Lone Pair…π Contacts and Structure Signatures of r(UNCG) Tetraloops, Z-Turns, and Z-Steps: A WebFR3D Survey
Abstract
1. Introduction
2. Background: r(UNCG) Tetraloops and Lone Pair…π Contacts
2.1. The Structural Signature of r(UNCG) Tetraloops Has Evolved
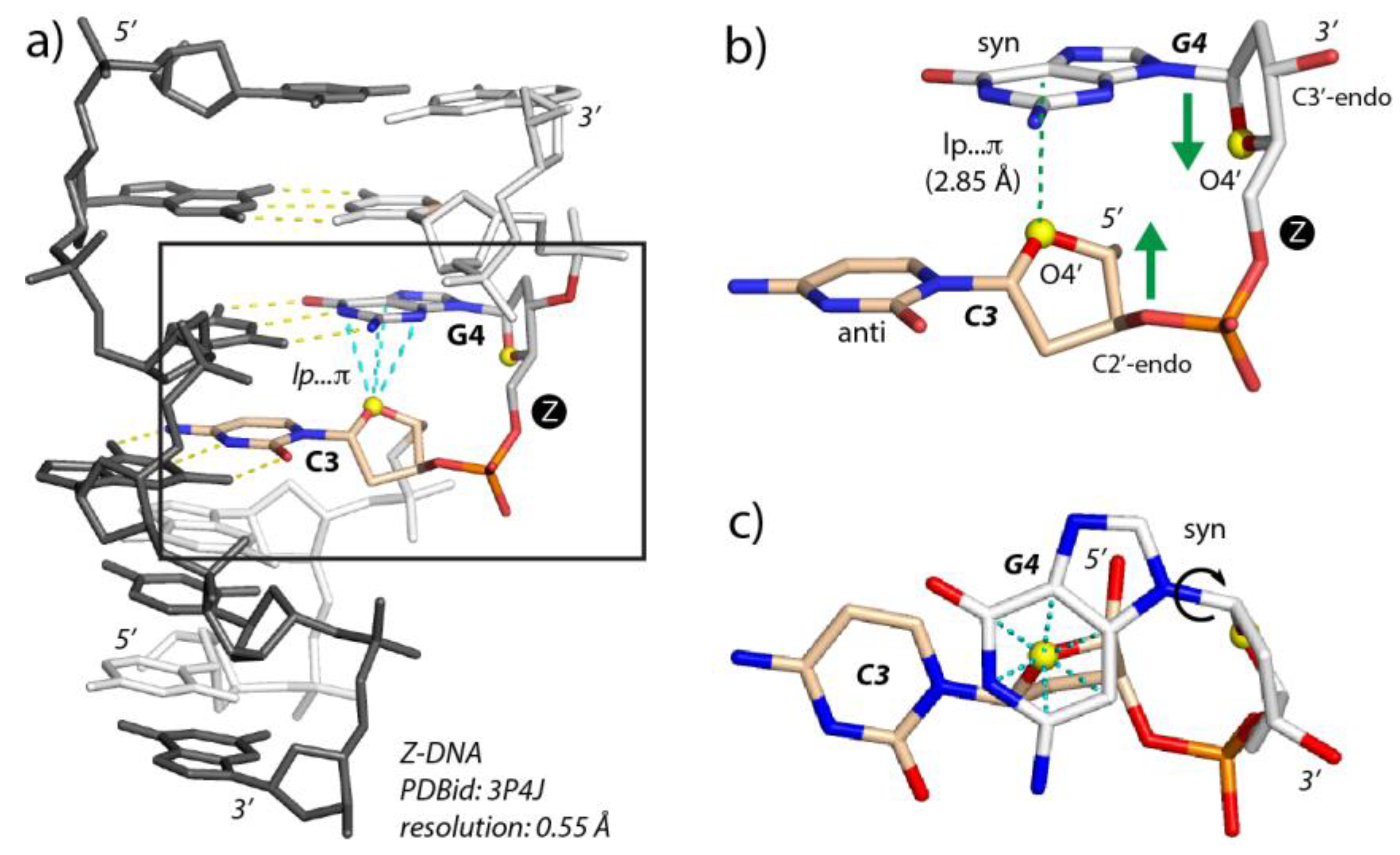
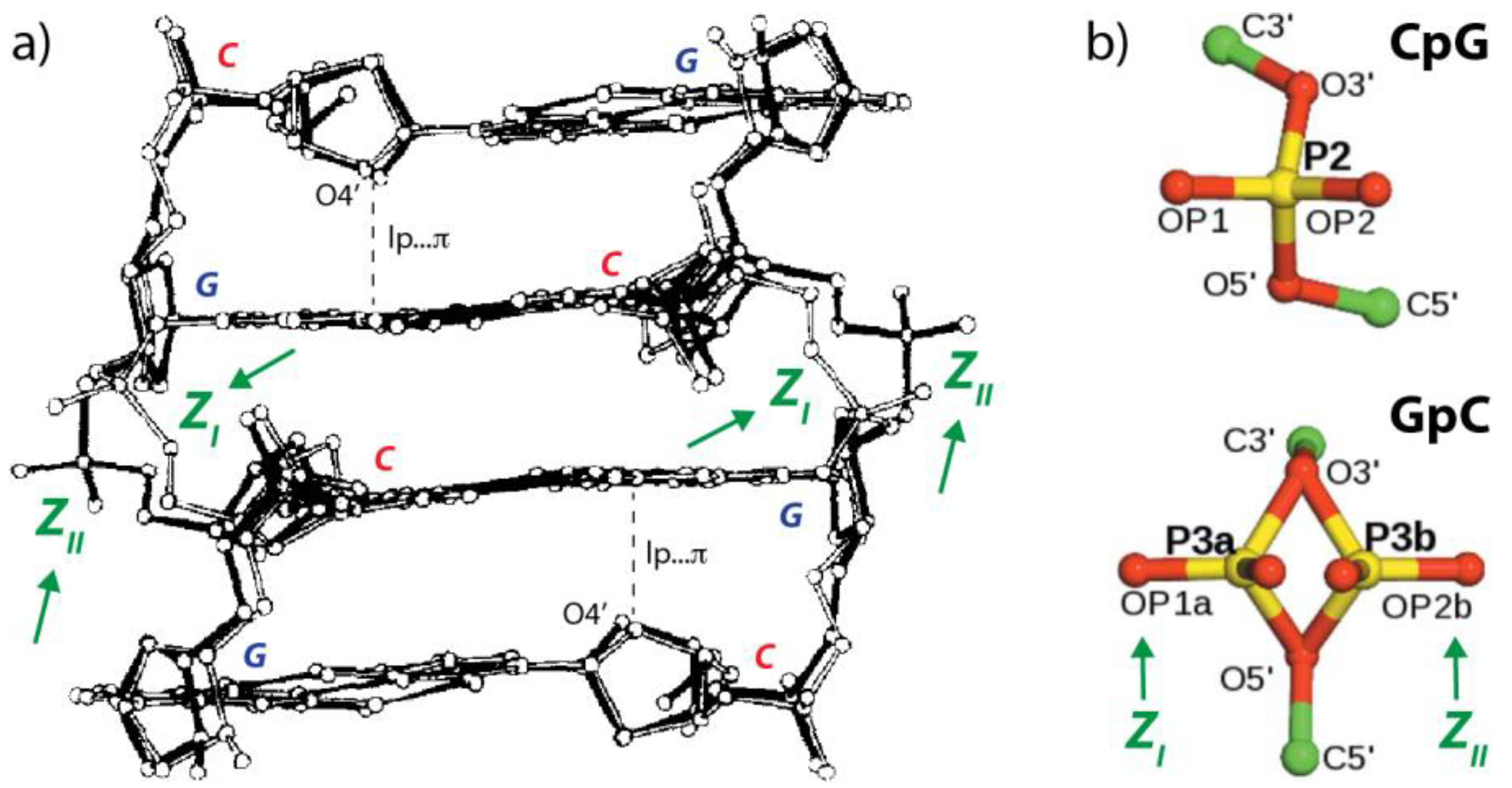
2.2. r(UNCG) Tetraloops, Although Called Z-Turns, Comprise a CpG Z-Step with a Lone Pair…π Contact
2.3. r(UNCG) and Z-DNA Molecular Dynamics Simulation Challenges
3. Results
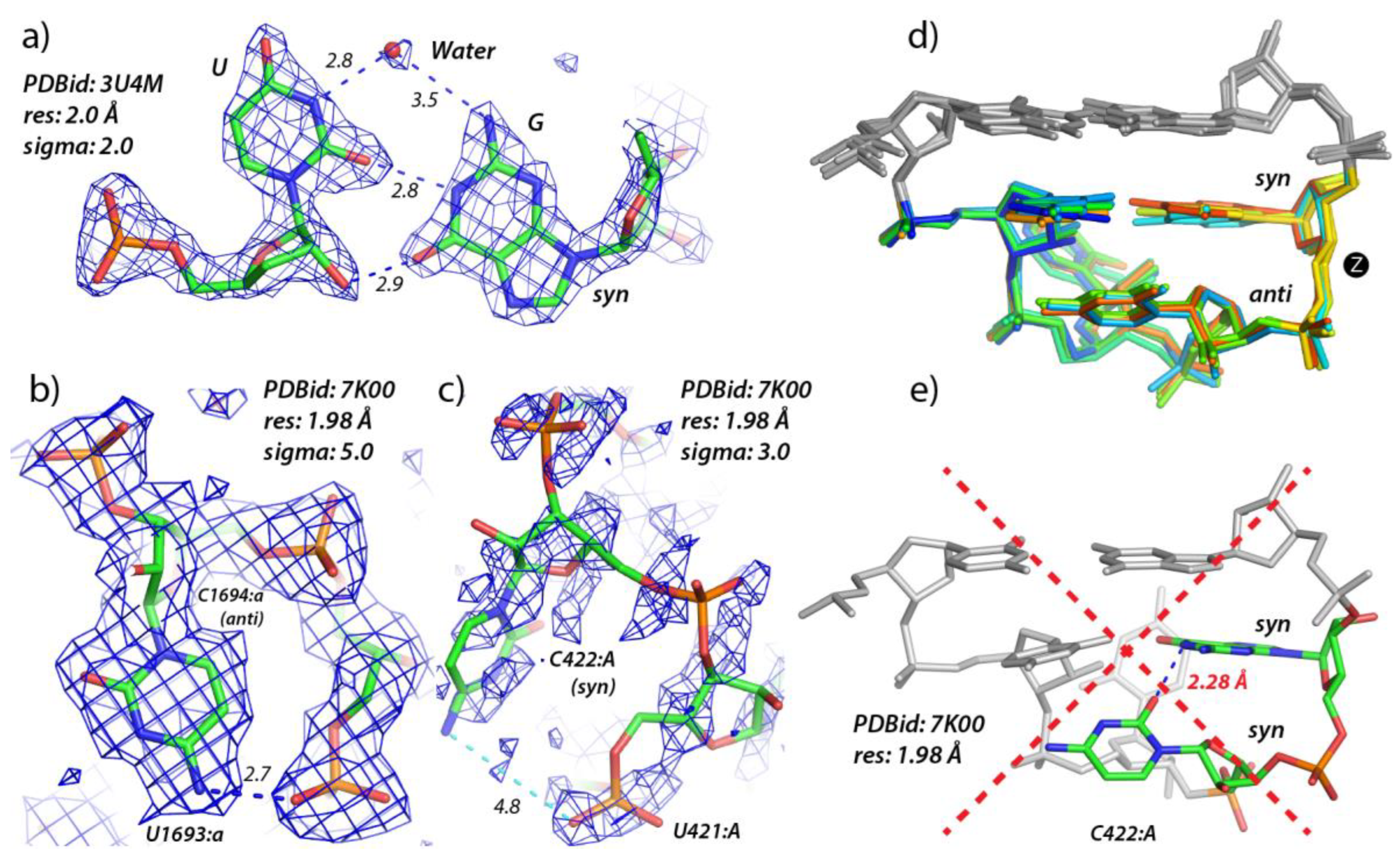
3.1. Defining lp…π Contacts
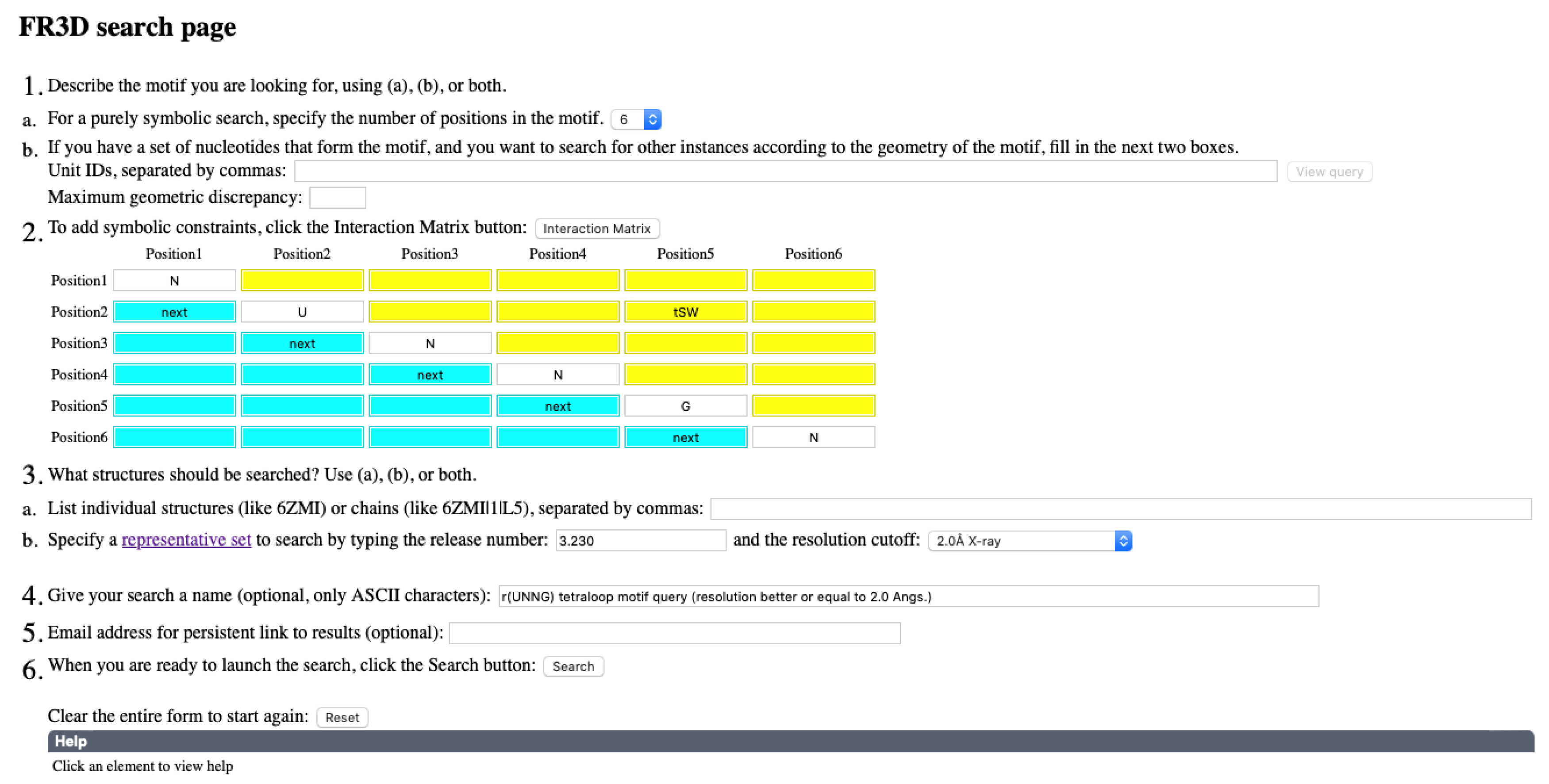
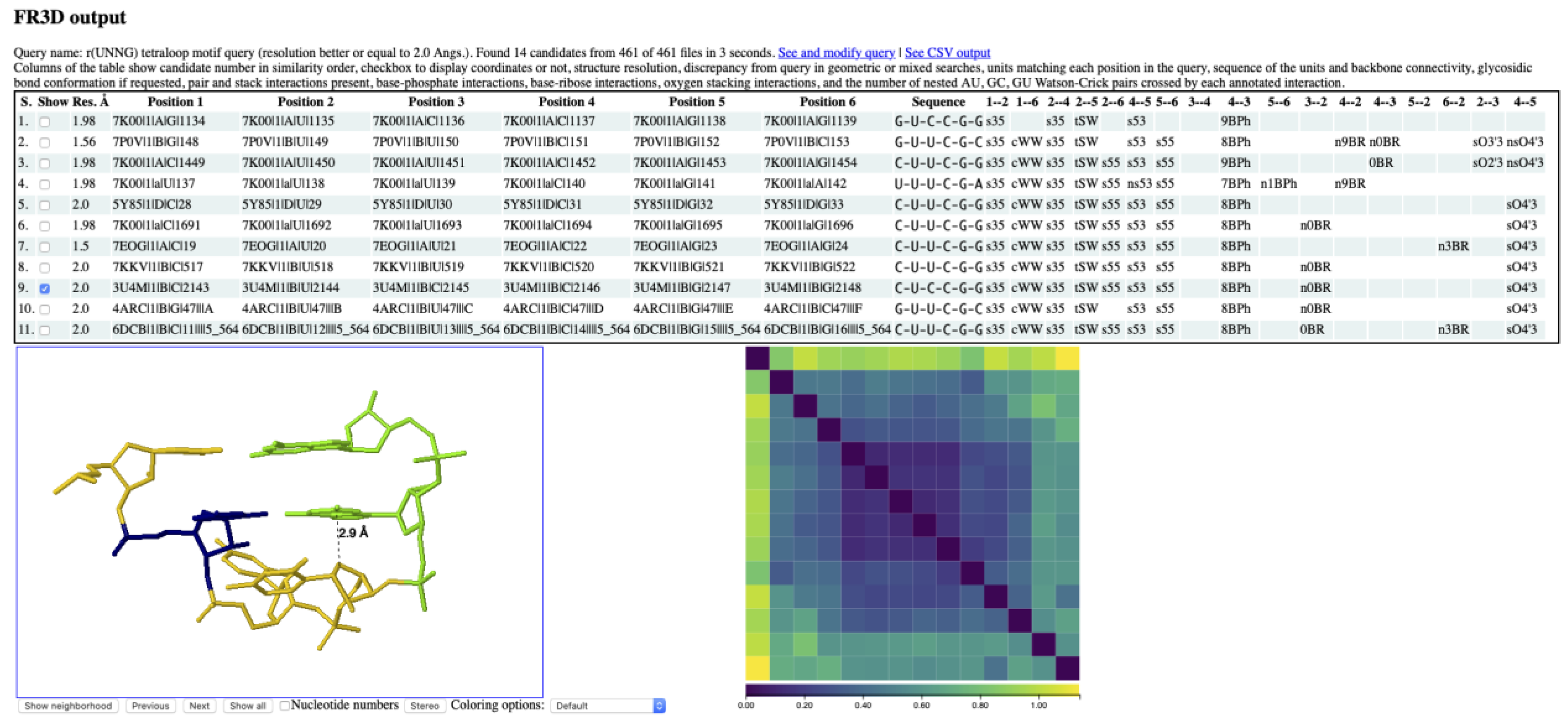
3.2. Finding lp…π Contacts with WebFR3D
3.3. Z-Steps and Zanti-Steps Identified by WebFR3D
3.4. r(UNNG) Z-Turn Signatures Derived from X-ray and Cryo-EM Structures
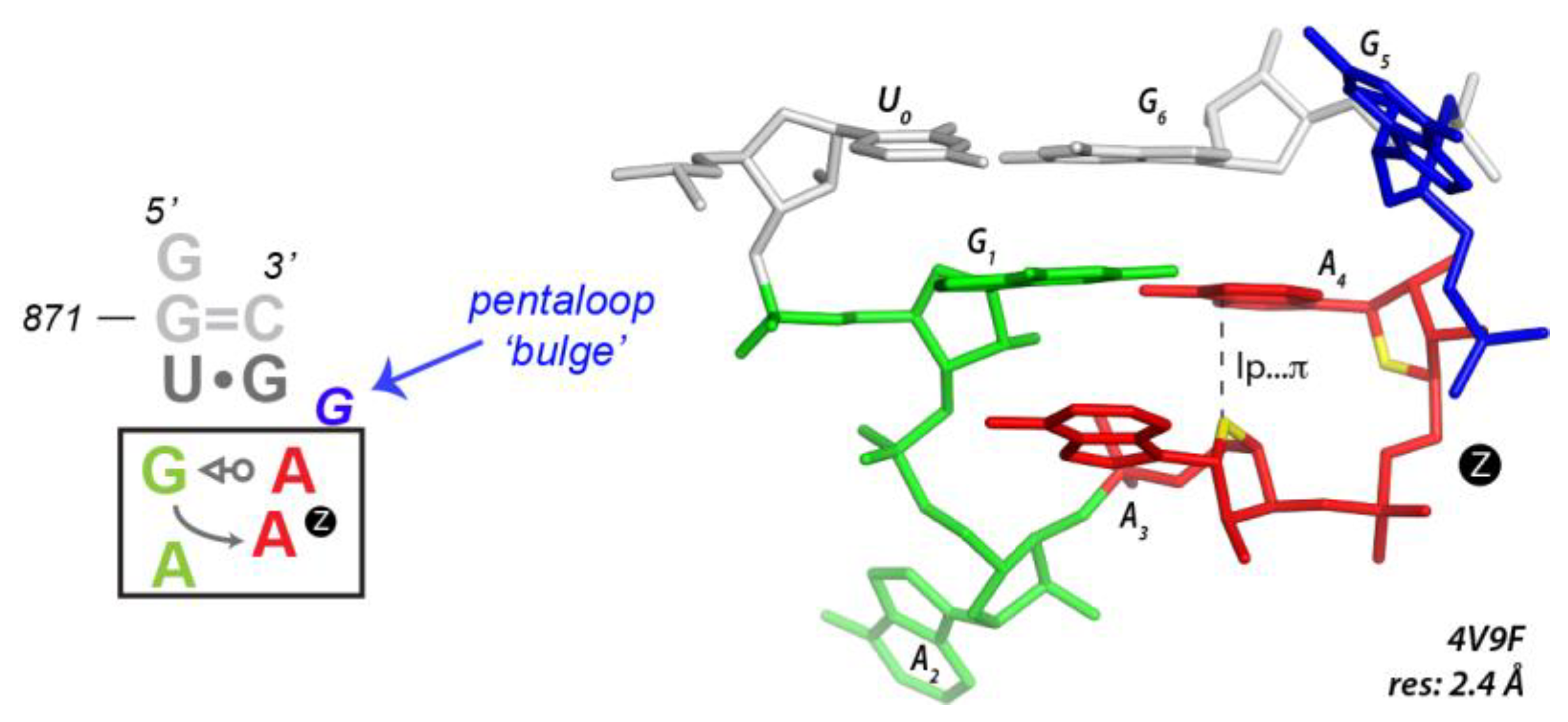
3.5. Finding Unusual r(GNNA) Z-Turns
3.6. UNNG versus CNNG Z-Turns: Isosteric or Not?
4. Discussion
4.1. Outliers: Validating, Correcting, or Discarding
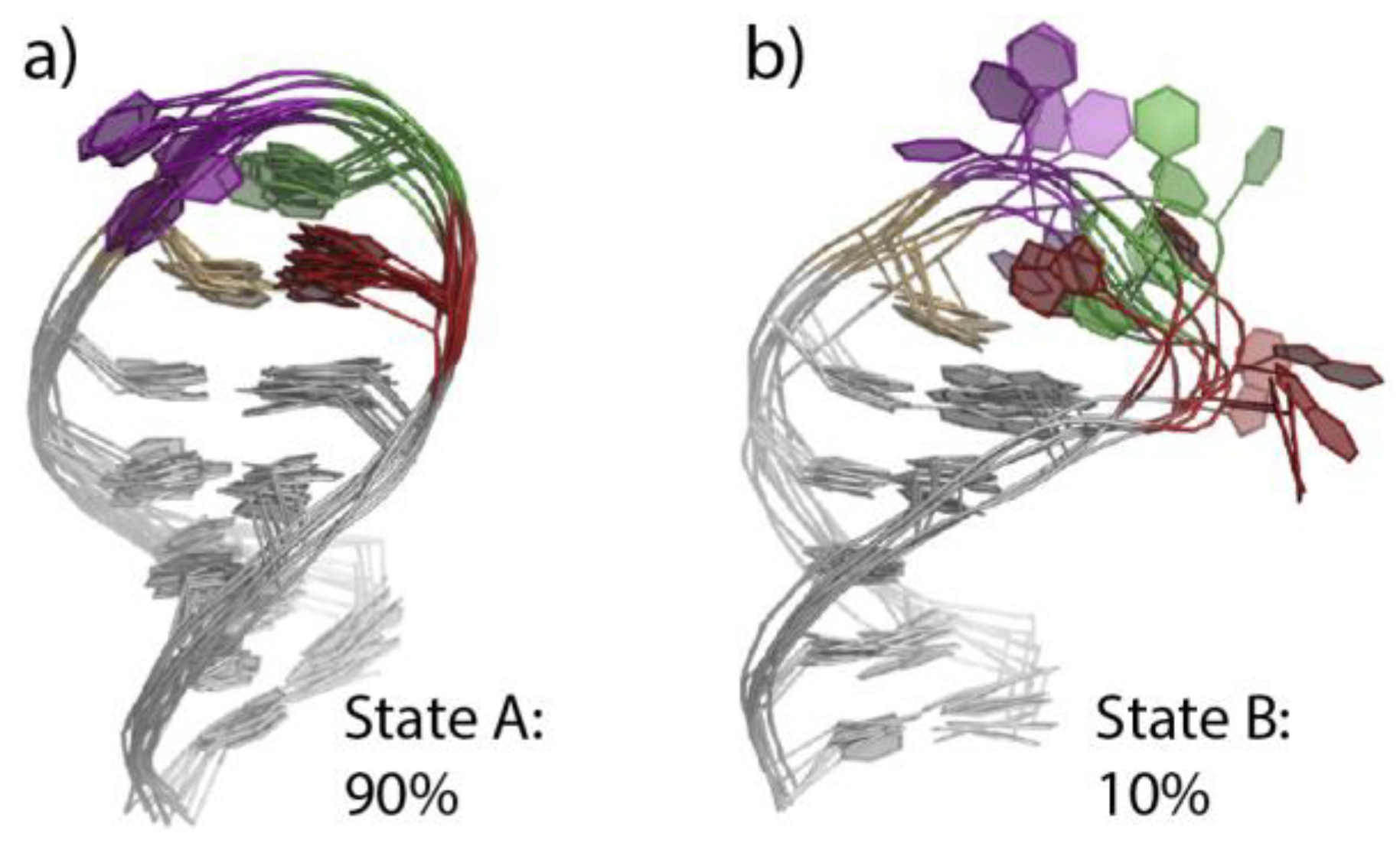
4.2. Use of Structural Signatures for MD Simulations
4.3. WebFR3D Limitations and Strategy
5. Conclusions
6. Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Auffinger, P.; Westhof, E. An extended structural signature for the tRNA anticodon loop. RNA 2001, 7, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Leontis, N.B.; Altman, R.B.; Berman, H.M.; Brenner, S.E.; Brown, J.W.; Engelke, D.R.; Harvey, S.C.; Holbrook, S.R.; Jossinet, F.; Lewis, S.E.; et al. The RNA Ontology Consortium: An open invitation to the RNA community. RNA 2006, 12, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.J.; Olson, W.K. 3DNA: A versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008, 3, 1213–1227. [Google Scholar] [CrossRef] [PubMed]
- Sarver, M.; Zirbel, C.L.; Stombaugh, J.; Mokdad, A.; Leontis, N.B. FR3D: Finding local and composite recurrent structural motifs in RNA 3D structures. J. Math. Biol. 2008, 56, 215–252. [Google Scholar] [CrossRef] [PubMed]
- Petrov, A.I.; Zirbel, C.L.; Leontis, N.B. WebFR3D—A server for finding, aligning and analyzing recurrent RNA 3D motifs. Nucleic Acids Res. 2011, 39, W50–W55. [Google Scholar] [CrossRef]
- Lu, X.J.; Bussemaker, H.J.; Olson, W.K. DSSR: An integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res. 2015, 43, e142. [Google Scholar] [CrossRef]
- Sweeney, B.A.; Roy, P.; Leontis, N.B. An introduction to recurrent nucleotide interactions in RNA. Wiley Interdiscip. Rev. RNA 2015, 6, 17–45. [Google Scholar] [CrossRef]
- Parlea, L.G.; Sweeney, B.A.; Hosseini-Asanjan, M.; Zirbel, C.L.; Leontis, N.B. The RNA 3D Motif Atlas: Computational methods for extraction, organization and evaluation of RNA motifs. Methods 2016, 103, 99–119. [Google Scholar] [CrossRef]
- Berman, H.M.; Lawson, C.L.; Schneider, B. Developing community resources for nucleic acid structures. Life 2022, 12, 540. [Google Scholar] [CrossRef]
- Roy, P.; Bhattacharyya, D. Contact networks in RNA: A structural bioinformatics study with a new tool. J. Comput. Aided Mol. Des. 2022, 36, 131–140. [Google Scholar] [CrossRef]
- Leontis, N.B.; Lescoute, A.; Westhof, E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006, 16, 279–287. [Google Scholar] [CrossRef] [PubMed]
- D’Ascenzo, L.; Leonarski, F.; Vicens, Q.; Auffinger, P. ‘Z-DNA like’ fragments in RNA: A recurring structural motif with implications for folding, RNA/protein recognition and immune response. Nucleic Acids Res. 2016, 44, 5944–5956. [Google Scholar] [CrossRef] [PubMed]
- D’Ascenzo, L.; Leonarski, F.; Vicens, Q.; Auffinger, P. Revisiting GNRA and UNCG folds: U-turns versus Z-turns in RNA hairpin loops. RNA 2017, 23, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Egli, M.; Sarkhel, S. Lone pair-aromatic interactions: To stabilize or not to stabilize. Acc. Chem. Res. 2007, 40, 197–205. [Google Scholar] [CrossRef]
- Chawla, M.; Chermak, E.; Zhang, Q.; Bujnicki, J.M.; Oliva, R.; Cavallo, L. Occurrence and stability of lone pair-π stacking interactions between ribose and nucleobases in functional RNAs. Nucleic Acids Res. 2017, 45, 11019–11032. [Google Scholar] [CrossRef] [PubMed]
- Kozelka, J. Lone pair-π interactions in biological systems: Occurrence, function, and physical origin. Eur. Biophys. J. 2017, 46, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Kruse, H.; Mrazikova, K.; D’Ascenzo, L.; Sponer, J.; Auffinger, P. Short but weak: The Z-DNA lone-pair…π conundrum challenges standard carbon Van der Waals radii. Angew. Chem. Int. Ed. Engl. 2020, 59, 16553–16560. [Google Scholar] [CrossRef]
- Cheong, C.; Varani, G.; Tinoco, I. Solution structure of an unusually stable RNA hairpin, 5’GGAC(UUCG)GUCC. Nature 1990, 346, 680–681. [Google Scholar] [CrossRef]
- Varani, G.; Cheong, C.; Tinoco, I., Jr. Structure of an unusually stable RNA hairpin. Biochemistry 1991, 30, 3280–3289. [Google Scholar] [CrossRef]
- Richardson, J.S.; Schneider, B.; Murray, L.W.; Kapral, G.J.; Immormino, R.M.; Headd, J.J.; Richardson, D.C.; Ham, D.; Hershkovits, E.; Williams, L.D.; et al. RNA backbone: Consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution). RNA 2008, 14, 465–481. [Google Scholar] [CrossRef]
- Sheehy, J.P.; Davis, A.R.; Znosko, B.M. Thermodynamic characterization of naturally occurring RNA tetraloops. RNA 2010, 16, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Allain, F.H.-T.; Varani, G. Structure of the P1 helix from group I self-splicing introns. J. Mol. Biol. 1995, 250, 333–353. [Google Scholar] [CrossRef] [PubMed]
- Nozinovic, S.; Furtig, B.; Jonker, H.R.; Richter, C.; Schwalbe, H. High-resolution NMR structure of an RNA model system: The 14-mer cUUCGg tetraloop hairpin RNA. Nucleic Acids Res. 2010, 38, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Ennifar, E.; Nikulin, A.; Tishchenko, S.; Serganov, A.; Nevskaya, N.; Garber, M.; Ehresmann, B.; Ehresmann, C.; Nikonov, S.; Dumas, P. The crystal structure of UUCG tetraloop. J. Mol. Biol. 2000, 304, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Borkar, A.N.; Vallurupalli, P.; Camilloni, C.; Kay, L.E.; Vendruscolo, M. Simultaneous NMR characterisation of multiple minima in the free energy landscape of an RNA UUCG tetraloop. Phys. Chem. Chem. Phys. 2017, 19, 2797–2804. [Google Scholar] [CrossRef]
- Nichols, P.J.; Henen, M.A.; Born, A.; Strotz, D.; Guntert, P.; Vogeli, B. High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints. Commun. Biol. 2018, 1, 61. [Google Scholar] [CrossRef]
- Bottaro, S.; Nichols, P.J.; Vogeli, B.; Parrinello, M.; Lindorff-Larsen, K. Integrating NMR and simulations reveals motions in the UUCG tetraloop. Nucleic Acids Res. 2020, 48, 5839–5848. [Google Scholar] [CrossRef]
- Orioli, S.; Larsen, A.H.; Bottaro, S.; Lindorff-Larsen, K. How to learn from inconsistencies: Integrating molecular simulations with experimental data. Prog. Mol. Biol. Transl. Sci. 2020, 170, 123–176. [Google Scholar]
- Liu, B.; Shi, H.; Al-Hashimi, H.M. Developments in solution-state NMR yield broader and deeper views of the dynamic ensembles of nucleic acids. Curr. Opin. Struct. Biol. 2021, 70, 16–25. [Google Scholar] [CrossRef]
- Nichols, P.J.; Bevers, S.; Henen, M.; Kieft, J.S.; Vicens, Q.; Vogeli, B. Recognition of non-CpG repeats in Alu and ribosomal RNAs by the Z-RNA binding domain of ADAR1 induces A-Z junctions. Nat. Commun. 2021, 12, 793. [Google Scholar] [CrossRef]
- D’Ascenzo, L.; Vicens, Q.; Auffinger, P. Identification of receptors for UNCG and GNRA Z-turns and their occurrence in rRNA. Nucleic Acids Res. 2018, 46, 7989–7997. [Google Scholar] [CrossRef] [PubMed]
- Brzezinski, K.; Brzuszkiewicz, A.; Dauter, M.; Kubicki, M.; Jaskolski, M.; Dauter, Z. High regularity of Z-DNA revealed by ultra high-resolution crystal structure at 0.55 Å. Nucleic Acids Res. 2011, 39, 6238–6248. [Google Scholar] [CrossRef] [PubMed]
- Sokoloski, J.E.; Godfrey, S.A.; Dombrowski, S.E.; Bevilacqua, P.C. Prevalence of syn nucleobases in the active sites of functional RNAs. RNA 2011, 17, 1775–1787. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.P.; Dauter, M.; Dauter, Z. Phosphates in the Z-DNA dodecamer are flexible, but their P-SAD signal is sufficient for structure solution. Acta Cryst. 2014, D70, 1790–1800. [Google Scholar] [CrossRef]
- Martinez, C.R.; Iverson, B.L. Rethinking the term “pi-stacking”. Chem. Sci. 2012, 3, 2191–2201. [Google Scholar] [CrossRef]
- Carter-Fenk, K.; Herbert, J.M. Reinterpreting π-stacking. Phys. Chem. Chem. Phys. 2020, 22, 24870–24886. [Google Scholar] [CrossRef]
- Mrazikova, K.; Sponer, J.; Mlynsky, V.; Auffinger, P.; Kruse, H. Short-range imbalances in the AMBER Lennard-Jones potential for (deoxy)ribose…nucleobase lone-pair…π contacts in nucleic acids. J. Chem. Inf. Model. 2021, 61, 5644–5657. [Google Scholar] [CrossRef]
- Drew, H.R.; Dickerson, R.E. Conformation and dynamics in a Z-DNA tetramer. J. Mol. Biol. 1981, 152, 723–736. [Google Scholar] [CrossRef]
- Wang, A.H.J.; Quigley, G.J.; Kolpak, F.J.; Vandermarel, G.; Vanboom, J.H.; Rich, A. Left-handed double helical DNA—Variations in the backbone conformation. Science 1981, 211, 171–176. [Google Scholar] [CrossRef]
- Gessner, R.V.; Frederick, C.A.; Quigley, G.J.; Rich, A.; Wang, A.H.J. The molecular structure of the left-handed Z-DNA double helix at 1.0 Å atomic resolution. Geometry, conformation, and ionic interactions of d(CGCGCG). J. Biol. Chem. 1989, 264, 7912–7935. [Google Scholar] [CrossRef]
- Svozil, D.; Kalina, J.; Omelka, M.; Schneider, B. DNA conformations and their sequence preferences. Nucleic Acids Res. 2008, 36, 3690–3706. [Google Scholar] [CrossRef] [PubMed]
- Saenger, W. Principles of Nucleic Acid Structure; Springer: New York, NY, USA, 1984. [Google Scholar]
- Banas, P.; Hollas, D.; Zgarbova, M.; Jurecka, P.; Orozco, M.; Cheatham III, T.E.; Sponer, J.; Otyepka, M. Performance of molecular mechanics force fields for RNA simulations: Stability of UUCG and GNRA hairpins. J. Chem. Theory. Comput. 2010, 6, 3836–3849. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.A.; Garcia, A.E. High-resolution reversible folding of hyperstable RNA tetraloops using molecular dynamics simulations. Proc. Natl. Acad. Sci. USA 2013, 110, 16820–16825. [Google Scholar] [CrossRef]
- Kührova, P.; Banas, P.; Best, R.B.; Sponer, J.; Otyepka, M. Computer folding of RNA tetraloops? Are we there yet? J. Chem. Theory Comput. 2013, 9, 2115–2125. [Google Scholar] [CrossRef] [PubMed]
- Bergonzo, C.; Henriksen, N.M.; Roe, D.R.; Cheatham, T.E., 3rd. Highly sampled tetranucleotide and tetraloop motifs enable evaluation of common RNA force fields. RNA 2015, 21, 1578–1590. [Google Scholar] [CrossRef]
- Haldar, S.; Kuhrova, P.; Banas, P.; Spiwok, V.; Sponer, J.; Hobza, P.; Otyepka, M. Insights into stability and folding of GNRA and UNCG tetraloops revealed by microsecond molecular dynamics and well-tempered metadynamics. J. Chem. Theory Comput. 2015, 11, 3866–3877. [Google Scholar] [CrossRef] [PubMed]
- Giambasu, G.M.; York, D.M.; Case, D.A. Structural fidelity and NMR relaxation analysis in a prototype RNA hairpin. RNA 2015, 21, 963–974. [Google Scholar] [CrossRef]
- Bottaro, S.; Banas, P.; Sponer, J.; Bussi, G. Free energy landscape of GAGA and UUCG RNA tetraloops. J. Phys. Chem. Lett. 2016, 7, 4032–4038. [Google Scholar] [CrossRef]
- Aytenfisu, A.H.; Spasic, A.; Grossfield, A.; Stern, H.A.; Mathews, D.H. Revised RNA dihedral parameters for the Amber force field improve RNA molecular dynamics. J. Chem. Theory Comput. 2017, 13, 900–915. [Google Scholar] [CrossRef]
- Kuhrova, P.; Best, R.B.; Bottaro, S.; Bussi, G.; Sponer, J.; Otyepka, M.; Banas, P. Computer folding of RNA tetraloops: Identification of key force field deficiencies. J. Chem. Theory Comput. 2016, 12, 4534–4548. [Google Scholar] [CrossRef]
- Sponer, J.; Bussi, G.; Krepl, M.; Banas, P.; Bottaro, S.; Cunha, R.A.; Gil-Ley, A.; Pinamonti, G.; Poblete, S.; Jurecka, P.; et al. RNA structural dynamics as captured by molecular simulations: A comprehensive overview. Chem. Rev. 2018, 118, 4177–4338. [Google Scholar] [CrossRef] [PubMed]
- Zgarbova, M.; Sponer, J.; Jurecka, P. Z-DNA as a touchstone for additive empirical force fields and a refinement of the alpha/gamma DNA torsions for AMBER. J. Chem. Theory Comput. 2021, 17, 6292–6301. [Google Scholar] [CrossRef] [PubMed]
- Mrazikova, K.; Mlynsky, V.; Kuhrova, P.; Pokorna, P.; Kruse, H.; Krepl, M.; Otyepka, M.; Banas, P.; Sponer, J. UUCG RNA tetraloop as a formidable force-field challenge for MD simulations. J. Chem. Theory. Comput. 2020, 16, 7601–7617. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.; Piana, S.; Dirks, R.M.; Shaw, D.E. RNA force field with accuracy comparable to state-of-the-art protein force fields. Proc. Natl. Acad. Sci. USA 2018, 115, E1346–E1355. [Google Scholar] [CrossRef]
- Cesari, A.; Bottaro, S.; Lindorff-Larsen, K.; Banas, P.; Sponer, J.; Bussi, G. Fitting corrections to an RNA force field using experimental data. J. Chem. Theory Comput. 2019, 15, 3425–3431. [Google Scholar] [CrossRef]
- Kuhrova, P.; Mlynsky, V.; Zgarbova, M.; Krepl, M.; Bussi, G.; Best, R.B.; Otyepka, M.; Sponer, J.; Banas, P. Improving the performance of the Amber RNA force field by tuning the hydrogen-bonding interactions. J. Chem. Theory Comput. 2019, 15, 3288–3305. [Google Scholar] [CrossRef]
- Chen, J.; Liu, H.; Cui, X.; Li, Z.; Chen, H.F. RNA-specific force field optimization with CMAP and reweighting. J. Chem. Inf. Model. 2022, 62, 372–385. [Google Scholar] [CrossRef]
- Mlynsky, V.; Janecek, M.; Kuhrova, P.; Frohlking, T.; Otyepka, M.; Bussi, G.; Banas, P.; Sponer, J. Toward Convergence in Folding Simulations of RNA Tetraloops: Comparison of Enhanced Sampling Techniques and Effects of Force Field Modifications. J. Chem Theory Comput 2022, 18, 2642–2656. [Google Scholar] [CrossRef]
- Kuhrova, P.; Mlynsky, V.; Zgarbova, M.; Krepl, M.; Bussi, G.; Best, R.B.; Otyepka, M.; Sponer, J.; Banas, P. Correction to “Improving the performance of the Amber RNA force field by tuning the hydrogen-bonding interactions”. J. Chem. Theory Comput. 2020, 16, 818–819. [Google Scholar] [CrossRef]
- Ferner, J.; Villa, A.; Duchardt, E.; Widjajakusuma, E.; Wohnert, J.; Stock, G.; Schwalbe, H. NMR and MD studies of the temperature-dependent dynamics of RNA YNMG-tetraloops. Nucleic Acids Res. 2008, 36, 1928–1940. [Google Scholar] [CrossRef]
- Zgarbova, M.; Luque, F.J.; Sponer, J.; Cheatham, T.E., 3rd; Otyepka, M.; Jurecka, P. Toward improved description of DNA backbone: Revisiting Epsilon and Zeta torsion force field parameters. J. Chem. Theory Comput. 2013, 9, 2339–2354. [Google Scholar] [CrossRef] [PubMed]
- Zgarbova, M.; Sponer, J.; Otyepka, M.; Cheatham, T.E., 3rd; Galindo-Murillo, R.; Jurecka, P. Refinement of the sugar-phosphate backbone torsion beta for AMBER force fields improves the description of Z- and B-DNA. J. Chem. Theory Comput. 2015, 11, 5723–5736. [Google Scholar] [CrossRef]
- Galindo-Murillo, R.; Robertson, J.C.; Zgarbova, M.; Sponer, J.; Otyepka, M.; Jurecka, P.; Cheatham, T.E., 3rd. Assessing the current state of Amber force field modifications for DNA. J. Chem. Theory Comput. 2016, 12, 4114–4127. [Google Scholar] [CrossRef] [PubMed]
- Fakharzadeh, A.; Zhang, J.; Roland, C.; Sagui, C. Novel eGZ-motif formed by regularly extruded guanine bases in a left-handed Z-DNA helix as a major motif behind CGG trinucleotide repeats. Nucleic Acids Res. 2022, 50, 4860–4876. [Google Scholar] [CrossRef] [PubMed]
- Kruse, H.; Sponer, J.; Auffinger, P. Comment on “Evaluating Unexpectedly Short Non-covalent Distances in X-ray Crystal Structures of Proteins with Electronic Structure Analysis”. J. Chem. Inf. Model. 2019, 59, 3605–3608. [Google Scholar] [CrossRef] [PubMed]
- Leontis, N.B.; Zirbel, C.L. Nonredundant 3D structure datasets for RNA knowledge extraction and benchmarking. In RNA 3D Structure Analysis and Prediction; Leontis, N.B., Westhof, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 281–298. [Google Scholar]
- Watson, Z.L.; Ward, F.R.; Meheust, R.; Ad, O.; Schepartz, A.; Banfield, J.F.; Cate, J.H. Structure of the bacterial ribosome at 2 Å resolution. Elife 2020, 9, e60482. [Google Scholar] [CrossRef]
- Liu, Z.; Gutierrez-Vargas, C.; Wei, J.; Grassucci, R.A.; Ramesh, M.; Espina, N.; Sun, M.; Tutuncuoglu, B.; Madison-Antenucci, S.; Woolford, J.L., Jr.; et al. Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit. Proc. Natl. Acad. Sci. USA 2016, 113, 12174–12179. [Google Scholar] [CrossRef] [PubMed]
- Knappenberger, A.J.; Reiss, C.W.; Strobel, S.A. Structures of two aptamers with differing ligand specificity reveal ruggedness in the functional landscape of RNA. Elife 2018, 7, e36381. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Naschberger, A.; Mortezaei, N.; Herrmann, J.M.; Amunts, A. Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi. Nat. Commun. 2020, 11, 5187. [Google Scholar] [CrossRef]
- Casanal, A.; Lohkamp, B.; Emsley, P. Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 2020, 29, 1069–1078. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Dauter, Z.; Wlodawer, A.; Minor, W.; Jaskolski, M.; Rupp, B. Avoidable errors in deposited macromolecular structures: An impediment to efficient data mining. IUCrJ 2014, 1, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Minor, W.; Dauter, Z.; Helliwell, J.R.; Jaskolski, M.; Wlodawer, A. Safeguarding structural data repositories against bad apples. Structure 2016, 24, 216–220. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Leonarski, F.; D’Ascenzo, L.; Auffinger, P. Mg2+ ions: Do they bind to nucleobase nitrogens? Nucleic Acids Res. 2017, 45, 987–1004. [Google Scholar] [CrossRef]
- Wlodawer, A. Stereochemistry and validation of macromolecular structures. Methods Mol. Biol. 2017, 1607, 595–610. [Google Scholar]
- Wlodawer, A.; Dauter, Z.; Porebski, P.J.; Minor, W.; Stanfield, R.; Jaskolski, M.; Pozharski, E.; Weichenberger, C.X.; Rupp, B. Detect, correct, retract: How to manage incorrect structural models. FEBS J. 2018, 285, 444–466. [Google Scholar] [CrossRef]
- Rupp, B.; Wlodawer, A.; Minor, W.; Helliwell, J.R.; Jaskolski, M. Correcting the record of structural publications requires joint effort of the community and journal editors. FEBS J. 2016, 283, 4452–4457. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Roversi, P.; Tronrud, D.E. Ten things I ‘hate’ about refinement. Acta Crystallogr. D Struct. Biol. 2021, 77, 1497–1515. [Google Scholar] [CrossRef]
- Hillen, H.S.; Lavdovskaia, E.; Nadler, F.; Hanitsch, E.; Linden, A.; Bohnsack, K.E.; Urlaub, H.; Richter-Dennerlein, R. Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling. Nat. Commun. 2021, 12, 3672. [Google Scholar] [CrossRef]
- Czudnochowski, N.; Ashley, G.W.; Santi, D.V.; Alian, A.; Finer-Moore, J.; Stroud, R.M. The mechanism of pseudouridine synthases from a covalent complex with RNA, and alternate specificity for U2605 versus U2604 between close homologs. Nucleic Acids Res. 2014, 42, 2037–2048. [Google Scholar] [CrossRef] [PubMed]
- Herbert, A. A genetic instruction code based on DNA conformation. Trends Genet. 2019, 35, 887–890. [Google Scholar] [CrossRef] [PubMed]
- Herbert, A.; Karapetyan, S.; Poptsova, M.; Vasquez, K.M.; Vicens, Q.; Vögeli, B. Special Issue: A, B and Z: The structure, function and genetics of Z-DNA and Z-RNA. Int. J. Mol. Sci. 2021, 22, 7686. [Google Scholar] [CrossRef] [PubMed]
- Stombaugh, J.; Zirbel, C.L.; Westhof, E.; Leontis, N.B. Frequency and isostericity of RNA base pairs. Nucleic Acids Res. 2009, 37, 2294–2312. [Google Scholar] [CrossRef]
- Boerema, A.P.; Aibara, S.; Paul, B.; Tobiasson, V.; Kimanius, D.; Forsberg, B.O.; Wallden, K.; Lindahl, E.; Amunts, A. Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nat. Plants 2018, 4, 212–217. [Google Scholar] [CrossRef]
- Auffinger, P.; Louise-May, S.; Westhof, E. Molecular dynamics simulations of the anticodon hairpin of tRNA(asp): Structuring effects of C-H…O hydrogen bonds and of long-range hydration forces. J. Am. Chem. Soc. 1996, 118, 1181–1189. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zirbel, C.L.; Auffinger, P. Lone Pair…π Contacts and Structure Signatures of r(UNCG) Tetraloops, Z-Turns, and Z-Steps: A WebFR3D Survey. Molecules 2022, 27, 4365. https://doi.org/10.3390/molecules27144365
Zirbel CL, Auffinger P. Lone Pair…π Contacts and Structure Signatures of r(UNCG) Tetraloops, Z-Turns, and Z-Steps: A WebFR3D Survey. Molecules. 2022; 27(14):4365. https://doi.org/10.3390/molecules27144365
Chicago/Turabian StyleZirbel, Craig L., and Pascal Auffinger. 2022. "Lone Pair…π Contacts and Structure Signatures of r(UNCG) Tetraloops, Z-Turns, and Z-Steps: A WebFR3D Survey" Molecules 27, no. 14: 4365. https://doi.org/10.3390/molecules27144365
APA StyleZirbel, C. L., & Auffinger, P. (2022). Lone Pair…π Contacts and Structure Signatures of r(UNCG) Tetraloops, Z-Turns, and Z-Steps: A WebFR3D Survey. Molecules, 27(14), 4365. https://doi.org/10.3390/molecules27144365





