Critical Quality Control Methods for a Novel Anticoagulant Candidate LFG-Na by HPSEC-MALLS-RID and Bioactivity Assays
Abstract
:1. Introduction
2. Results and Discussion
2.1. Determination of the Refractive Index Increment (dn/dc)
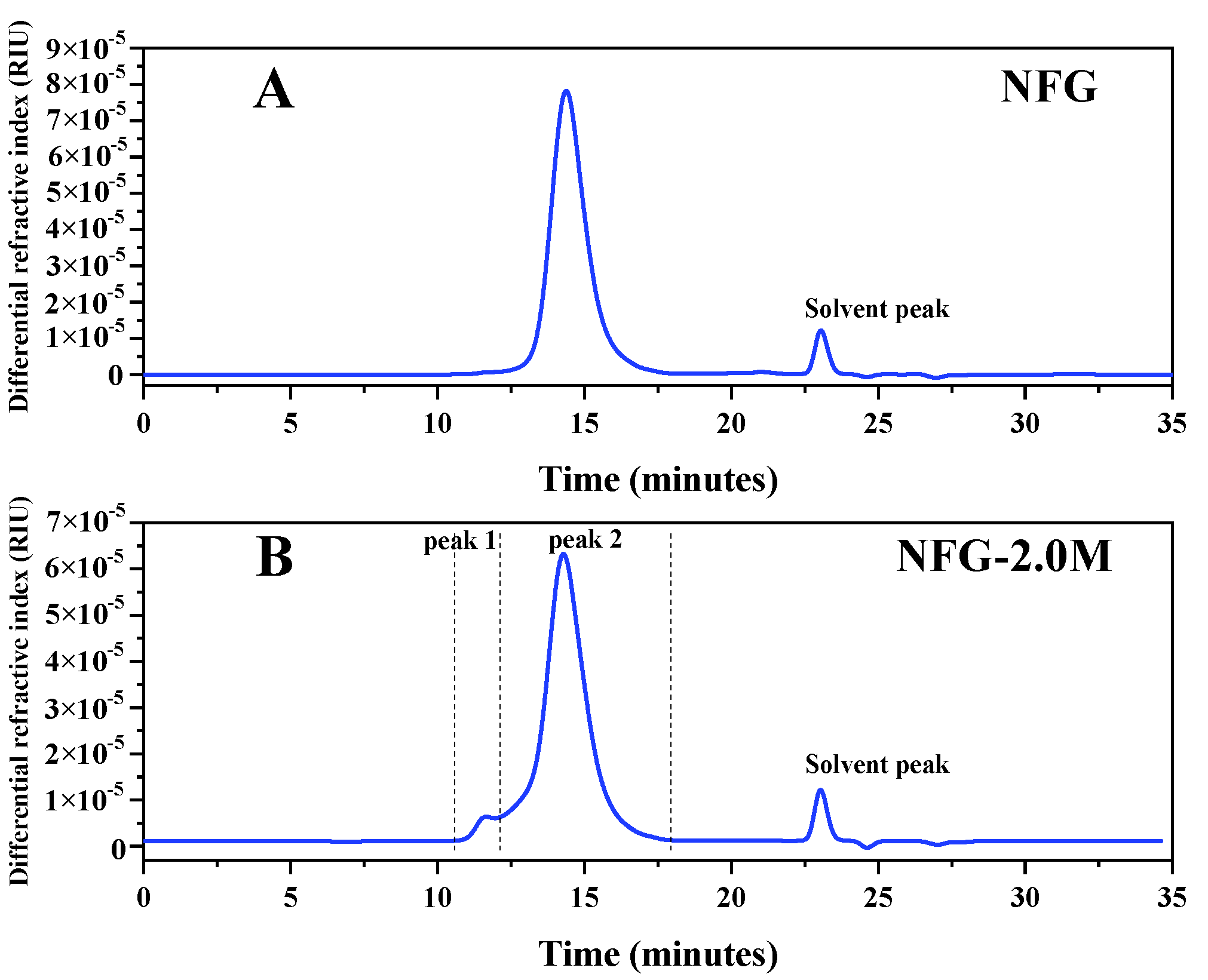
2.2. The Starting Material NFG, NFG-2.0M and Their Molecular Weights
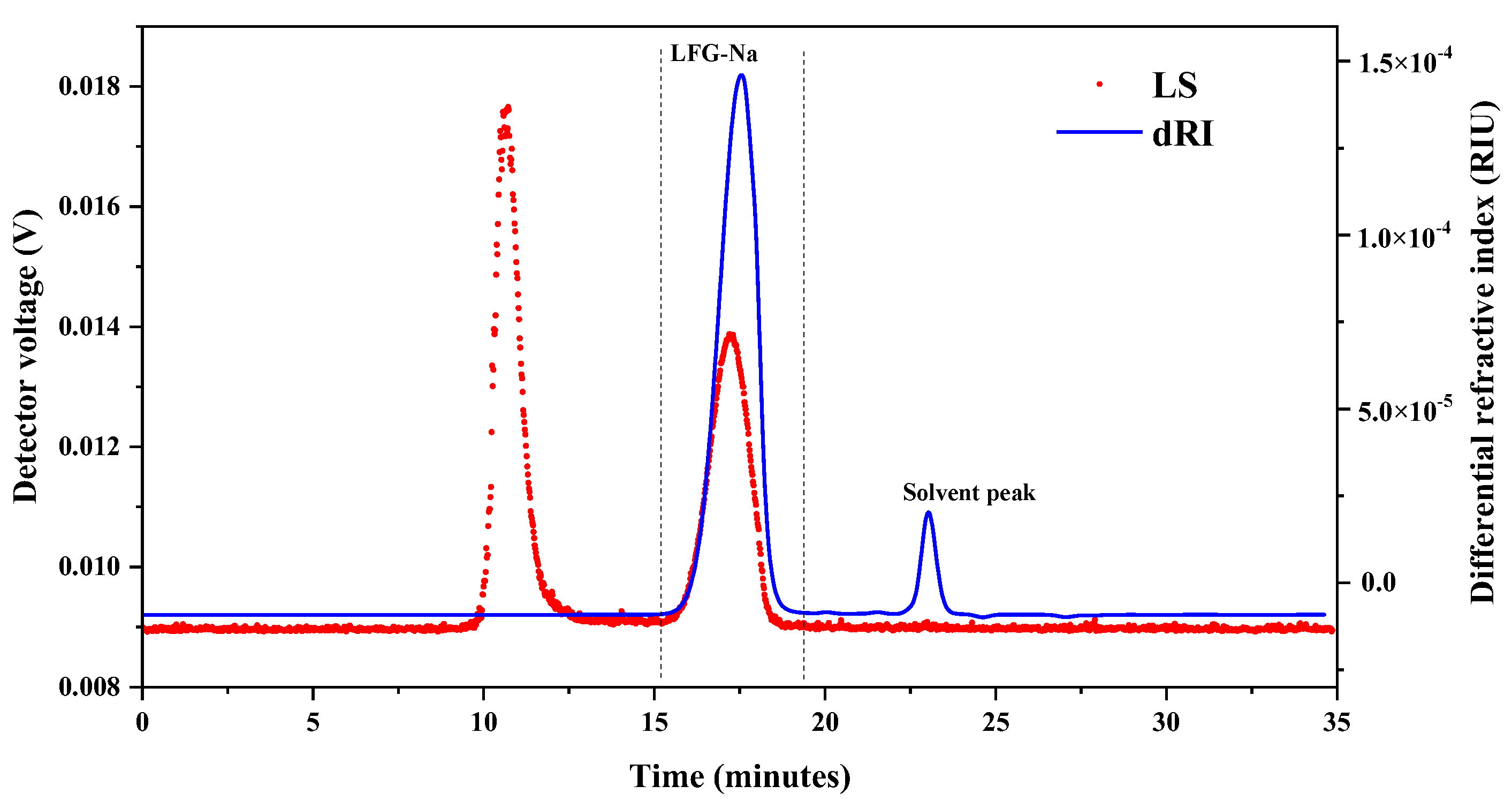
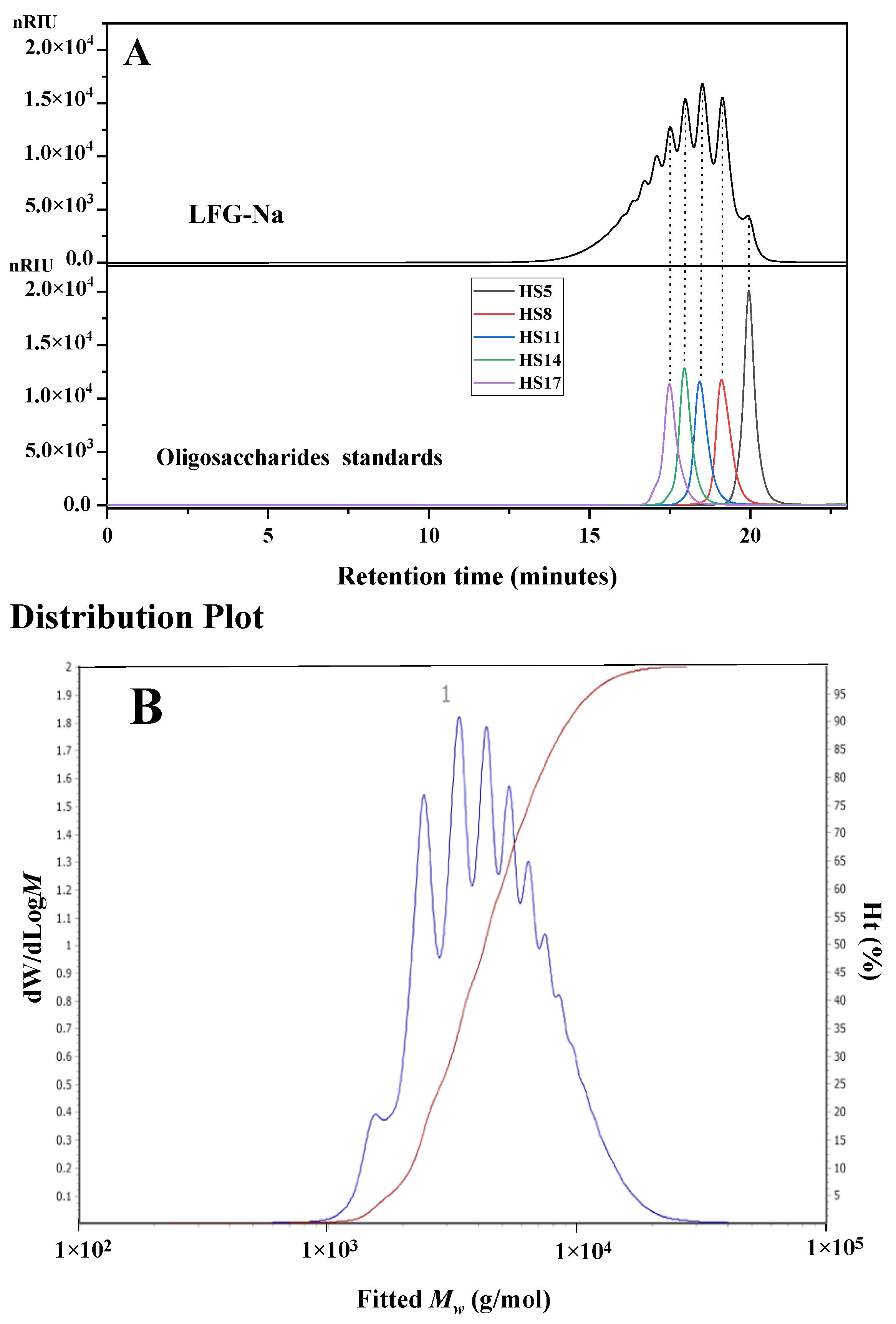
2.3. Determination of Molecular Weight and Molecular Weight Distribution of LFG-Na Using HPSEC-MALLS-RID
2.4. Determination of Molecular Weight and Molecular Weight Distribution of LFG-Na Using HPGPC
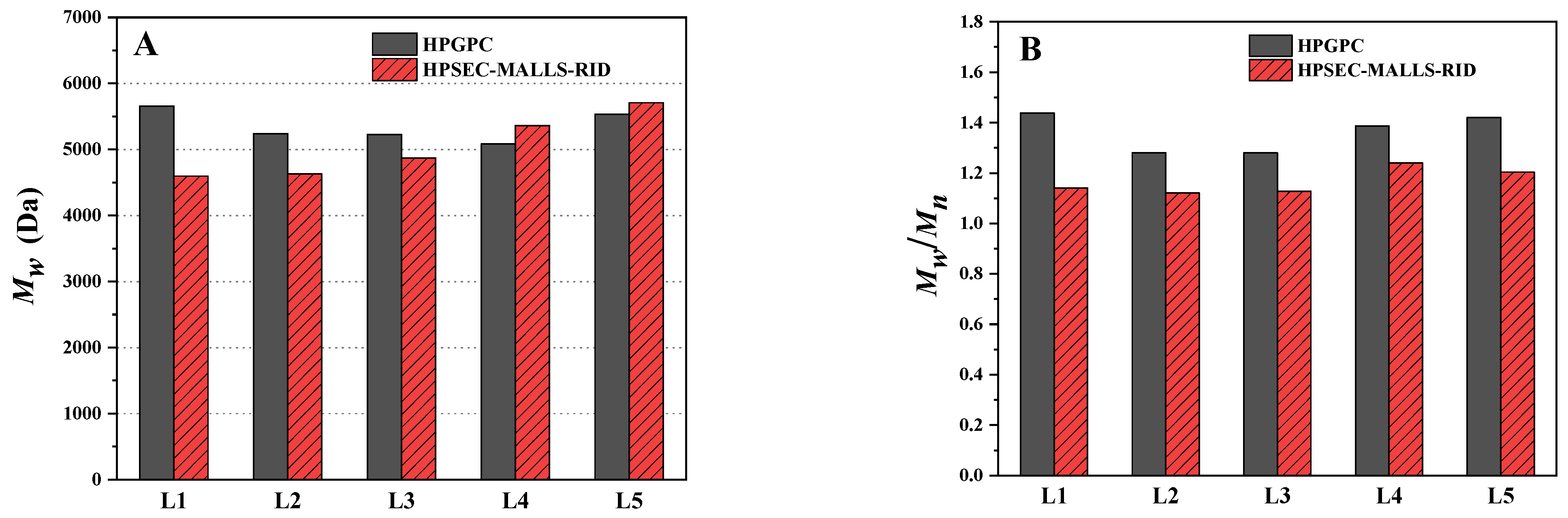
2.5. Comparison of the Determination Using HPGPC and HPSEC-MALLS-RID
2.6. Comparison of the Chemical Characteristics of LFG-Na
2.7. Comparison of Biological Potency
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Preparation of the Starting Material NFG
3.3. Preparation of LFG
3.4. HPSEC-MALLS-RID Analysis
3.4.1. dn/dc Measurement of NFG and LFG-Na
3.4.2. The Method of HPSEC-MALLS-RID
3.5. Determination of Molecular Weight and Homogeneity of LFG-Na by HPGPC
3.6. Chemical Characteristics and NMR Analysis of LFG-Na
3.7. Activity Assays of LFG-Na In Vitro
3.7.1. APTT Assay
3.7.2. Potency for Anti-iXase
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
References
- Mackman, N. Triggers, targets and treatments for thrombosis. Nature 2008, 451, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Timmis, A.; Townsend, N.; Gale, C.; Grobbee, R.; Maniadakis, N.; Flather, M.; Wilkins, E.; Wright, L.; Vos, R.; Bax, J.; et al. European Society of Cardiology: Cardiovascular Disease Statistics 2017. Eur. Heart J. 2018, 39, 508–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, P.; Gao, Y.; Zheng, M.; Xu, T.; Schoenhagen, P.; Jin, Z. Recent progress and market analysis of anticoagulant drugs. J. Thorac. Dis. 2018, 10, 2011–2025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearon, C.; Akl, E.A.; Comerota, A.J.; Prandoni, P.; Bounameaux, H.; Goldhaber, S.Z.; Nelson, M.E.; Wells, P.S.; Gould, M.K.; Dentali, F.; et al. Antithrombotic therapy for VTE disease: Antithrombotic Therapy and Prevention of Thrombosis, 9th ed: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines. Chest 2012, 141 (Suppl. S2), e419S–e496S. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, B.I.; Quinlan, D.J.; Eikelboom, J.W. Novel oral factor Xa and thrombin inhibitors in the management of thromboembolism. Annu. Rev. Med. 2011, 62, 41–57. [Google Scholar] [CrossRef]
- Fareed, J.; Thethi, I.; Hoppensteadt, D. Old versus new oral anticoagulants: Focus on pharmacology. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 79–99. [Google Scholar] [CrossRef]
- Lesko, L.J. Anticoagulants: What is new and what is the standard? Clin. Pharmacol. Ther. 2016, 100, 126–128. [Google Scholar] [CrossRef]
- Shoeb, M.; Fang, M.C. Assessing bleeding risk in patients taking anticoagulants. J. Thromb. Thrombolysis 2013, 35, 312–319. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Zhao, L.; Gao, N.; Yin, R.; Li, S.; Sun, H.; Zhou, L.; Zhao, G.; Purcell, S.W.; Zhao, J. From multi-target anticoagulants to DOACs, and intrinsic coagulation factor inhibitors. Blood Rev. 2020, 39, 100615. [Google Scholar] [CrossRef]
- Zhou, L.; Gao, N.; Sun, H.; Xiao, C.; Yang, L.; Lin, L.; Yin, R.; Li, Z.; Zhang, H.; Ji, X.; et al. Effects of Native Fucosylated Glycosaminoglycan, Its Depolymerized Derivatives on Intrinsic Factor Xase, Coagulation, Thrombosis, and Hemorrhagic Risk. Thromb. Haemost. 2020, 120, 607–619. [Google Scholar] [CrossRef]
- Ahmad, S.S.; Rawala-Sheikh, R.; Walsh, P.N. Components and assembly of the factor X activating complex. Semin. Thromb. Hemost. 1992, 18, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Gao, N.; Ren, L.; Liu, S.; Lin, L.; Zheng, W.; Zhou, L.; Yin, R.; Zhao, J. The components and activities analysis of a novel anticoagulant candidate dHG-5. Eur. J. Med. Chem. 2020, 207, 112796. [Google Scholar] [CrossRef] [PubMed]
- Pomin, V.H. Holothurian fucosylated chondroitin sulfate. Mar. Drugs 2014, 12, 232–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonseca, R.J.; Santos, G.R.; Mourao, P.A. Effects of polysaccharides enriched in 2,4-disulfated fucose units on coagulation, thrombosis and bleeding. Practical and conceptual implications. Thromb. Haemost. 2009, 102, 829–836. [Google Scholar] [CrossRef]
- Li, H.; Yuan, Q.; Lv, K.; Ma, H.; Gao, C.; Liu, Y.; Zhang, S.; Zhao, L. Low-molecular-weight fucosylated glycosaminoglycan and its oligosaccharides from sea cucumber as novel anticoagulants: A review. Carbohydr. Polym. 2021, 251, 117034. [Google Scholar] [CrossRef]
- Yin, R.; Zhou, L.; Gao, N.; Li, Z.; Zhao, L.; Shang, F.; Wu, M.; Zhao, J. Oligosaccharides from depolymerized fucosylated glycosaminoglycan: Structures and minimum size for intrinsic factor Xase complex inhibition. J. Biol. Chem. 2018, 293, 14089–14099. [Google Scholar] [CrossRef] [Green Version]
- Luo, L.; Wu, M.; Xu, L.; Lian, W.; Xiang, J.; Lu, F.; Gao, N.; Xiao, C.; Wang, S.; Zhao, J. Comparison of physicochemical characteristics and anticoagulant activities of polysaccharides from three sea cucumbers. Mar. Drugs 2013, 11, 399–417. [Google Scholar] [CrossRef] [Green Version]
- Li, S.-P.; Wu, D.-T.; Lv, G.-P.; Zhao, J. Carbohydrates analysis in herbal glycomics. TrAC Trends Anal. Chem. 2013, 52, 155–169. [Google Scholar] [CrossRef]
- Kim, C.; Deratani, A.; Bonfils, F. Determination of the Refractive Index Increment of Natural and Synthetic Poly(Cis-1,4-Isoprene) Solutions and Its Effect on Structural Parameters. J. Liq. Chromatogr. Relat. Technol. 2009, 33, 37–45. [Google Scholar] [CrossRef]
- Gao, N.; Chen, R.; Mou, R.; Xiang, J.; Zhou, K.; Li, Z.; Zhao, J. Purification, structural characterization and anticoagulant activities of four sulfated polysaccharides from sea cucumber Holothuria fuscopunctata. Int. J. Biol. Macromol. 2020, 164, 3421–3428. [Google Scholar] [CrossRef]
- Cheong, K.L.; Wu, D.T.; Zhao, J.; Li, S.P. A rapid and accurate method for the quantitative estimation of natural polysaccharides and their fractions using high performance size exclusion chromatography coupled with multi-angle laser light scattering and refractive index detector. J. Chromatogr. A 2015, 1400, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gong, X.; Wang, Z.; Pan, C.; Zhao, Y.; Gao, X.; Liu, W. Multiple fingerprint profiles and chemometrics analysis of polysaccharides from Sarcandra glabra. Int. J. Biol. Macromol. 2019, 123, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wen, D.; Gao, N.; Xiao, C.; Yang, L.; Xu, L.; Lian, W.; Peng, W.; Jiang, J.; Zhao, J. Anticoagulant and antithrombotic evaluation of native fucosylated chondroitin sulfates and their derivatives as selective inhibitors of intrinsic factor Xase. Eur. J. Med. Chem. 2015, 92, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Luo, L.; Cai, Y.; Yang, W.; Lin, L.; Li, Z.; Gao, N.; Purcell, S.W.; Wu, M.; Zhao, J. Structural Elucidation and Biological Activity of a Highly Regular Fucosylated Glycosaminoglycan from the Edible Sea Cucumber Stichopus herrmanni. J. Agric. Food. Chem. 2017, 65, 9315–9323. [Google Scholar] [CrossRef]
- Gao, N.; Lu, F.; Xiao, C.; Yang, L.; Chen, J.; Zhou, K.; Wen, D.; Li, Z.; Wu, M.; Jiang, J.; et al. beta-Eliminative depolymerization of the fucosylated chondroitin sulfate and anticoagulant activities of resulting fragments. Carbohydr. Polym. 2015, 127, 427–437. [Google Scholar] [CrossRef]
- Liu, J.; Shang, F.; Yang, Z.; Wu, M.; Zhao, J. Structural analysis of a homogeneous polysaccharide from Achatina fulica. Int. J. Biol. Macromol. 2017, 98, 786–792. [Google Scholar] [CrossRef]
- Shang, F.; Gao, N.; Yin, R.; Lin, L.; Xiao, C.; Zhou, L.; Li, Z.; Purcell, S.W.; Wu, M.; Zhao, J. Precise structures of fucosylated glycosaminoglycan and its oligosaccharides as novel intrinsic factor Xase inhibitors. Eur. J. Med. Chem. 2018, 148, 423–435. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, T.; Sun, H.; Lin, L.; Gao, N.; Wang, W.; Li, S.; Zhao, J. Pharmacokinetics and Pharmacodynamics of a Depolymerized Glycosaminoglycan from Holothuria fuscopunctata, a Novel Anticoagulant Candidate, in Rats by Bioanalytical Methods. Mar. Drugs 2021, 19, 212. [Google Scholar] [CrossRef]

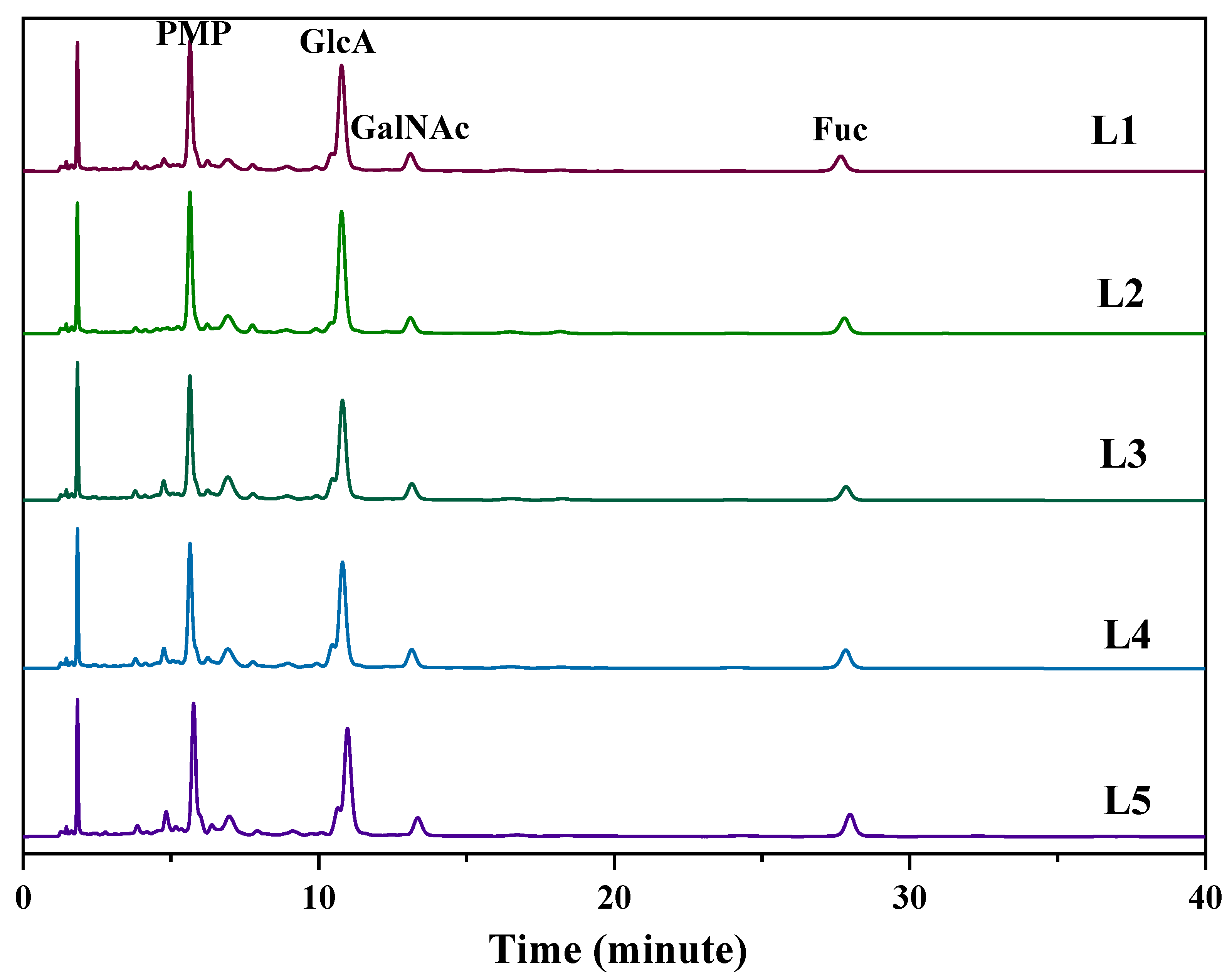
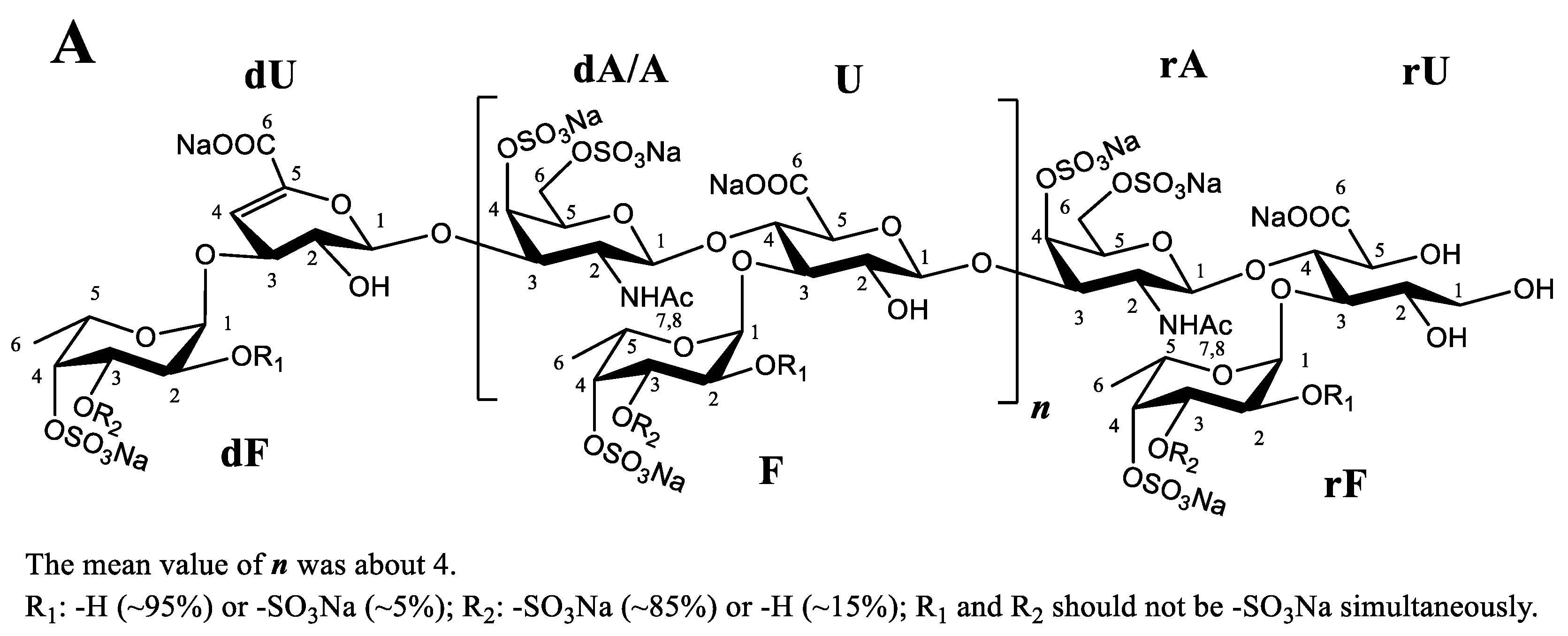
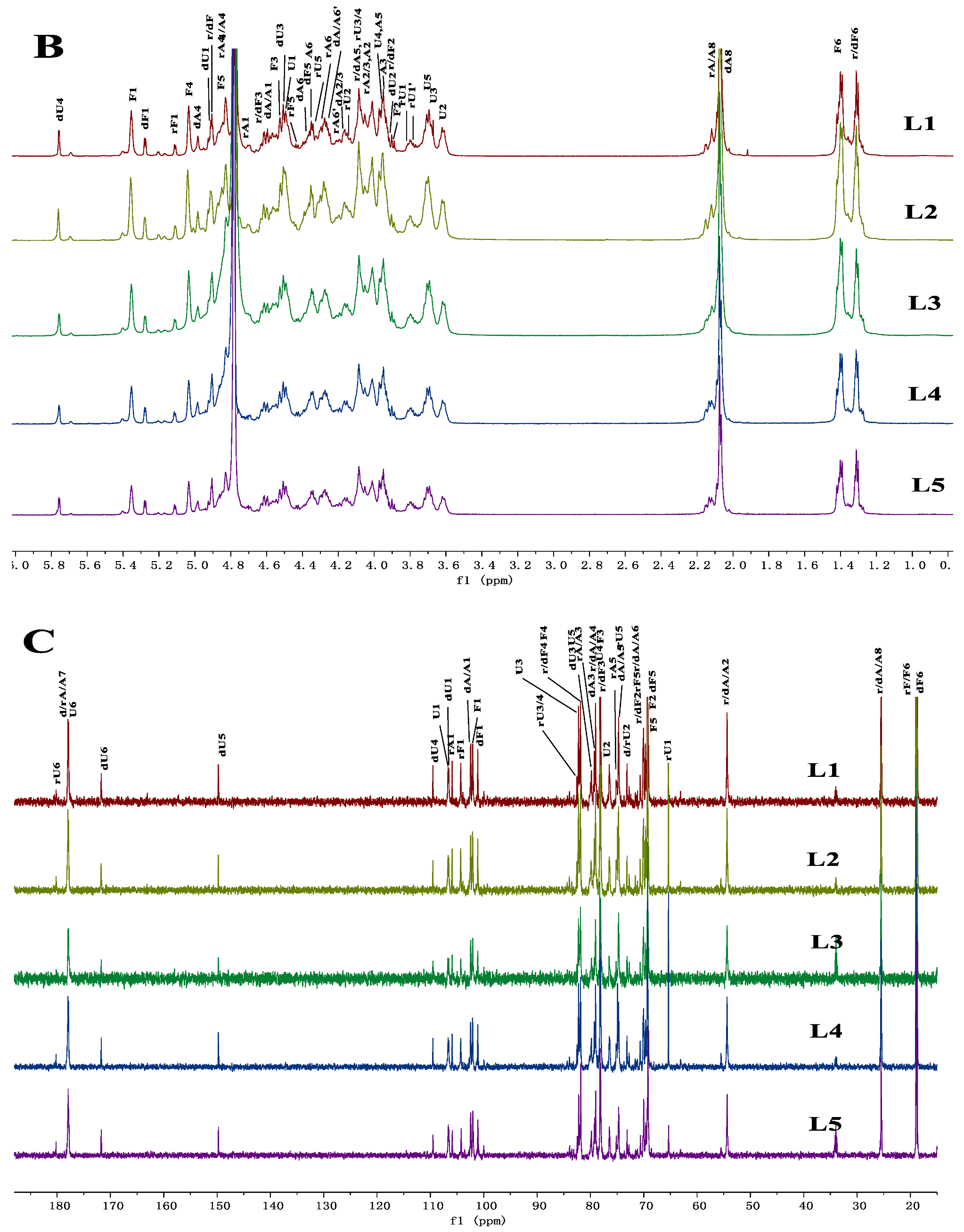
| Sample | Batch | dn/dc (mL/g) | Mean Value (mL/g) |
|---|---|---|---|
| LFG-Na | |||
| L1 | 0.1277 | 0.1248 ± 0.0046 | |
| L2 | 0.1269 | ||
| L3 | 0.1237 | ||
| L4 | 0.1283 | ||
| L5 | 0.1173 | ||
| NFG | |||
| N1 | 0.1168 | 0.1163 ± 0.0028 | |
| N2 | 0.1154 | ||
| N3 | 0.1187 | ||
| N4 | 0.1113 | ||
| N5 | 0.1162 | ||
| N6 | 0.1192 | ||
| NFG-2.0M | |||
| NM1 | 0.1162 | 0.1166 ± 0.0008 | |
| NM2 | 0.1172 | ||
| NM3 | 0.1157 | ||
| NM4 | 0.1173 |
| Sample | Batch | ||
|---|---|---|---|
| NFG | |||
| N1 | 60,000 | 1.184 | |
| N2 | 70,940 | 1.239 | |
| N3 | 61,350 | 1.180 | |
| N4 | 58,270 | 1.175 | |
| N5 | 74,280 | 1.241 | |
| N6 | 64,260 | 1.105 | |
| NFG-2.0M | |||
| NM1 | 84,840 | 1.407 | |
| NM2 | 86,950 | 1.360 | |
| NM3 | 77,680 | 1.297 | |
| NM4 | 84,320 | 1.360 |
| Batch | HPGPC | HPSEC-MALLS-RID | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Parameters | Molecular Weight Distribution | Molecular Parameters | Molecular Weight Distribution | |||||||
| M1500 | M1500~8000 | M8000 | M1500 | M1500~8000 | M8000 | |||||
| L1 | 5657 | 1.438 | 1.25% | 85.34% | 13.41% | 4596 (±0.546%) | 1.141 (±0.897%) | 0 | 94.192% | 5.808% |
| L2 | 5241 | 1.281 | 0.57% | 86.80% | 12.63% | 4629 (±0.519%) | 1.121 (±0.851%) | 0 | 94.539% | 5.461% |
| L3 | 5227 | 1.280 | 0.53% | 86.60% | 12.86% | 4870 (±0.583%) | 1.128 (±0.921%) | 0 | 93.024% | 6.976% |
| L4 | 5082 | 1.387 | 2.52% | 82.91% | 14.57% | 5361 (±0.479%) | 1.240 (±0.699%) | 0 | 87.329% | 12.671% |
| L5 | 5531 | 1.420 | 1.91% | 81.48% | 16.61% | 5708 (±0.541%) | 1.204 (±0.854%) | 0 | 85.055% | 14.945% |
| Sugar Ring | H | δa | 1H-1H Couplings b | Correlated Signals | C | δ | Correlated Signals | |||
|---|---|---|---|---|---|---|---|---|---|---|
| COSY | TOCSY | ROESY | HSQC | HMBC | ||||||
| rU | H-1 | 3.800 | J(1,1′) = 11.33 | H1′, H2 | H1′, H2, H3/4 | H2, H3 | C-1 | 65.40 | H-1 | H-1 |
| H-1′ | 3.761 | J(1,2) = 6.12 | H1, H2 | H1, H2, H3/4 | H3 | H-1′ | H-1′ | |||
| H-2 | 4.146 | J(1′,2) = 3.96 | H1/1′, H3 | H1/1′, H3/4 | H1/1′, H3 | C-2 | 72.67 | H-2 | -- | |
| H-3 | 4.072 | -- | H2 | H1/1′, H2 | H2; rF1 | C-3 | 82.54 | H-3 | -- | |
| H-4 | 4.052 | -- | H5 | H1/1′, H2 | -- | C-4 | 82.54 | H-4 | -- | |
| H-5 | 4.329 | -- | H4 | H4 | -- | C-5 | 74.78 | H-5 | -- | |
| -- | -- | -- | -- | -- | -- | C-6 | 180.22 | -- | H5 | |
| rF | H-1 | 5.116 | J(1,2) = 4.02 | H2 | H2, H3, H4 | H2; rU3 | C-1 | 104.33 | H-1 | H1, H5; rU3 |
| H-2 | 3.885 | -- | H1, H3 | H1, H3, H4 | H1, H3, H4 | C-2 | 69.36 | H-2 | H2, H3, H4 | |
| H-3 | 4.610 | J(3,4) =2.94 | H2, H4 | H1, H2, H4 | H2, H3, H4, H5 | C-3 | 78.10 | H-3 | H1, H2, H4 | |
| H-4 | 4.909 | -- | H3 | H1, H2, H3 | H3, H5, H6 | C-4 | 81.83 | H-4 | H3, H5 | |
| H-5 | 4.428 | -- | H6 | H6 | H3, H4, H6 | C-5 | 69.71 | H-5 | H1, H4 | |
| H-6 | 1.359 | -- | H5 | H5 | H4, H5 | C-6 | 18.94 | H-6 | H4, H5, H6 | |
| rA | H-1 | 4.695 | J(1,2) = 7.04 | H2 | H2, H3 | H2, H3, H5; rU4 | C-1 | 104.33 | H-1 | rU4 |
| H-2 | 3.974 | -- | H1, H3 | H1, H3, H4 | H3, H1 | C-2 | 54.22 | H-2 | H3, H4 | |
| H-3 | 4.173 | -- | H2, H4 | H1, H2, H4 | H1, H2, H4; U1 | C-3 | 78.74 | H-3 | H1, H4; U1 | |
| H-4 | 4.829 | -- | H3 | H2, H3, H5 | H3, H5, H6 | C-4 | 79.25 | H-4 | H3, H5, H6′ | |
| H-5 | 4.092 | -- | H6, H6′ | H4, H6/6′ | H1, H4, H6/6′ | C-5 | 75.06 | H-5 | H6/6′ | |
| H-6 | 4.276 | -- | H5, H6′ | H5, H6′ | H4, H5, H6′ | C-6 | 70.52 | H-6/6′ | H5 | |
| H-6′ | 4.199 | -- | H5, H6 | H5, H6 | H4, H5, H6 | C-7 | 177.97 | -- | H2, H8 | |
| H-8 | 2.090 | -- | -- | -- | -- | C-8 | 25.26 | H-8 | H8 | |
| U | H-1 | 4.491 | J(1,2) = 8.88 | H2 | H2, H3, H4, H5 | H2, H3, H5; rA/A3 | C-1 | 106.71 | H-1 | H2, H3; A3 |
| H-2 | 3.616 | J(2,3) = 7.74 | H1, H3 | H1, H3, H4, H5 | H4, H1 | C-2 | 76.45 | H-2 | H2, H3 | |
| H-3 | 3.690 | -- | H2, H4 | H1, H2, H4, H5 | H1, H2, H4; F1 | C-3 | 82.00 | H-3 | H2, H4, H5; F1 | |
| H-4 | 4.021 | -- | H3, H5 | H1, H2, H3, H5 | H2, H3/5; A1 | C-4 | 78.07 | H-4 | H3, H5 | |
| H-5 | 3.706 | -- | H4 | H1, H2, H3, H4 | H1, H3 | C-5 | 79.84 | H-5 | H4, H5 | |
| -- | -- | -- | -- | -- | -- | C-6 | 177.87 | -- | H4, H5 | |
| A | H-1 | 4.485 | J(1,2) = 8.52 | H2 | H2, H3 | H2, H3, H5; U4 | C-1 | 102.34 | H-1 | H2; U4 |
| H-2 | 4.021 | -- | H1, H3 | H1, H3, H4 | H3, H1 | C-2 | 54.24 | H-2 | H3, H4 | |
| H-3 | 3.895 | -- | H2, H4 | H1, H2, H4 | H1, H2, H4; U1 | C-3 | 79.11 | H-3 | H1, H4; U1 | |
| H-4 | 4.751 | -- | H3 | H2, H3, H5 | H3, H5, H6 | C-4 | 78.90 | H-4 | H3, H5, H6′ | |
| H-5 | 3.932 | -- | H6, H6′ | H4, H6/6′ | H1, H4, H6/6′ | C-5 | 74.62 | H-5 | H6/6′ | |
| H-6 | 4.287 | -- | H5, H6′ | H5, H6′ | H4, H5, H6′ | C-6 | 69.88 | H-6/6′ | H5 | |
| H-6′ | 4.183 | -- | H5, H6 | H5, H6 | H4, H5, H6 | C-7 | 177.84 | -- | H2, H8 | |
| H-8 | 1.998 | -- | -- | -- | -- | C-8 | 25.38 | H-8 | H8 | |
| F | H-1 | 5.356 | J(1,2) = 3.72 | H2 | H2, H3, H4 | H2, H3, H4, H5; U3 | C-1 | 102.06 | H-1 | H3; U3 |
| H-2 | 3.950 | J(2,3) = 10.08 | H1, H3 | H1, H3, H4 | H1, H3, H4 | C-2 | 69.24 | H-2 | H2, H3, H4 | |
| H-3 | 4.521 | -- | H2, H4 | H1, H2, H4 | H2, H3, H4, H5 | C-3 | 78.28 | H-3 | H1, H2, H4 | |
| H-4 | 5.040 | -- | H3 | H1, H2, H3 | H3, H5, H6 | C-4 | 82.20 | H-4 | H3, H5 | |
| H-5 | 4.849 | -- | H6 | H6 | H3, H4, H6 | C-5 | 69.28 | H-5 | H1, H4 | |
| H-6 | 1.404 | -- | H5 | H5 | H4, H5 | C-6 | 18.89 | H-6 | H4, H5, H6 | |
| dA | H-1 | 4.571 | J(1,2) = 8.52 | H2 | H2, H3 | H2, H3, H5, U4 | C-1 | 102.51 | H-1 | H2; U4 |
| H-2 | 4.163 | -- | H1, H3 | H1, H3, H4 | H3, H1 | C-2 | 54.37 | H-2 | H3, H4 | |
| H-3 | 4.166 | -- | H2, H4 | H1, H2, H4 | H1, H2, H4, dA1 | C-3 | 78.69 | H-3 | H1, H4; dU1 | |
| H-4 | 4.988 | -- | H3 | H2, H3, H5 | H3, H5, H6 | C-4 | 78.91 | H-4 | H3, H5, H6′ | |
| H-5 | 4.088 | -- | H6, H6′ | H4, H6/6′ | H1, H4, H6/6′ | C-5 | 74.99 | H-5 | H6/6′ | |
| H-6 | 4.371 | -- | H5, H6′ | H5, H6′ | H4, H5, H6′ | C-6 | 70.16 | H-6/6′ | H5 | |
| H-6′ | 4.263 | -- | H5, H6 | H5, H6 | H4, H5, H6 | C-7 | 178.04 | -- | H2, H8 | |
| H-8 | 2.065 | -- | -- | -- | -- | C-8 | 25.38 | H-8 | H8 | |
| ∆U | H-1 | 4.922 | J(1,2) = 7.20 | H2 | H2, H3, H4 | H1, H3, H4, dA3 | C-1 | 105.91 | H-1 | H1, H2; dA3 |
| H-2 | 3.911 | J(2,3) = 8.22 | H1, H3 | H1, H3, H4 | H3 | C-2 | 73.10 | H-2 | H2, H3, H4 | |
| H-3 | 4.491 | -- | H2, H4 | H1, H2, H4 | H1, H2, H4, dF1 | C-3 | 79.90 | H-3 | H2; dF1 | |
| H-4 | 5.760 | -- | H3 | H1, H2, H3 | H1, H3 | C-4 | 109.54 | H-4 | H3, H4 | |
| -- | -- | -- | -- | -- | -- | C-5 | 149.79 | -- | H4, H3 | |
| -- | -- | -- | -- | -- | -- | C-6 | 171.76 | -- | H4 | |
| dF | H-1 | 5.282 | J(1,2) = 3.96 | H2 | H2, H3, H4 | H2, dU3 | C-1 | 101.15 | H-1 | H5; dU3 |
| H-2 | 3.951 | J(2,3) = 10.88 | H1, H3 | H1, H3, H4 | H1, H3, H4 | C-2 | 69.24 | H-2 | H2, H3, H4 | |
| H-3 | 4.610 | J(3,4) = 3.06 | H2, H4 | H1, H2, H4 | H2, H3, H4, H5 | C-3 | 78.10 | H-3 | H1, H2, H4 | |
| H-4 | 4.909 | -- | H3 | H1, H2, H3 | H3, H5, H6 | C-4 | 81.83 | H-4 | H3, H5 | |
| H-5 | 4.356 | -- | H6 | H6 | H3, H4, H6 | C-5 | 70.03 | H-5 | H1, H4 | |
| H-6 | 1.301 | -- | H5 | H5 | H4, H5 | C-6 | 18.71 | H-6 | H4, H5, H6 | |
| Batch | APTT b (μg/mL) | Anti-iXase c (U/mg) | |
|---|---|---|---|
| L1 | 4596 | 8.89 ± 0.08 | 88.7 |
| L2 | 4629 | 8.84 ± 0.03 | 85.9 |
| L3 | 4870 | 8.64 ± 0.06 | 89.8 |
| L4 | 5361 | 7.92 ± 0.14 | 94.1 |
| L5 | 5708 | 7.36 ± 0.11 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, S.; Wang, Y.; Wu, J.; Wang, S.; Wei, H.; Zhang, Y.; Zhou, J.; Shi, Y. Critical Quality Control Methods for a Novel Anticoagulant Candidate LFG-Na by HPSEC-MALLS-RID and Bioactivity Assays. Molecules 2022, 27, 4522. https://doi.org/10.3390/molecules27144522
Zheng S, Wang Y, Wu J, Wang S, Wei H, Zhang Y, Zhou J, Shi Y. Critical Quality Control Methods for a Novel Anticoagulant Candidate LFG-Na by HPSEC-MALLS-RID and Bioactivity Assays. Molecules. 2022; 27(14):4522. https://doi.org/10.3390/molecules27144522
Chicago/Turabian StyleZheng, Shunliang, Yi Wang, Jiashuo Wu, Siyao Wang, Huaifu Wei, Yongchun Zhang, Jianbo Zhou, and Yue Shi. 2022. "Critical Quality Control Methods for a Novel Anticoagulant Candidate LFG-Na by HPSEC-MALLS-RID and Bioactivity Assays" Molecules 27, no. 14: 4522. https://doi.org/10.3390/molecules27144522
APA StyleZheng, S., Wang, Y., Wu, J., Wang, S., Wei, H., Zhang, Y., Zhou, J., & Shi, Y. (2022). Critical Quality Control Methods for a Novel Anticoagulant Candidate LFG-Na by HPSEC-MALLS-RID and Bioactivity Assays. Molecules, 27(14), 4522. https://doi.org/10.3390/molecules27144522






