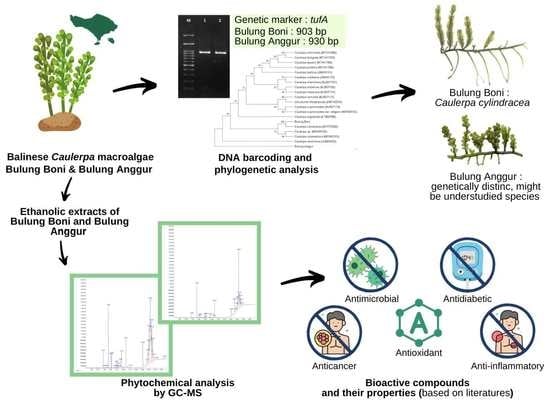
Phytochemical Analysis and Molecular Identification of Green Macroalgae Caulerpa spp. from Bali, Indonesia
Abstract
:1. Introduction
2. Results and Discussion
2.1. Morphological Characterization
2.2. Phytochemical Analysis
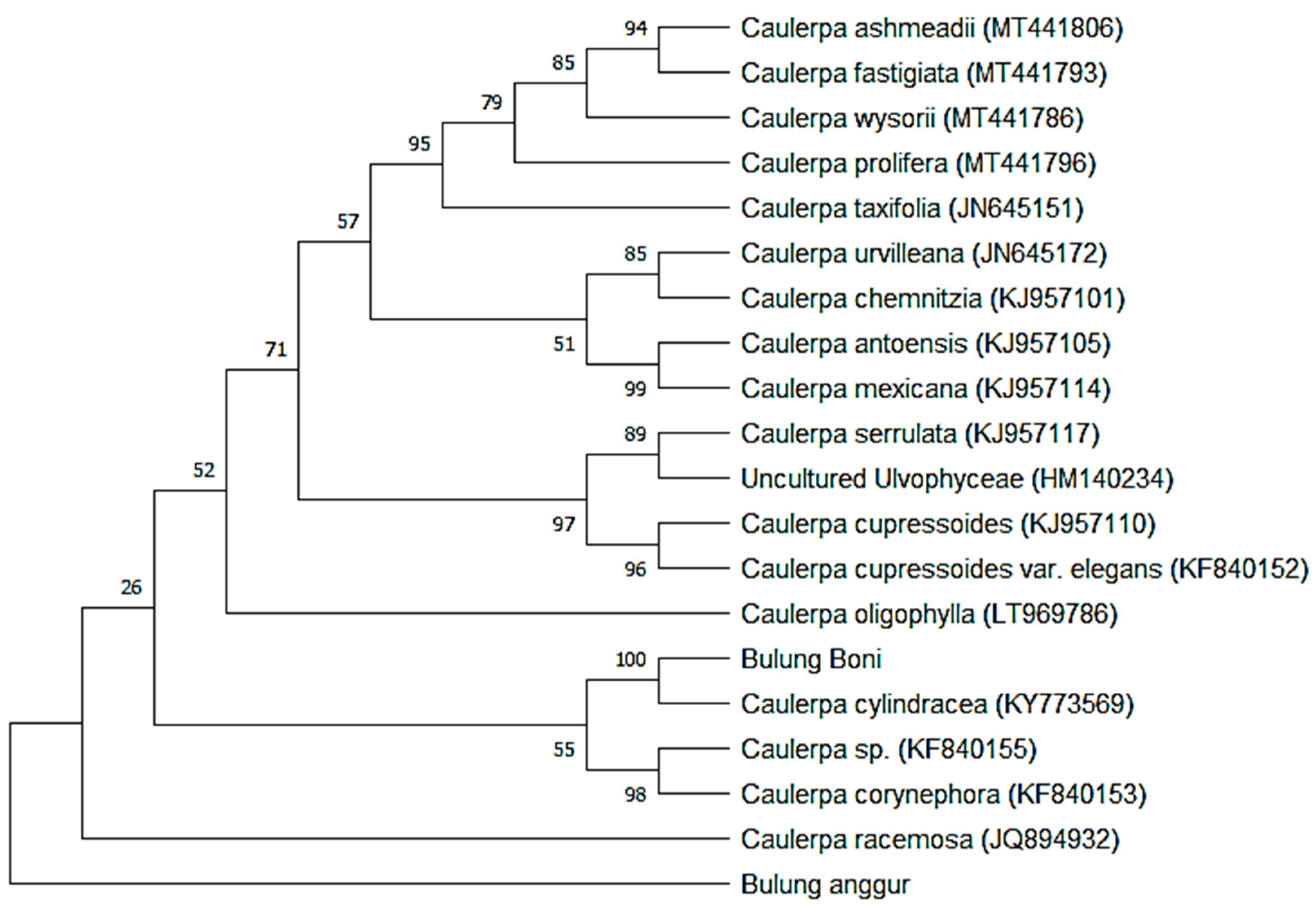
2.3. Molecular Identification
3. Materials and Methods
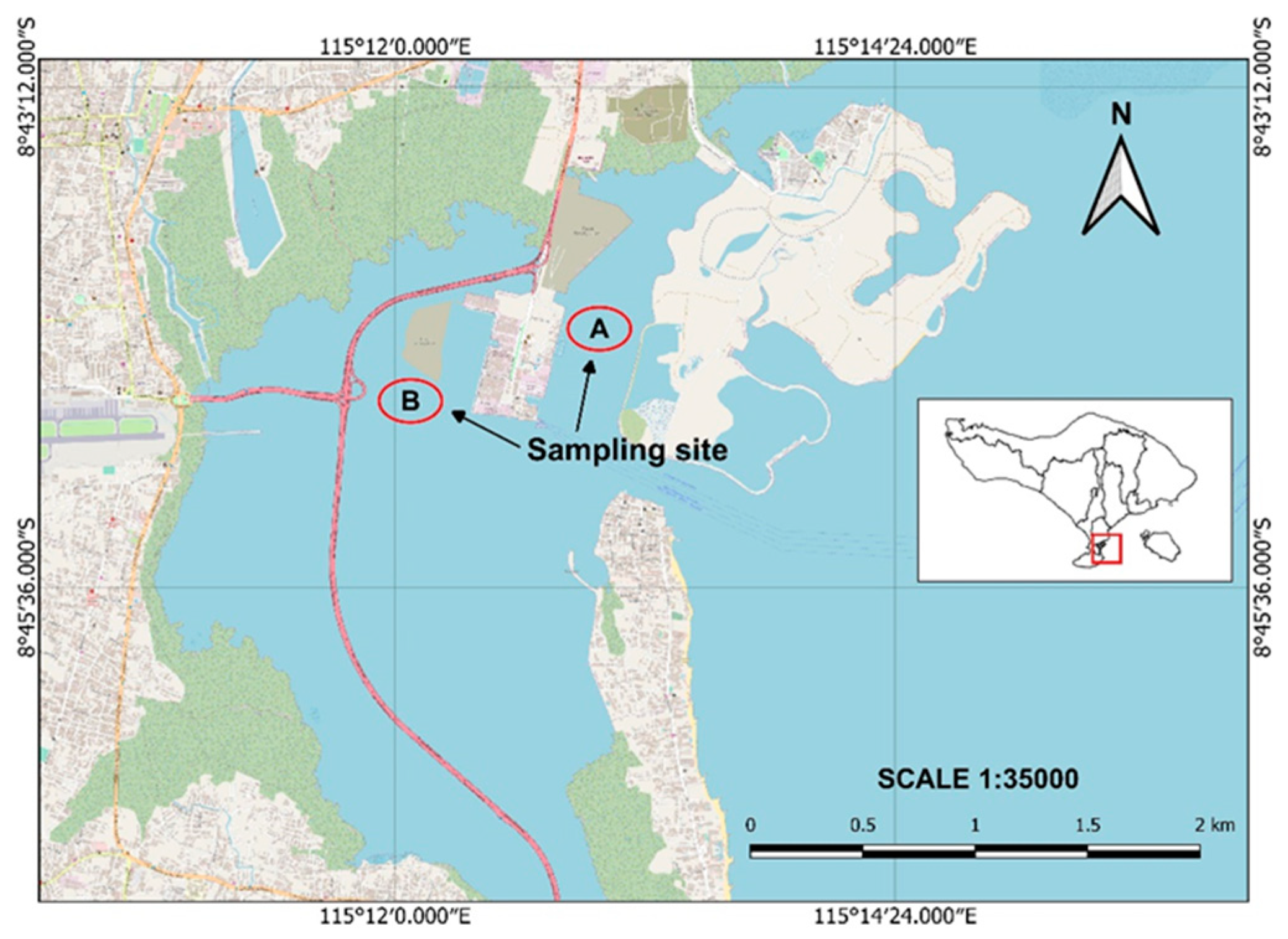
3.1. Sample Collection
3.2. Maceration and GC-MS Analysis
3.3. DNA Extraction and PCR Amplification
3.4. Computational Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bjerregaard, R.; Valderrama, D.; Sims, N.; Radulovich, R.; Diana, J.; Capron, M.; Forster, J.; Goudey, C.; Yarish, C.; Hopkins, K.; et al. Seaweed Aquaculture for Food Security, Income Generation and Environmental Health in Tropical Developing Countries; World Bank Group: Washington, DC, USA, 2016. [Google Scholar] [CrossRef]
- Kolanjinathan, K.; Ganesh, P.; Saranraj, P. Pharmacological Importance of Seaweeds: A Review. World J. Fish Mar. Sci. 2014, 6, 1–15. [Google Scholar] [CrossRef]
- Ragunath, C.; Kumar, Y.A.S.; Kanivalan, I.; Radhakrishnan, S. Phytochemical Screening and Gc-Ms Analysis of Bioactive Constituents in the Methanolic Extract of Caulerpa Racemosa (Forssk.) j. Agardh and Padina Boergesenii Allender & Kraft. Curr. Appl. Sci. Technol. 2020, 20, 380–393. [Google Scholar] [CrossRef]
- Julyasih, M.K.; Suata, K.; Wirawan, I.; Mantik Astawa, I. Seaweed Extracts Improve Lipid Profile of Wistar Rat. Indones. J. Biomed. Sci. 2011, 5, 224796. [Google Scholar]
- Sasadara, M.M.V.; Wirawan, I.G.P.; Jawi, I.M.; Sritamin, M.; Dewi, N.N.A.; Adi, A.A.A.M. Anti-Inflammatory Effect of Red Macroalgae Bulung sangu (Gracilaria Sp.) Extract in UVB-Irradiated Mice. Pakistan J. Biol. Sci. 2021, 24, 80–89. [Google Scholar] [CrossRef]
- Wirawan, I.G.P.; Malida, M.; Sasadara, V.; Wijaya, I.N. DNA Barcoding in Molecular Identification and Phylogenetic Relationship of Beneficial Wild Balinese Red Algae, Bulung sangu (Gracilaria Sp.). Bali Med. J. 2021, 10, 82–88. [Google Scholar] [CrossRef]
- Wiraguna, G.P.; Anak Agung Pangkahila, W.M.A. Antioxidant Properties of Topical Caulerpa Sp. Extract on UVB-Induced Photoaging in Mice. Dermatol. Rep. 2018, 10, 20–25. [Google Scholar] [CrossRef] [Green Version]
- Coneva, V.; Chitwood, D.H. Plant Architecture without Multicellularity: Quandaries over Patterning and the Soma-Germline Divide in Siphonous Algae. Front. Plant Sci. 2015, 6, 287. [Google Scholar] [CrossRef] [Green Version]
- Famà, P.; Wysor, B.; Kooistra, W.H.C.F.; Zuccarello, G.C. Molecular Phylogeny of the Genus Caulerpa (Caulerpales, Chlorophyta) Inferred from Chloroplast TufA Gene. J. Phycol. 2002, 38, 1040–1050. [Google Scholar] [CrossRef] [Green Version]
- Zubia, M.; Draisma, S.G.A.; Morrissey, K.L.; Varela-álvarez, E.; de Clerck, O. Concise Review of the Genus Caulerpa J.V. Lamouroux. J. Appl. Phycol. 2020, 32, 23–39. [Google Scholar] [CrossRef]
- Farnsworth, E.J.; Chu, M.; Kress, W.J.; Neill, A.K.; Best, J.H.; Pickering, J.; Stevenson, R.D.; Courtney, G.W.; Vandyk, J.K.; Ellison, A.M. Next-Generation Field Guides. Bioscience 2013, 63, 891–899. [Google Scholar] [CrossRef] [Green Version]
- Belton, G.S.; van Reine, W.F.P.; Huisman, J.M.; Draisma, S.G.A.; Gurgel, C.F.D. Resolving Phenotypic Plasticity and Species Designation in the Morphologically Challenging Caulerpa racemosa-peltata Complex (Chlorophyta, Caulerpaceae). J. Phycol. 2014, 50, 32–54. [Google Scholar] [CrossRef] [PubMed]
- Estrada, J.L.; Bautista, N.S.; Dionisio-Sese, M.L. Morphological Variation of Two Common Sea Grapes (Caulerpa Lentillifera and Caulerpa Racemosa) from Selected Regions in the Philippines. Biodiversitas 2020, 21, 1823–1832. [Google Scholar] [CrossRef]
- Letchuman, S. Short Introduction of DNA Barcoding. Int. J. Res. 2018, 5, 673–686. [Google Scholar]
- Stam, W.T.; Olsen, J.L.; Zaleski, S.F.; Murray, S.N.; Brown, K.R.; Walters, L.J. A Forensic and Phylogenetic Survey of Caulerpa Species (Caulerpales, Chlorophyta) from the Florida Coast, Local Aquarium Shops, and e-Commerce: Establishing a Proactive Baseline for Early Detection. J. Phycol. 2006, 42, 1113–1124. [Google Scholar] [CrossRef]
- Saunders, G.W.; Kucera, H. An Evaluation of RbcL, TufA, UPA, LSU and ITS as DNA Barcode Markers for the Marine Green Macroalgae. Cryptogam. Algol. 2010, 31, 487–528. [Google Scholar]
- Kazi, M.A.; Reddy, C.R.K.; Jha, B. Molecular Phylogeny and Barcoding of Caulerpa (Bryopsidales) Based on the TufA, RbcL, 18S RDNA and ITS RDNA Genes. PLoS ONE 2013, 8, e82438. [Google Scholar] [CrossRef] [Green Version]
- Sauvage, T.; Payri, C.; Draisma, S.G.A.; van Reine, W.F.P.H.; Verbruggen, H.; Belton, G.S.; Frederico, C.; Gurgel, D.; Gabriel, D.; Sherwood, A.R.; et al. Molecular Diversity of the Caulerpa racemosa—Caulerpa peltata Complex (Caulerpaceae, Bryopsidales) in New Caledonia, with New Australasian Records for C. racemosa Var. Cylindracea. Phycologia 2013, 52, 222, Erratum in Phycologia 2013, 52, 6–13. [Google Scholar] [CrossRef] [Green Version]
- Dumilag, R.V.; Aguinaldo, Z.Z.A.; Alcoriza, V.A.M.; Balucanag, M.P.S.B.; Dulalia, A.R.T.; Sayasa, A.R. DNA Barcodes of Caulerpa Species (Caulerpaceae, Chlorophyta) from the Northern Philippines. Philipp. J. Sci. 2019, 148, 337–347. [Google Scholar]
- Karthick, P.; Murthy, K.N.; Ramesh, C.; Narayana, S.; Mohanraju, R. Molecular Authentication of Green Algae Caulerpa (Caulerpales, Chlorophyta) Based on ITS and TufA Genes from Andaman Islands, India. Indian J. Exp. Biol. 2020, 58, 109–114. [Google Scholar]
- Darmawan, M.; Zamani, N.P.; Irianto, H.E.; Madduppa, H. Molecular Characterization of Caulerpa Racemosa (Caulerpales, Chlorophyta) from Indonesia Based on the Plastid TufA Gene. Squalen Bull. Mar. Fish. Postharvest Biotechnol. 2021, 16, 101–109. [Google Scholar] [CrossRef]
- Vieira, C.; De Ramon N’Yeurt, A.; Rasoamanendrika, F.A.; D’Hondt, S.; Tran, L.A.T.; Van den Spiegel, D.; Kawai, H.; De Clerck, O. Marine Macroalgal Biodiversity of Northern Madagascar: Morpho-Genetic Systematics and Implications of Anthropic Impacts for Conservation; Springer: Dordrecht, The Netherlands, 2021; Volume 30, ISBN 0123456789. [Google Scholar]
- Du, G.; Wu, F.; Mao, Y.; Guo, S.; Xue, H.; Bi, G. DNA Barcoding Assessment of Green Macroalgae in Coastal Zone around Qingdao, China. J. Ocean Univ. China 2014, 13, 97–103. [Google Scholar] [CrossRef]
- Kumar, J.G.S.; Umamaheswari, S.; Kavimani, S.; Ilavarasan, R. Pharmacological Potential of Green Algae Caulerpa: A Review. Int. J. Pharm. Sci. Res. 2019, 10, 1014. [Google Scholar] [CrossRef]
- Belton, G.S.; Draisma, S.G.A.; van Reine, W.F.P.H.; Huisman, J.M.; Gurgel, C.F.D. A Taxonomic Reassessment of Caulerpa (Chlorophyta, Caulerpaceae) in Southern Australia, Based on TufA and RbcL Sequence Data. Phycologia 2019, 58, 234–253. [Google Scholar] [CrossRef]
- Santamaría, J.; Golo, R.; Cebrian, E.; García, M.; Vergés, A. Stressful Conditions Give Rise to a Novel and Cryptic Filamentous Form of Caulerpa Cylindracea. Front. Mar. Sci. 2021, 8, 548679. [Google Scholar] [CrossRef]
- Delzer, G.C.; McKenzie, S.W. Five-Day Biochemical Oxygen Demand. In U.S. Geological Survey Techniques of Water-Resources Investigations; USGS: Reston, VA, USA, 2003; Volume 7, pp. 1–21. [Google Scholar]
- Saraswati, N.L.G.R.A.; Arthana, I.W.; Hendrawan, I.G. Analisis Kualitas Perairan Pada Wilayah Perairan Pulau Serangan Bagian Utara Berdasarkan Baku Mutu Air Laut. J. Mar. Aquat. Sci. 2017, 3, 163. [Google Scholar] [CrossRef]
- Gengmao, Z.; Yu, H.; Xing, S.; Shihui, L.; Quanmei, S.; Changhai, W. Salinity Stress Increases Secondary Metabolites and Enzyme Activity in Safflower. Ind. Crops Prod. 2015, 64, 175–181. [Google Scholar] [CrossRef]
- Mei, X.; Wang, X.; Li, G. Pathogenicity and Volatile Nematicidal Metabolites from Duddingtonia Flagrans against Meloidogyne Incognita. Microorganisms 2021, 9, 2268. [Google Scholar] [CrossRef]
- Albeshri, A.; Baeshen, N.A.; Bouback, T.A.; Aljaddawi, A.A. A Review of Rhazya Stricta Decne Phytochemistry, Bioactivities, Pharmacological Activities, Toxicity, and Folkloric Medicinal Uses. Plants 2021, 10, 2508. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, R.; Kayani, W.K.; Ahmed, T.; Malik, F.; Hussain, S.; Ashfaq, M.; Ali, H.; Rubnawaz, S.; Green, B.D.; Calderwood, D.; et al. Assessment of Antidiabetic Potential and Phytochemical Profiling of Rhazya Stricta Root Extracts. BMC Complement. Med. Ther. 2020, 20, 293. [Google Scholar] [CrossRef]
- Zhou, G.; Shi, Q.S.; Huang, X.M.; Xie, X.B. The Three Bacterial Lines of Defense against Antimicrobial Agents. Int. J. Mol. Sci. 2015, 16, 21711–21733. [Google Scholar] [CrossRef] [Green Version]
- DeLeon, S.; Clinton, A.; Fowler, H.; Everett, J.; Horswill, A.R.; Rumbaugh, K.P. Synergistic Interactions of Pseudomonas Aeruginosa and Staphylococcus Aureus in an In Vitro Wound Model. Infect. Immun. 2014, 82, 4718–4728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuah, X.; Okechukwu, P.; Amini, F.; Teo, S. Eicosane, Pentadecane and Palmitic Acid: The Effects in In Vitro Wound Healing Studies. Asian Pac. J. Trop. Biomed. 2018, 8, 490–499. [Google Scholar] [CrossRef]
- Faridha Begum, I.; Mohankumar, R.; Jeevan, M.; Ramani, K. GC–MS Analysis of Bio-Active Molecules Derived from Paracoccus Pantotrophus FMR19 and the Antimicrobial Activity Against Bacterial Pathogens and MDROs. Indian J. Microbiol. 2016, 56, 426–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.R.; Lian, X.Z.; Guo, H.Y.; McGuire, P.M.; De Li, R.; Wang, R.; Yu, F.H. Isolation and Characterization of Methyl Esters and Derivatives from Euphorbia Kansui (Euphorbiaceae) and Their Inhibitory Effects on the Human SGC-7901 Cells. J. Pharm. Pharm. Sci. 2005, 8, 528–535. [Google Scholar] [PubMed]
- Cueto, G.M.; Zerba, E.; Picollo, M.I. Biological Effect of 1-Dodecanol in Teneral and Post-Teneral Rhodnius prolixus and Triatoma infestans (Hemiptera: Reduviidae). Mem. Inst. Oswaldo Cruz 2005, 100, 59–61. [Google Scholar] [CrossRef] [PubMed]
- Togashi, N.; Shiraishi, A.; Nishizaka, M.; Matsuoka, K.; Endo, K.; Hamashima, H.; Inoue, Y. Antibacterial Activity of Long-Chain Fatty Alcohols against Staphylococcus aureus. Molecules 2007, 12, 139–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raman, V.; LA, S.; Pardha, S.; Rao, N.; Krishna, N.B.; Sudhakar, M.; Radhakrishnan, T. Antibacterial, Antioxidant Activity and GC-MS Analysis of Eupatorium odoratum. Asian J. Pharm. Clin. Res. 2012, 5, 99–106. [Google Scholar]
- Komansilan, A.; Abadi, A.L.; Yanuwiadi, B.; Kaligis, D.A. Isolation and Identification of Biolarvicide from Soursop (Annona muricata Linn) Seeds to Mosquito (Aedes aegypti) Larvae. Int. J. Eng. Technol. 2012, 12, 28–32. [Google Scholar]
- Huang, Y.P.; Chang, Y.T.; Hsieh, S.L.; Sandnes, F.E. An Adaptive Knowledge Evolution Strategy for Finding Near-Optimal Solutions of Specific Problems. Expert Syst. Appl. 2011, 38, 3806–3818. [Google Scholar] [CrossRef] [Green Version]
- Kimura, J.; Maki, N. New Loliolide Derivatives from the Brown Alga Undaria pinnatifida. J. Nat. Prod. 2002, 65, 57–58. [Google Scholar] [CrossRef]
- Kuniyoshi, M. Germination Inhibitors from the Brown Alga Sargassum crassifolium (Phaeophyta, Sargassaceae). Bot. Mar. 1985, 28, 501–504. [Google Scholar] [CrossRef]
- Yuan, Z.H.; Han, L.J.; Fan, X.; Li, S.; Shi, D.Y.; Sun, J.; Ma, M.; Yang, Y.C.; Shi, J.G. Chemical Constituents from Red Alga Corallina pilulifera. Zhongguo Zhongyao Zazhi 2006, 31, 1787–1790. [Google Scholar] [PubMed]
- Percot, A.; Yalçin, A.; Aysel, V.; Erduǧan, H.; Dural, B.; Güven, K.C. Loliolide in Marine Algae. Nat. Prod. Res. 2009, 23, 460–465. [Google Scholar] [CrossRef]
- Grabarczyk, M.; Wińska, K.; Mączka, W.; Potaniec, B.; Anioł, M. Loliolide—The Most Ubiquitous Lactone. Folia Biol. Oecologica 2015, 11, 1–8. [Google Scholar] [CrossRef]
- Park, S.H.; Kim, D.S.; Kim, S.; Lorz, L.R.; Choi, E.; Lim, H.Y.; Hossain, M.A.; Jang, S.G.; Choi, Y.I.; Park, K.J.; et al. Loliolide Presents Antiapoptosis and Antiscratching Effects in Human Keratinocytes. Int. J. Mol. Sci. 2019, 20, 651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Julyasih, K.S.M.; Wirawan, I.G.P. Potential Effect of Macro Alga Caulerpa Sp. and Gracilaria Sp. Extract Lowering Malondialdehyde Level of Wistar Rats Fed High Cholesterol Diet. Int. J. Biosci. Biotechnol. 2017, 5, 71–79. [Google Scholar] [CrossRef]
- Rezanka, T.; Olsovska, J.; Sobotka, M.; Sigler, K. The Use of APCI-MS with HPLC and Other Separation Techniques for Identification of Carotenoids and Related Compounds. Curr. Anal. Chem. 2009, 5, 1–25. [Google Scholar] [CrossRef]
- Rahul, M.; Suresh, N.; Anil, T.; Arun, M.; Nivedita, G.; Mv, B. GC-MS Analysis of Phytocomponents of Seaweed, Caulerpa Racemosa. Int. J. Pharma Res. Rev. 2014, 3, 39. [Google Scholar]
- Sakunthala, M.; Paul, J.J.P. GC-MS Analysis of Ethanolic Extract of Caulerpa Mexicana Sonder Ex Kuetzing from Manapad Coast, Tamil Nadu, India. J. Emerg. Technol. Innov. Res. 2019, 6, 244–247. [Google Scholar]
- Baldauf, S.L.; Manhart, J.R.; Palmer, J.D. Different Fates of the Chloroplast TufA Gene Following Its Transfer to the Nucleus in Green Algae. Proc. Natl. Acad. Sci. USA 1990, 87, 5317–5321. [Google Scholar] [CrossRef] [Green Version]
- Vieira, H.H.; Bagatini, I.L.; Guinart, C.M.; Vieira, A.A.H. TufA Gene as Molecular Marker for Freshwater Chlorophyceae. Algae 2016, 31, 155–165. [Google Scholar] [CrossRef] [Green Version]
- Bentaallah, M.E.A.; Taibi, N.E.; Cantasano, N. Additional New Records of Caulerpa cylindracea Sonder 1845 along the West Algerian Coasts. Indian J. Geo-Mar. Sci. 2021, 50, 122–129. [Google Scholar]
- Boumediene, H.K.; Lotfi, B.T. First Record of Invasive Green Algae Caulerpa racemosa Var. Cylindracea in Oran Bay (Western Alegria). Indian J. Geo-Mar. Sci. 2019, 48, 335–342. [Google Scholar]
- Cantasano, N.; Pellicone, G.; Di Martino, V. The Spread of Caulerpa cylindracea in Calabria (Italy) and the Effects of Shipping Activities. Ocean Coast. Manag. 2017, 144, 51–58. [Google Scholar] [CrossRef]
- Grant, W.M. Molecular Phylogeography and Climate Change Biology of the Invasive Green Marine Macroalgae Caulerpa taxifolia and Caulerpa cylindracea in Australia. Ph.D. Thesis, The University of Adelaide, Adelaide, Australia, 2015; pp. 1–133. [Google Scholar]
- Fernández-García, C.; Wysor, B.; Riosmena-Rodríguez, R.; Peña-Salamanca, E.; Verbruggen, H. Species Richness Estimates for the Eastern Tropical Pacific. Phytotaxa 2016, 252, 185–204. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]

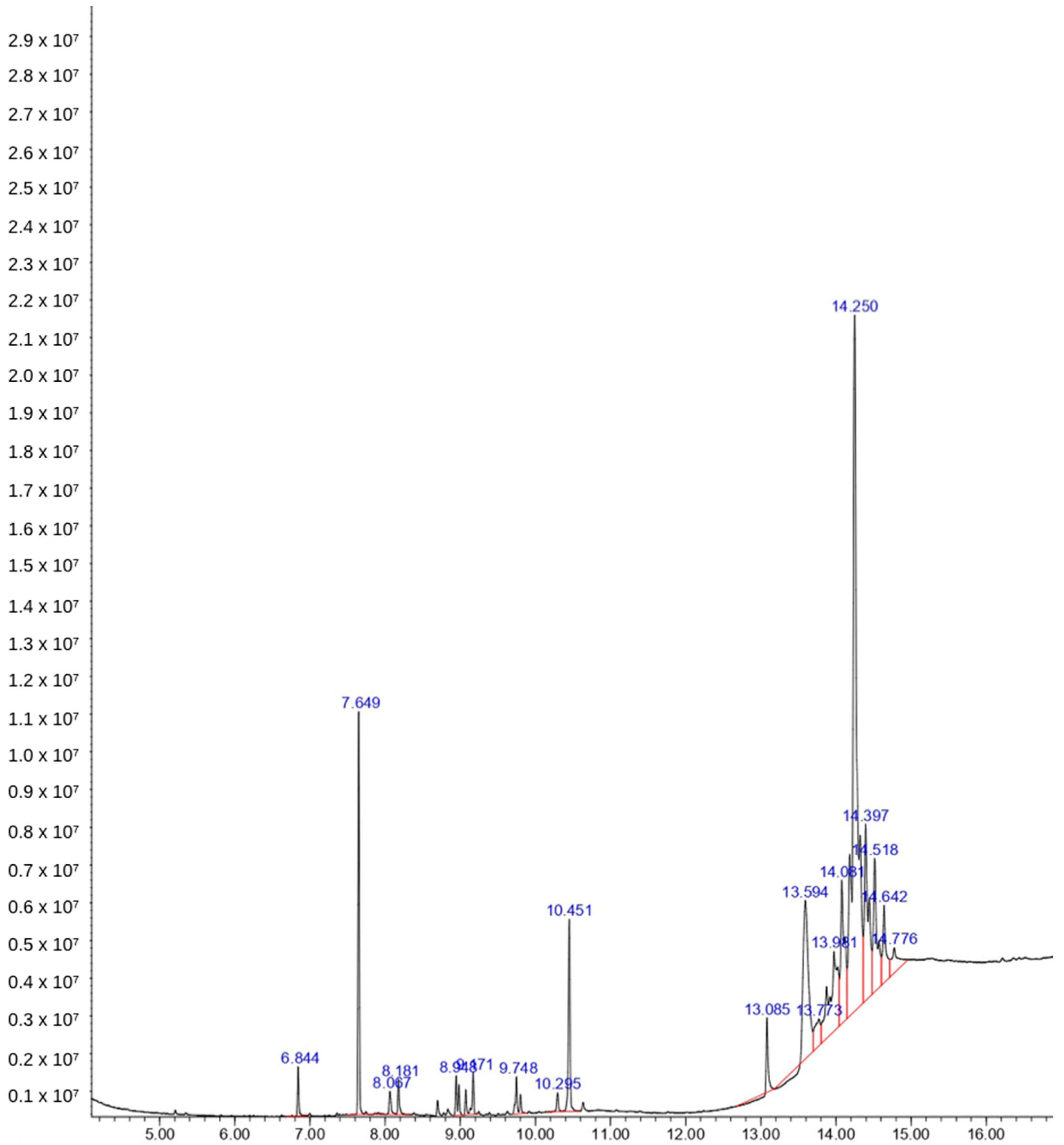


| Chemicals | Rt | AUC | Molecular Weight | Formula |
|---|---|---|---|---|
| Cyclododecane | 6.844 | 0.78 | 168.32 | C12H24 |
| Pentanoic acid | 7.649 | 6.48 | 102.13 | C5H10O2 |
| 3-Heptadecene | 8.067 | 0.69 | 238.5 | C17H34 |
| Eicosane | 8.181 | 0.65 | 282.5 | C20H42 |
| Neophytadiene | 8.948 | 1.08 | 278.5 | C20H38 |
| 11,13-Dimethyl-12-tetradecen-1-ol acetate | 9.748 | 1.09 | 282.5 | C18H34O2 |
| 1-Dodecanol | 10.451 | 3.92 | 186.33 | C12H26O |
| Androst-5,15-dien-3ol acetate | 13.594 | 9.03 | 314.5 | C21H30O2 |
| 2-Naphthalene-sulfonic acid | 13.981 | 7.89 | 924.9 | C39H31N7NaO13S3+ |
| Cyclohexanamine | 14.250 | 36.91 | 99.17 | C7H15N |
| 2-p-Nitrophenyl-1,3,4-Oxadiazol-5-one | 14.776 | 2.04 | 207.14 | C8H5N3O4 |
| Chemicals | Rt | AUC | Molecular Weight | Formula |
|---|---|---|---|---|
| 1-Decene | 6.843 | 1.24 | 140.27 | C10H20 |
| Pentadecene | 6.998 | 0.23 | 210.4 | C15H30 |
| Dodecane | 7.396 | 0.35 | 170.33 | C12H26 |
| 8-Heptadecene | 8.069 | 1.58 | 238.5 | C17H34 |
| Heptadecane | 8.179 | 1.10 | 240.5 | C17H36 |
| Loliolide | 8.697 | 0.16 | 196.24 | C11H16O3 |
| Neophytadiene | 8.947 | 0.65 | 278.5 | C20H38 |
| 1-Nonadecene | 9.154 | 0.93 | 266.5 | C19H38 |
| Hexadecanoic acid | 9.549 | 1.29 | 256.42 | C16H32O |
| Phytol | 10.431 | 3.53 | 296.5 | C20H40O |
| 1-Hydroxy-1-o-fluorophenyl-4-nitroimidazole-3-oxide | 13.771 | 0.90 | 100.08 | C4H5N3O2 |
| Terephthalic acid | 14.259 | 31.99 | 166.13 | C6H4(CO2H)2 |
| 1,2-Cyclohexanedicarboxylic acid | 14.399 | 10.69 | 172.18 | C8H12O4 |
| Species | Accession No | Bulung Boni | Bulung Anggur | ||
|---|---|---|---|---|---|
| Pairwise | ID | Pairwise | ID | ||
| Caulerpa racemose | JQ894932 | 0.013 | 94.10% | 1.200 | 39.60% |
| Caulerpa cylindracea | KY773569 | 0.000 | 94.30% | 1.250 | 40.80% |
| Caulerpa sp. | KF840155 | 0.010 | 97.70% | 1.250 | 41.00% |
| Caulerpa corynephora | KF840153 | 0.009 | 96.40% | 1.226 | 40.20% |
| Caulerpa serrulate | KJ957117 | 0.026 | 95.60% | 1.259 | 41.10% |
| Caulerpa cupressoides | KJ957110 | 0.027 | 95.40% | 1.262 | 41.00% |
| Caulerpa cupressoides var. elegans | KF840152 | 0.028 | 95.20% | 1.240 | 40.20% |
| Caulerpa urvilleana | JN645172 | 0.026 | 95.80% | 1.246 | 41.20% |
| Ulcultured Unvophyceae | HM140234 | 0.027 | 91.80% | 1.269 | 40.40% |
| Caulerpa antoensis | KJ957105 | 0.035 | 94.70% | 1.275 | 41.00% |
| Caulerpa mexicana | KJ957114 | 0.034 | 94.90% | 1.257 | 41.20% |
| Caulerpa chemnitzia | KJ957101 | 0.035 | 94.70% | 1.250 | 41.40% |
| Caulerpa wysorii | MT441786 | 0.037 | 88.20% | 1.289 | 40.30% |
| Caulerpa prolifera | MT441796 | 0.038 | 95.00% | 1.301 | 40.30% |
| Caulerpa ashmeadii | MT441806 | 0.039 | 94.70% | 1.241 | 40.60% |
| Caulerpa oligophylla | LT969786 | 0.021 | 87.90% | 1.226 | 37.00% |
| Caulerpa fastigiate | MT441793 | 0.041 | 92.30% | 1.234 | 40.30% |
| Caulerpa taxifolia | JN645151 | 0.044 | 94.20% | 1.278 | 40.60% |
| Bulung boni | - | - | 1.229 | 41.10% | |
| Bulung anggur | - | 1.229 | 41.10% | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wirawan, I.G.P.; Dewi, N.K.E.S.; Sasadara, M.M.V.; Sunyamurthi, I.G.N.A.; Jawi, I.M.; Wijaya, I.N.; Darmawati, I.A.P.; Suada, I.K.; Krisnandika, A.A.K. Phytochemical Analysis and Molecular Identification of Green Macroalgae Caulerpa spp. from Bali, Indonesia. Molecules 2022, 27, 4879. https://doi.org/10.3390/molecules27154879
Wirawan IGP, Dewi NKES, Sasadara MMV, Sunyamurthi IGNA, Jawi IM, Wijaya IN, Darmawati IAP, Suada IK, Krisnandika AAK. Phytochemical Analysis and Molecular Identification of Green Macroalgae Caulerpa spp. from Bali, Indonesia. Molecules. 2022; 27(15):4879. https://doi.org/10.3390/molecules27154879
Chicago/Turabian StyleWirawan, I Gede Putu, Ni Kadek Emi Sintha Dewi, Maria Malida Vernandes Sasadara, I Gde Nengah Adhilaksman Sunyamurthi, I Made Jawi, I Nyoman Wijaya, Ida Ayu Putri Darmawati, I Ketut Suada, and Anak Agung Keswari Krisnandika. 2022. "Phytochemical Analysis and Molecular Identification of Green Macroalgae Caulerpa spp. from Bali, Indonesia" Molecules 27, no. 15: 4879. https://doi.org/10.3390/molecules27154879
APA StyleWirawan, I. G. P., Dewi, N. K. E. S., Sasadara, M. M. V., Sunyamurthi, I. G. N. A., Jawi, I. M., Wijaya, I. N., Darmawati, I. A. P., Suada, I. K., & Krisnandika, A. A. K. (2022). Phytochemical Analysis and Molecular Identification of Green Macroalgae Caulerpa spp. from Bali, Indonesia. Molecules, 27(15), 4879. https://doi.org/10.3390/molecules27154879