Staphylococcus epidermidis Cicaria, a Novel Strain Derived from the Human Microbiome, and Its Efficacy as a Treatment for Hair Loss
Abstract
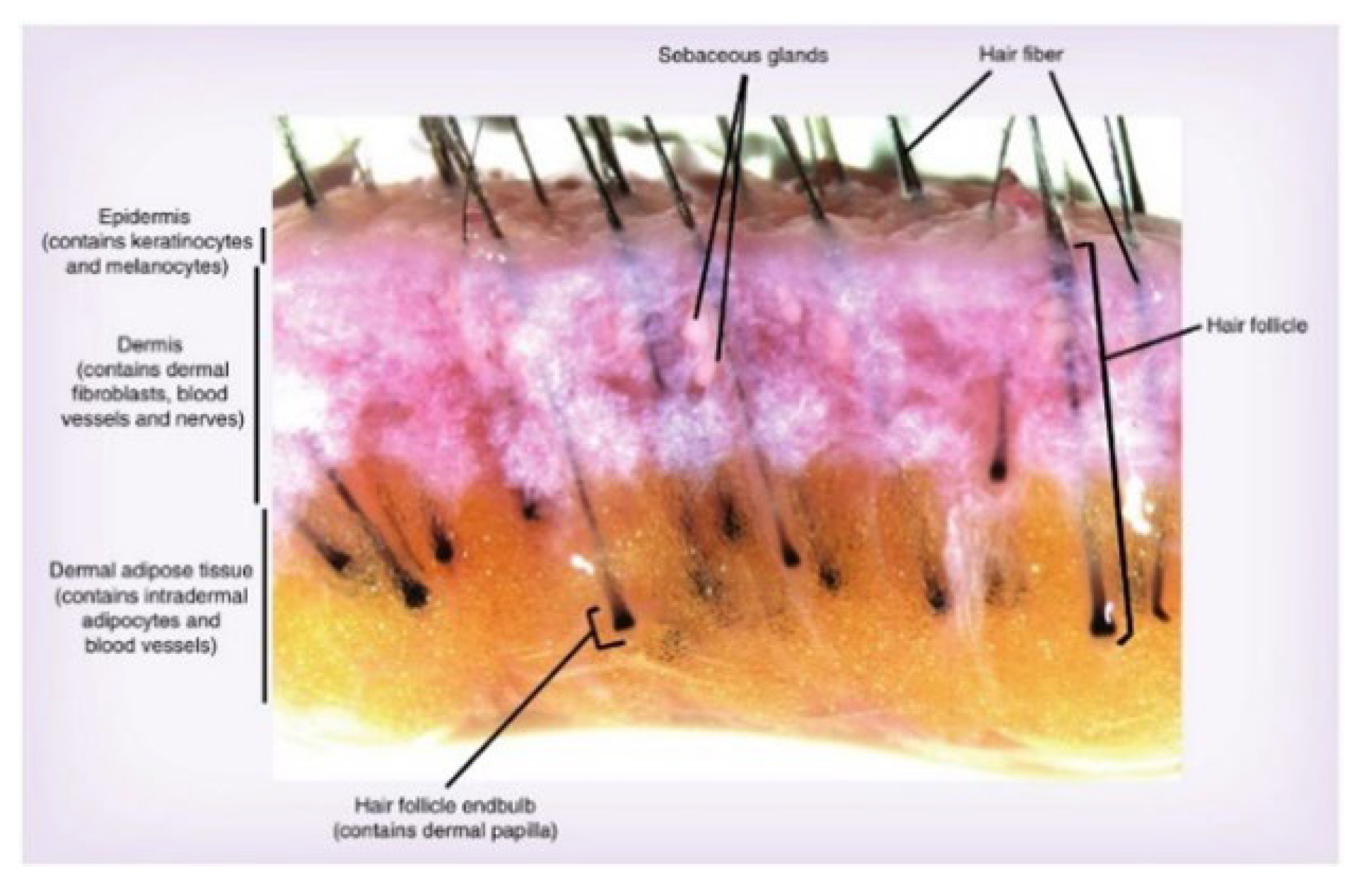
:1. Introduction
2. Results and Discussion
2.1. Scalp Microbiome Analysis of Subjects with and without Hair Loss
2.2. Selection of Target Strains of Hair Loss Suppression
2.2.1. Separation and Identification of Bacterial Strains
2.2.2. Isolated Bacterial Strains That Promote Epidermal Cell Growth Factor
2.3. Microbiome Analysis of before/after Clinical Trial of Shamppo Containing of Cicaria supernant
2.4. Identification and Analysis of Active Components in Cicaria
2.5. Evaluation of the Efficacy of Fractions
2.5.1. Analysis of Hair Follicle Length
2.5.2. Analysis of Hair Root Diameter
2.5.3. Analysis of Hair Follicle Growth Cycle Analysis
3. Materials and Methods
3.1. Recruitment of Subjects for Scalp Microbiome Analysis
3.2. Microbiome Analysis
3.3. Separation and Identification of Strains Involved in Hair Loss Suppression
3.4. Morphological Analysis of Cicaria
3.5. Assessment of Proliferation of Human Follicle Derma Papilla Cells
3.6. Evaluation of Growth Factor (VEGF, FGF7) Expression in Human Papilla Cells
3.7. Clinical Trial of Cicaria Supernant
3.8. Isolation of Active Components of Cicaria
3.9. Evaluation of the Fraction Efficacy
3.10. Culturing Human Papilla Cells and Analysis of Hair Growth Factors
3.11. Human Fibroblast Culture and Wound Recovery Analysis
3.12. Isolation and Cultivation of Hair Follicles
3.13. Measurement of Hair Follicle Length and Hair Root Diameter
3.14. Evaluation of Hair Follicle Growth Cycle
3.15. Statistical Analysis
- Rate of change in hair follicle length (%) = (Hair follicle length at each time point/Average hair follicle length at day 0) × 100—Average percentage of hair follicle length at day 0;
- Rate of change in hair root diameter (%) = (Hair root diameter at each time point/Average hair root diameter at day 0) × 100—Average percentage of hair root diameter at day 0;
- Rate of hair follicle growth cycle (%) = (Number of hairs in catagen/Total number of hairs) × 100.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marks, D.H.; Penzi, L.R.; Ibler, E.; Manatis-Lornell, A.; Hagigeorges, D.; Yasuda, M.; Drake, L.A.; Senna, M.M. The Medical and Psychosocial Associations of Alopecia: Recognizing Hair Loss as More Than a Cosmetic Concern. Am. J. Clin. Dermatol. 2019, 20, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-Y.; Hong, C.-H.; Lee, S.-J.; Sun, S.-H.; Lee, C.-M. The Efficacy and Safety Human Study of Narasoo Healing Shampoo and Hair Tonic for Scalp’s Lipids & Moisture and Hair Growth -One Center, one group pre-post comparison Pilot Human study. J. Korean Med. Ophthalmol. Otolaryngol. Dermatol. 2014, 27, 56–71. [Google Scholar]
- Loriaux, D.L.; Menard, R.; Taylor, A.; Pita, J.C.; Santen, R. Spironolactone and Endocrine Dysfunction. Ann. Intern. Med. 1976, 85, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A. The intersection of microbiome and host at the skin interface: Genomic- and metagenomic-based insights. Genome Res. 2015, 25, 1514–1520. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D.; Murray, P.R.; et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012, 22, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Paulino, L.C.; Tseng, C.-H.; Strober, B.E.; Blaser, M.J. Molecular Analysis of Fungal Microbiota in Samples from Healthy Human Skin and Psoriatic Lesions. J. Clin. Microbiol. 2006, 44, 2933–2941. [Google Scholar] [CrossRef] [PubMed]
- Scharschmidt, T.C.; Fischbach, M.A. What lives on our skin: Ecology, genomics and therapeutic opportunities of the skin microbiome. Drug Discov. Today Dis. Mech. 2013, 10, e83–e89. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Segre, J.A. Dialogue between skin microbiota and immunity. Science 2014, 346, 954–959. [Google Scholar] [CrossRef] [PubMed]
- Siracusa, M.C.; Saenz, S.A.; Hill, D.; Kim, B.S.; Headley, M.; Doering, T.A.; Wherry, E.J.; Jessup, H.K.; Siegel, L.A.; Kambayashi, T.; et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature 2011, 477, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Korn, T.; Bettelli, E.; Oukka, M.; Kuchroo, V.K. IL-17 and Th17 cells. Annu. Rev. Immunol. 2009, 27, 485–517. [Google Scholar] [CrossRef] [PubMed]
- Cua, D.J.; Tato, C.M. Innate IL-17-producing cells: The sentinels of the immune system. Nat. Rev. Immunol. 2010, 10, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Harkey, M. Anatomy and physiology of hair. Forensic Sci. Int. 1993, 63, 9–18. [Google Scholar] [CrossRef]
- Lin, Q.; Panchamukhi, A.; Li, P.; Shan, W.; Zhou, H.; Hou, L.; Chen, W. Malassezia and Staphylococcus dominate scalp microbiome for seborrheic dermatitis. Bioprocess Biosyst. Eng. 2021, 44, 965–975. [Google Scholar] [CrossRef] [PubMed]







| Group | Test Material | Concentration | Day 3 | Day 6 | Day 9 | Day 12 | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 Strand | 2 Rate (%) | Strand | Rate (%) | Strand | Rate (%) | Strand | Rate (%) | |||
| Normal | - | - | 2/9 | 22.22 | 6/9 | 66.67 | 6/9 | 66.67 | 7/9 | 77.78 |
| Positive control | Minoxidil | 0.2 ppm | 0/9 | 0.00 | 5/9 | 55.56 | 6/9 | 66.67 | 6/9 | 66.67 |
| Indicate material | Adenosine (Aldrich) | 25 ppm | 0/9 | 0.00 | 3/9 | 33.33 | 7/9 | 77.78 | 8/9 | 88.89 |
| Test material | Microbiome Adenosine | 5 ppm | 0/9 | 0.00 | 2/9 | 22.22 | 7/9 | 77.78 | 8/9 | 88.89 |
| 25 ppm | 2/9 | 22.22 | 4/9 | 44.45 | 5/9 | 55.56 | 5/9 | 55.56 | ||
| 100 ppm | 3/9 | 33.33 | 4/9 | 44.45 | 5/9 | 66.67 | 7/9 | 77.78 | ||
| Microbiome Biotin | 0.5 ppm | 0/9 | 0.00 | 6/9 | 66.67 | 6/9 | 66.67 | 7/9 | 77.78 | |
| Microbiome Culture solution | 5 ppm | 0/9 | 0.00 | 5/9 | 55.56 | 5/9 | 55.56 | 6/9 | 66.67 | |
| Gene Symbol | F/R | Sequence |
|---|---|---|
| VEGF | F | 5′-GTGCCCACTGAGGAGTTCAAC-3′ |
| R | 5′-CCCTATGTGCTGGCCTTGAT-3′ | |
| FGF7 | F | 5′-TCCTGCCAACTTTGCTCTACA-3′ |
| R | 5′-CAGGGCTGGAACAGTTCACAT-3′ | |
| β-actin | F | 5′-GGCCATCTCTTGCTCGAAGT-3′ |
| R | 5′-GAGACCTTCAACACCCCAGC-3′ |
| No | Component | Ratio (w/w) |
|---|---|---|
| 1 | CICARIA-W | 39.05 |
| 2 | Polyquaternium-10 | 0.2 |
| 3 | Sodium Citrate | 0.1 |
| 4 | Sodium Cocoyl Alaninate (solution) | 40 |
| 5 | Lauryl Hydroxysultaine (solution) | 9 |
| 6 | Coco-Betaine (solution) | 9 |
| 7 | Lauramide MIPA | 0.8 |
| 8 | Aspartic Acid | 0.5 |
| 9 | Caprylyl Glycol | 0.3 |
| 10 | Glyceryl Caprylate | 0.3 |
| 11 | Polyquaternium-22 | 0.5 |
| 12 | Citric Acid | 0.25 |
| Component | Contents (%) |
|---|---|
| S. epidermidis Cicaria supernant | 93.8 |
| Water | 4.0 |
| 1,2-Hexandiol | 2.0 |
| Biotin | 0.1 |
| Arginine | 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, H.; Kim, S.Y.; Kang, B.H.; Baek, C.; Kwon, J.E.; Jeang, J.W.; Heo, Y.M.; Kim, H.-B.; Heo, C.Y.; Kang, S.M.; et al. Staphylococcus epidermidis Cicaria, a Novel Strain Derived from the Human Microbiome, and Its Efficacy as a Treatment for Hair Loss. Molecules 2022, 27, 5136. https://doi.org/10.3390/molecules27165136
Jo H, Kim SY, Kang BH, Baek C, Kwon JE, Jeang JW, Heo YM, Kim H-B, Heo CY, Kang SM, et al. Staphylococcus epidermidis Cicaria, a Novel Strain Derived from the Human Microbiome, and Its Efficacy as a Treatment for Hair Loss. Molecules. 2022; 27(16):5136. https://doi.org/10.3390/molecules27165136
Chicago/Turabian StyleJo, HyungWoo, Seon Yu Kim, Byung Ha Kang, Chaeyun Baek, Jeong Eun Kwon, Jin Woo Jeang, Young Mok Heo, Hye-Been Kim, Chan Yeong Heo, So Min Kang, and et al. 2022. "Staphylococcus epidermidis Cicaria, a Novel Strain Derived from the Human Microbiome, and Its Efficacy as a Treatment for Hair Loss" Molecules 27, no. 16: 5136. https://doi.org/10.3390/molecules27165136
APA StyleJo, H., Kim, S. Y., Kang, B. H., Baek, C., Kwon, J. E., Jeang, J. W., Heo, Y. M., Kim, H.-B., Heo, C. Y., Kang, S. M., Shin, B. H., Nam, D. Y., Lee, Y.-G., Kang, S. C., & Lee, D.-G. (2022). Staphylococcus epidermidis Cicaria, a Novel Strain Derived from the Human Microbiome, and Its Efficacy as a Treatment for Hair Loss. Molecules, 27(16), 5136. https://doi.org/10.3390/molecules27165136








