Appl. Microbiol. Biotechnol. 2009, 82, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Chen, Y.H.; Lin, Y.C.; Tsai, K.C.; Chiu, H.T. Functional characterization and substrate specificity of spinosyn rhamnosyltransferase by in vitro reconstitution of spinosyn biosynthetic enzymes. J. Biol. Chem. 2009, 284, 7352–7363. [Google Scholar] [CrossRef] [PubMed]
- Grzegorzewicz, A.E.; Ma, Y.; Jones, V.; Crick, D.; Liav, A.; McNeil, M.R. Development of a microtitre plate-based assay for lipid-linked glycosyltransferase products using the mycobacterial cell wall rhamnosyltransferase WbbL. Microbiology 2008, 154 Pt 12, 3724–3730. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sivendran, S.; Jones, V.; Sun, D.; Wang, Y.; Grzegorzewicz, A.E.; Scherman, M.S.; Napper, A.D.; McCammon, J.A.; Lee, R.E.; Diamond, S.L.; et al. Identification of triazinoindol-benzimidazolones as nanomolar inhibitors of the Mycobacterium tuberculosis enzyme TDP-6-deoxy-d-xylo-4-hexopyranosid-4-ulose 3,5-epimerase (RmlC). Bioorg. Med. Chem. 2010, 18, 896–908. [Google Scholar] [CrossRef]
- Mills, J.A.; Motichka, K.; Jucker, M.; Wu, H.P.; Uhlik, B.C.; Stern, R.J.; Scherman, M.S.; Vissa, V.D.; Pan, F.; Kundu, M.; et al. Inactivation of the mycobacterial rhamnosyltransferase, which is needed for the formation of the arabinogalactan-peptidoglycan linker, leads to irreversible loss of viability. J. Biol. Chem. 2004, 279, 43540–43546. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Ramirez, M.; Medina, G.; Gonzalez-Valdez, A.; Grosso-Becerra, V.; Soberon-Chavez, G. The Pseudomonas aeruginosa rmlBDAC operon, encoding dTDP-L-rhamnose biosynthetic enzymes, is regulated by the quorum-sensing transcriptional regulator RhlR and the alternative sigma factor sigmaS. Microbiology 2012, 158 Pt 4, 908–916. [Google Scholar] [CrossRef]
- Poon, K.K.; Westman, E.L.; Vinogradov, E.; Jin, S.; Lam, J.S. Functional characterization of MigA and WapR: Putative rhamnosyltransferases involved in outer core oligosaccharide biosynthesis of Pseudomonas aeruginosa. J. Bacteriol. 2008, 190, 1857–1865. [Google Scholar] [CrossRef]
- Rocchetta, H.L.; Burrows, L.L.; Lam, J.S. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 1999, 63, 523–553. [Google Scholar] [CrossRef] [PubMed]
- Rocchetta, H.L.; Burrows, L.L.; Pacan, J.C.; Lam, J.S. Three rhamnosyltransferases responsible for assembly of the A-band D-rhamnan polysaccharide in Pseudomonas aeruginosa: A fourth transferase, WbpL, is required for the initiation of both A-band and B-band lipopolysaccharide synthesis. Mol. Microbiol. 1998, 28, 1103–1119. [Google Scholar] [CrossRef]
- Melamed, J.; Kocev, A.; Torgov, V.; Veselovsky, V.; Brockhausen, I. Biosynthesis of the Pseudomonas aeruginosa common polysaccharide antigen by D-Rhamnosyltransferases WbpX and WbpY. Glycoconj. J. 2022, 39, 393–411. [Google Scholar] [CrossRef]
- Zhu, K.; Rock, C.O. RhlA converts beta-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the beta-hydroxydecanoyl-beta-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. J. Bacteriol. 2008, 190, 3147–3154. [Google Scholar] [CrossRef]
- Ochsner, U.A.; Fiechter, A.; Reiser, J. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J. Biol. Chem. 1994, 269, 19787–19795. [Google Scholar] [CrossRef]
- Rahim, R.; Ochsner, U.A.; Olvera, C.; Graninger, M.; Messner, P.; Lam, J.S.; Soberon-Chavez, G. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol. 2001, 40, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Deziel, E.; Lepine, F.; Milot, S.; Villemur, R. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 2003, 149 Pt 8, 2005–2013. [Google Scholar] [CrossRef] [PubMed]
- Tavares, L.F.; Silva, P.M.; Junqueira, M.; Mariano, D.C.; Nogueira, F.C.; Domont, G.B.; Freire, D.M.; Neves, B.C. Characterization of rhamnolipids produced by wild-type and engineered Burkholderia kururiensis. Appl. Microbiol. Biotechnol. 2013, 97, 1909–1921. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Luo, J.; Li, S. Bacteria-Catalyzed Arginine Glycosylation in Pathogens and Host. Front. Cell. Infect. Microbiol. 2020, 10, 185. [Google Scholar] [CrossRef]
- He, C.; Liu, N.; Li, F.; Jia, X.; Peng, H.; Liu, Y.; Xiao, Y. Complex Structure of Pseudomonas aeruginosa Arginine Rhamnosyltransferase EarP with Its Acceptor Elongation Factor P. J. Bacteriol. 2019, 201, e00128-19. [Google Scholar] [CrossRef]
- Lassak, J.; Keilhauer, E.C.; Fürst, M.; Wuichet, K.; Gödeke, J.; Starosta, A.L.; Chen, J.M.; Søgaard-Andersen, L.; Rohr, J.; Wilson, D.N.; et al. Arginine-rhamnosylation as new strategy to activate translation elongation factor P. Nat. Chem. Biol. 2015, 11, 266–270. [Google Scholar] [CrossRef]
- Yanagisawa, T.; Takahashi, H.; Suzuki, T.; Masuda, A.; Dohmae, N.; Yokoyama, S. Neisseria meningitidis Translation Elongation Factor P and Its Active-Site Arginine Residue Are Essential for Cell Viability. PLoS ONE 2016, 11, e0147907. [Google Scholar] [CrossRef]
- Geno, K.A.; Gilbert, G.L.; Song, J.Y.; Skovsted, I.C.; Klugman, K.P.; Jones, C.; Konradsen, H.B.; Nahm, M.H. Pneumococcal Capsules and Their Types: Past, Present, and Future. Clin. Microbiol. Rev. 2015, 28, 871–899. [Google Scholar] [CrossRef]
- Park, S.; Nahm, M.H. L-rhamnose is often an important part of immunodominant epitope for pneumococcal serotype 23F polysaccharide antibodies in human sera immunized with PPV23. PLoS ONE 2013, 8, e83810. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Legnani, L.; Ronchi, S.; Fallarini, S.; Lombardi, G.; Campo, F.; Panza, L.; Lay, L.; Poletti, L.; Toma, L.; Ronchetti, F.; et al. Synthesis, molecular dynamics simulations, and biology of a carba-analogue of the trisaccharide repeating unit of Streptococcus pneumoniae 19F capsular polysaccharide. Org. Biomol. Chem. 2009, 7, 4428–4436. [Google Scholar] [CrossRef] [PubMed]
- Aanensen, D.M.; Mavroidi, A.; Bentley, S.D.; Reeves, P.R.; Spratt, B.G. Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 2007, 189, 7856–7876. [Google Scholar] [CrossRef]
- James, D.B.; Gupta, K.; Hauser, J.R.; Yother, J. Biochemical activities of Streptococcus pneumoniae serotype 2 capsular glycosyltransferases and significance of suppressor mutations affecting the initiating glycosyltransferase Cps2E. J. Bacteriol. 2013, 195, 5469–5478. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, S.; Xiong, C.; Jin, G.; Chen, Z.; Gu, G.; Guo, Z. Biochemical studies of a beta-1,4-rhamnoslytransferase from Streptococcus pneumonia serotype 23F. Org. Biomol. Chem. 2019, 17, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Ardissone, S.; Noel, K.D.; Klement, M.; Broughton, W.J.; Deakin, W.J. Synthesis of the flavonoid-induced lipopolysaccharide of Rhizobium Sp. strain NGR234 requires rhamnosyl transferases encoded by genes rgpF and wbgA. Mol. Plant-Microbe Interact. 2011, 24, 1513–1521. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Sugano, Y.; Shoda, M. Novel glycosyltransferase genes involved in the acetan biosynthesis of Acetobacter xylinum. Biochem. Biophys. Res. Commun. 2002, 295, 230–235. [Google Scholar] [CrossRef]
- Dong, S.; McPherson, S.A.; Wang, Y.; Li, M.; Wang, P.; Turnbough, C.J.; Pritchard, D.G. Characterization of the enzymes encoded by the anthrose biosynthetic operon of Bacillus anthracis. J. Bacteriol. 2010, 192, 5053–5062. [Google Scholar] [CrossRef][Green Version]
- Zorzoli, A.; Meyer, B.H.; Adair, E.; Torgov, V.I.; Veselovsky, V.V.; Danilov, L.L.; Uhrin, D.; Dorfmueller, H.C. Group A, B, C, and G Streptococcus Lancefield antigen biosynthesis is initiated by a conserved α-d-GlcNAc-β-1,4-L-rhamnosyltransferase. J. Biol. Chem. 2019, 294, 15237–15256. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Mukai, T.; Naka, T.; Fujiwara, N.; Maeda, Y.; Kai, M.; Mizuno, S.; Yano, I.; Makino, M. Novel rhamnosyltransferase involved in biosynthesis of serovar 4-specific glycopeptidolipid from Mycobacterium avium complex. J. Bacteriol. 2010, 192, 5700–5708. [Google Scholar] [CrossRef]
- Tsunashima, H.; Miyake, K.; Motono, M.; Iijima, S. Organization of the capsule biosynthesis gene locus of the oral streptococcus Streptococcus anginosus. J. Biosci. Bioeng. 2012, 113, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Gong, W.; Sun, C.; Zhao, J.; Wang, H.; Chen, Z.; Xiao, M.; Gu, G. Sequential One-Pot Three-Enzyme Synthesis of the Tetrasaccharide Repeating Unit of Group B Streptococcus Serotype VIII Capsular Polysaccharide. Chin. J. Chem. 2022, 40, 1039–1044. [Google Scholar] [CrossRef]
- Li, Q.; Hobbs, M.; Reeves, P.R. The variation of dTDP-L-rhamnose pathway genes in Vibrio cholerae. Microbiology 2003, 149 Pt 9, 2463–2474. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Chen, R.; Xie, K.; Chen, D.; Guo, B.; Liu, X.; Liu, J.; Zhang, M.; Dai, J. A regiospecific rhamnosyltransferase from Epimedium pseudowushanense catalyzes the 3-O-rhamnosylation of prenylflavonols. Org. Biomol. Chem. 2018, 16, 452–458. [Google Scholar] [CrossRef]
- Zong, G.; Fei, S.; Liu, X.; Li, J.; Gao, Y.; Yang, X.; Wang, X.; Shen, Y. Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-β-L-rhamnose and rhamnosylation mechanism. Plant J. 2019, 99, 257–269. [Google Scholar]
- Takenaka, Y.; Kato, K.; Ogawa-Ohnishi, M.; Tsuruhama, K.; Kajiura, H.; Yagyu, K.; Takeda, A.; Takeda, Y.; Kunieda, T.; Hara-Nishimura, I.; et al. Pectin RG-I rhamnosyltransferases represent a novel plant-specific glycosyltransferase family. Nat. Plants 2018, 4, 669–676. [Google Scholar] [CrossRef]
- Lu, C.; Dong, Y.; Ke, K.; Zou, K.; Wang, Z.; Xiao, W.; Pei, J.; Zhao, L. Modification to increase the thermostability and catalytic efficiency of α-L-rhamnosidase from Bacteroides thetaiotaomicron and high-level expression. Enzym. Microb. Technol. 2022, 158, 110040. [Google Scholar] [CrossRef]
- Xu, L.; Liu, X.; Li, Y.; Yin, Z.; Jin, L.; Lu, L.; Qu, J.; Xiao, M. Enzymatic rhamnosylation of anticancer drugs by an α-L-rhamnosidase from Alternaria sp. L1 for cancer-targeting and enzyme-activated prodrug therapy. Appl. Microbiol. Biotechnol. 2019, 103, 7997–8008. [Google Scholar] [CrossRef]
- Hakomori, S. Tumor-associated carbohydrate antigens defining tumor malignancy: Basis for development of anti-cancer vaccines. Adv. Exp. Med. Biol. 2001, 491, 369–402. [Google Scholar]
- Hakomori, S. Aberrant glycosylation in cancer cell membranes as focused on glycolipids: Overview and perspectives. Cancer Res. 1985, 45, 2405–2414. [Google Scholar]
- Guo, Z.; Wang, Q. Recent development in carbohydrate-based cancer vaccines. Curr. Opin. Chem. Biol. 2009, 13, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.M.; Danishefsky, S.J. A Vision for Vaccines Built from Fully Synthetic Tumor-Associated Antigens: From the Laboratory to the Clinic. J. Am. Chem. Soc. 2013, 135, 14462–14472. [Google Scholar] [CrossRef]
- Grabenstein, J.D.; Klugman, K.P. A century of pneumococcal vaccination research in humans. Clin. Microbiol. Infect. 2012, 18 (Suppl. 5), 15–24. [Google Scholar] [CrossRef]
- Holmberg, L.A.; Sandmaier, B.M. Vaccination with Theratope (STn-KLH) as treatment for breast cancer. Expert. Rev. Vaccines 2004, 3, 655–663. [Google Scholar] [CrossRef]
- Franco, A. Glycoconjugates as vaccines for cancer immunotherapy: Clinical trials and future directions. Anticancer Agents Med. Chem. 2008, 8, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhou, Z.; Tang, S.; Guo, Z. Carbohydrate-monophosphoryl lipid a conjugates are fully synthetic self-adjuvanting cancer vaccines eliciting robust immune responses in the mouse. ACS Chem. Biol. 2012, 7, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wang, Q.; Zhang, J.; Wu, Q.; Guo, Z. Synthesis and Evaluation of Protein Conjugates of GM3 Derivatives Carrying Modified Sialic Acids as Highly Immunogenic Cancer Vaccine Candidates. Medchemcomm 2011, 2, 524–530. [Google Scholar] [CrossRef][Green Version]
- Oyelaran, O.; McShane, L.M.; Dodd, L.; Gildersleeve, J.C. Profiling human serum antibodies with a carbohydrate antigen microarray. J. Proteome Res. 2009, 8, 4301–4310. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, B.; Ma, Z.; Wei, M.; Liu, J.; Li, D.; Zhang, H.; Wang, P.G.; Chen, M. L-Rhamnose Enhances the Immunogenicity of Melanoma-Associated Antigen A3 for Stimulating Antitumor Immune Responses. Bioconjug. Chem. 2016, 27, 1112–1118. [Google Scholar] [CrossRef]
- Sarkar, S.; Lombardo, S.A.; Herner, D.N.; Talan, R.S.; Wall, K.A.; Sucheck, S.J. Synthesis of a single-molecule L-rhamnose-containing three-component vaccine and evaluation of antigenicity in the presence of anti-L-rhamnose antibodies. J. Am. Chem. Soc. 2010, 132, 17236–17246. [Google Scholar] [CrossRef]
- Sarkar, S.; Salyer, A.C.; Wall, K.A.; Sucheck, S.J. Synthesis and immunological evaluation of a MUC1 glycopeptide incorporated into L-rhamnose displaying liposomes. Bioconjug. Chem. 2013, 24, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Rao, X.; Cai, L.; Liu, X.; Wang, H.; Wu, W.; Zhu, C.; Chen, M.; Wang, P.G.; Yi, W. Targeting Tumor Cells by Natural Anti-Carbohydrate Antibodies Using Rhamnose-Functionalized Liposomes. ACS Chem. Biol. 2016, 11, 1205–1209. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.K.; Vartak, A.; Karmakar, P.; Sucheck, S.J.; Wall, K.A. Augmenting Vaccine Immunogenicity through the Use of Natural Human Anti-rhamnose Antibodies. ACS Chem. Biol. 2018, 13, 2130–2142. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Hong, H.; Wang, J.; Li, C.; Zhou, Z.; Wu, Z. Rhamnose modified bovine serum albumin as a carrier protein promotes the immune response against sTn antigen. Chem. Commun. 2020, 56, 13959–13962. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gu, L.; Zhang, W.; Motari, E.; Cai, L.; Styslinger, T.J.; Wang, P.G. L-rhamnose antigen: A promising alternative to alpha-gal for cancer immunotherapies. ACS Chem. Biol. 2011, 6, 185–191. [Google Scholar] [CrossRef]
- Ou, C.; Prabhu, S.K.; Zhang, X.; Zong, G.; Yang, Q.; Wang, L.X. Synthetic Antibody-Rhamnose Cluster Conjugates Show Potent Complement-Dependent Cell Killing by Recruiting Natural Antibodies. Chemistry 2022, 28, e202200146. [Google Scholar] [CrossRef]
- Zhou, K.; Hong, H.; Lin, H.; Gong, L.; Li, D.; Shi, J.; Zhou, Z.; Xu, F.; Wu, Z. Chemical Synthesis of Antibody-Hapten Conjugates Capable of Recruiting the Endogenous Antibody to Magnify the Fc Effector Immunity of Antibody for Cancer Immunotherapy. J. Med. Chem. 2022, 65, 323–332. [Google Scholar] [CrossRef]
- De Coen, R.; Nuhn, L.; Perera, C.; Arista-Romero, M.; Risseeuw, M.D.P.; Freyn, A.; Nachbagauer, R.; Albertazzi, L.; Van Calenbergh, S.; Spiegel, D.A.; et al. Synthetic Rhamnose Glycopolymer Cell-Surface Receptor for Endogenous Antibody Recruitment. Biomacromolecules 2020, 21, 793–802. [Google Scholar] [CrossRef]
- Hilinski, M.K.; Mrozowski, R.M.; Clark, D.E.; Lannigan, D.A. Analogs of the RSK inhibitor SL0101: Optimization of in vitro biological stability. Bioorg. Med. Chem. Lett. 2012, 22, 3244–3247. [Google Scholar] [CrossRef]
- Chun, J.; Ha, I.J.; Kim, Y.S. Antiproliferative and apoptotic activities of triterpenoid saponins from the roots of Platycodon grandiflorum and their structure-activity relationships. Planta Med. 2013, 79, 639–645. [Google Scholar] [CrossRef]
- Ha, I.J.; Kang, M.; Na, Y.C.; Park, Y.; Kim, Y.S. Preparative separation of minor saponins from Platycodi Radix by high-speed counter-current chromatography. J. Sep. Sci. 2011, 34, 2559–2565. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, J.; Gu, G.; Li, G.; Cui, C.; Sun, B.; Lou, H. In situ RBL receptor visualization and its mediated anticancer activity for solasodine rhamnosides. Chembiochem 2011, 12, 2418–2420. [Google Scholar] [CrossRef] [PubMed]
- Garnier, P.; Wang, X.T.; Robinson, M.A.; van Kasteren, S.; Perkins, A.C.; Frier, M.; Fairbanks, A.J.; Davis, B.G. Lectin-directed enzyme activated prodrug therapy (LEAPT): Synthesis and evaluation of rhamnose-capped prodrugs. J. Drug Target. 2010, 18, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Tomsik, P.; Stoklasova, A.; Micuda, S.; Niang, M.; Suba, P.; Knizek, J.; Rezacova, M. Evaluation of the antineoplastic activity of L-rhamnose in vitro. A comparison with 2-deoxyglucose. Acta Med. 2008, 51, 113–119. [Google Scholar]
- Ma, Y.; Pan, F.; McNeil, M. Formation of dTDP-rhamnose is essential for growth of mycobacteria. J. Bacteriol. 2002, 184, 3392–3395. [Google Scholar] [CrossRef]
- Xiao, G.; Alphey, M.S.; Tran, F.; Pirrie, L.; Milbeo, P.; Zhou, Y.; Bickel, J.K.; Kempf, O.; Kempf, K.; Naismith, J.H.; et al. Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design. Bioorg. Med. Chem. 2021, 50, 116477. [Google Scholar] [CrossRef]
- Alphey, M.S.; Pirrie, L.; Torrie, L.S.; Boulkeroua, W.A.; Gardiner, M.; Sarkar, A.; Maringer, M.; Oehlmann, W.; Brenk, R.; Scherman, M.S.; et al. Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa. ACS Chem. Biol. 2013, 8, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Loranger, M.W.; Forget, S.M.; McCormick, N.E.; Syvitski, R.T.; Jakeman, D.L. Synthesis and evaluation of L-rhamnose 1C-phosphonates as nucleotidylyltransferase inhibitors. J. Org. Chem. 2013, 78, 9822–9833. [Google Scholar] [CrossRef]
- Kantardjieff, K.A.; Kim, C.Y.; Naranjo, C.; Waldo, G.S.; Lekin, T.; Segelke, B.W.; Zemla, A.; Park, M.S.; Terwilliger, T.C.; Rupp, B. Mycobacterium tuberculosis RmlC epimerase (Rv3465): A promising drug-target structure in the rhamnose pathway. Acta Crystallogr. D Biol. Crystallogr. 2004, 60 Pt 5, 895–902. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).