Construction of ssDNA-Attached LR-Chimera Involving Z-DNA for ZBP1 Binding Analysis
Abstract
1. Introduction
2. Results and Discussion
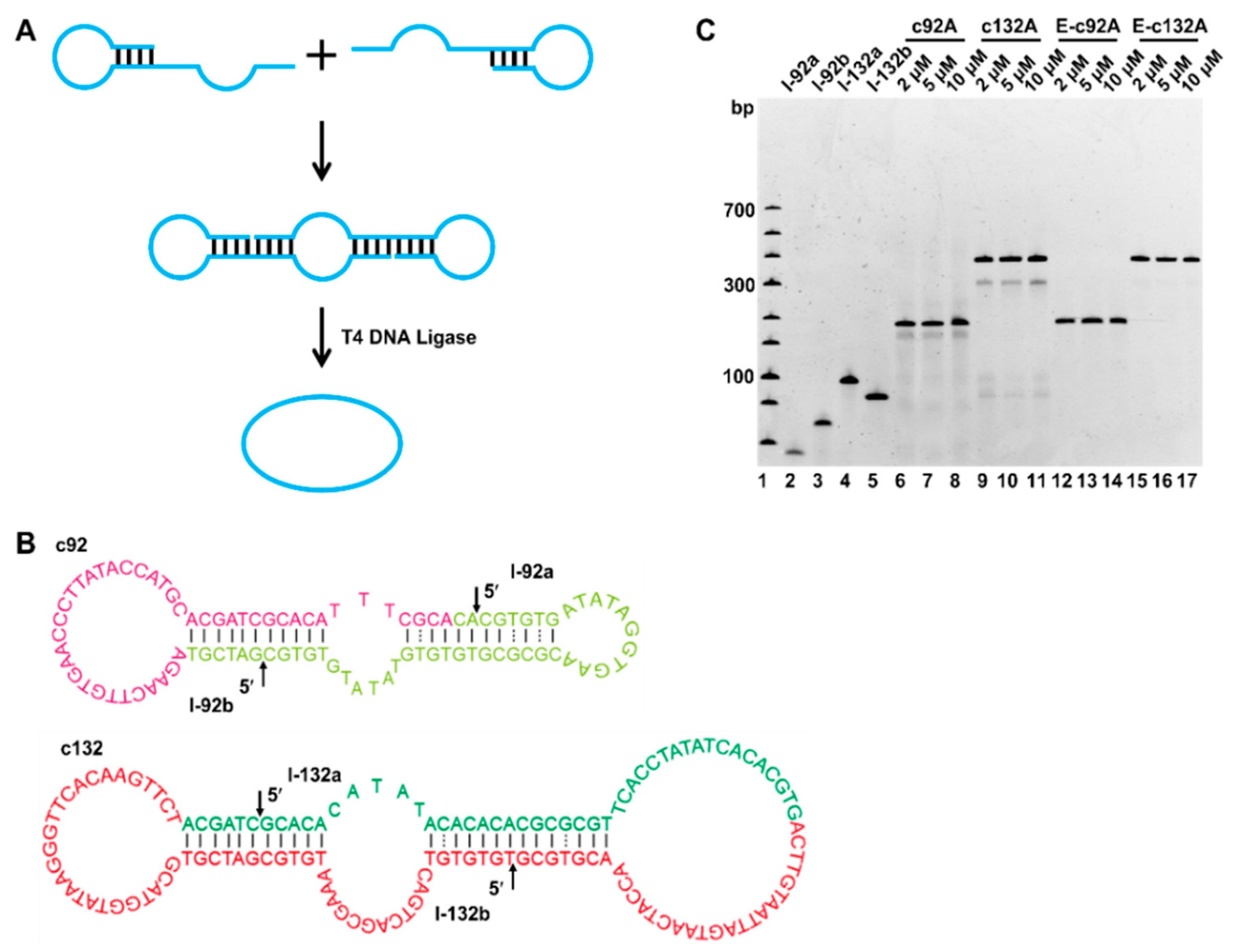
2.1. Preparation of Circular ssDNA by Two DNA Hairpins
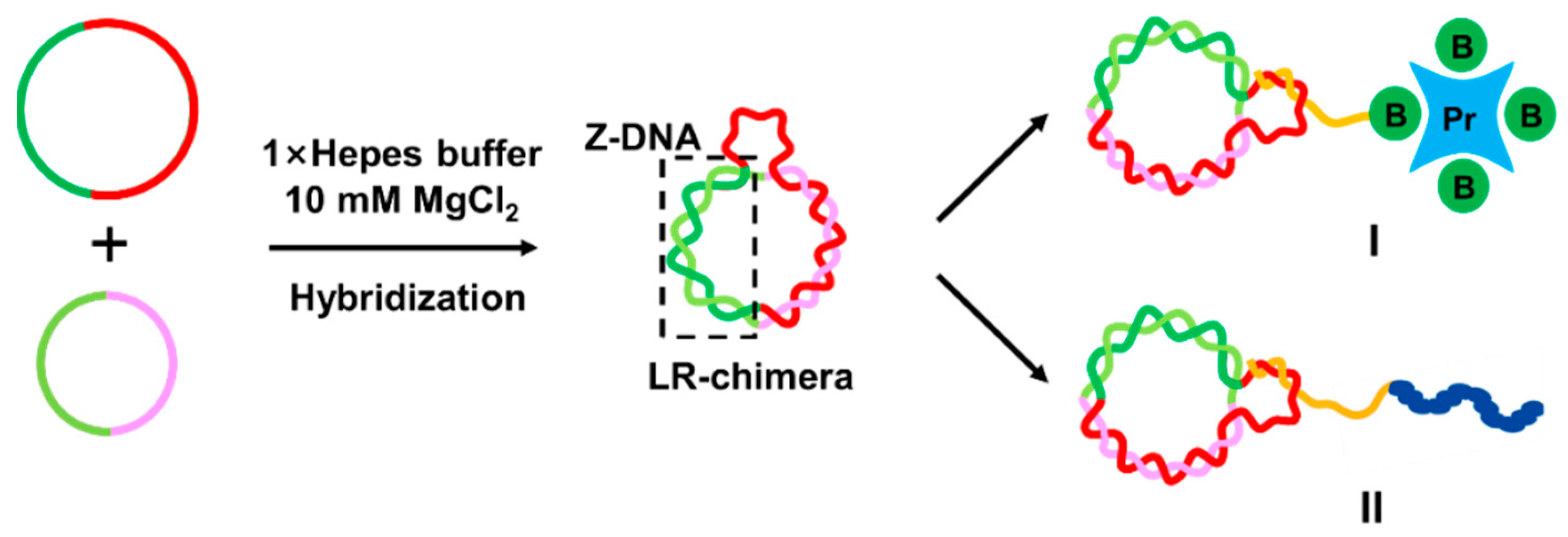
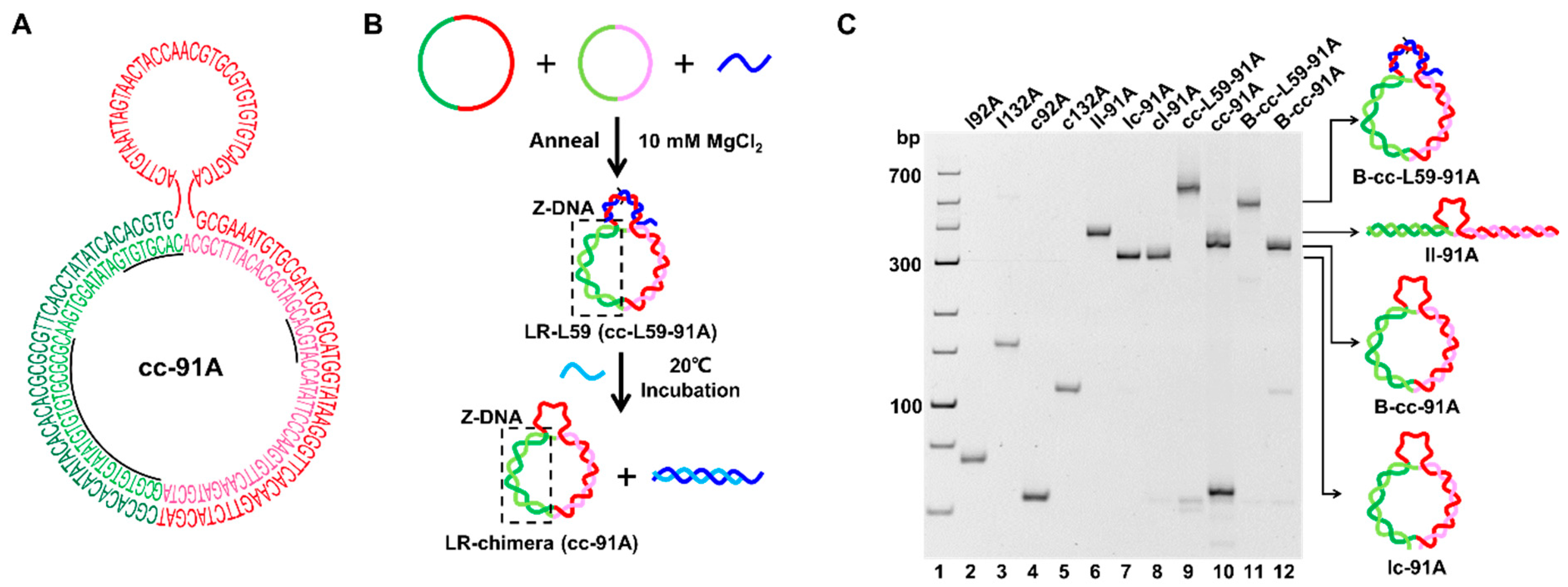
2.2. Preparation of LR-Chimera Involving Partly an APP Sequence and an ssDNA Part for Extra Hybridization
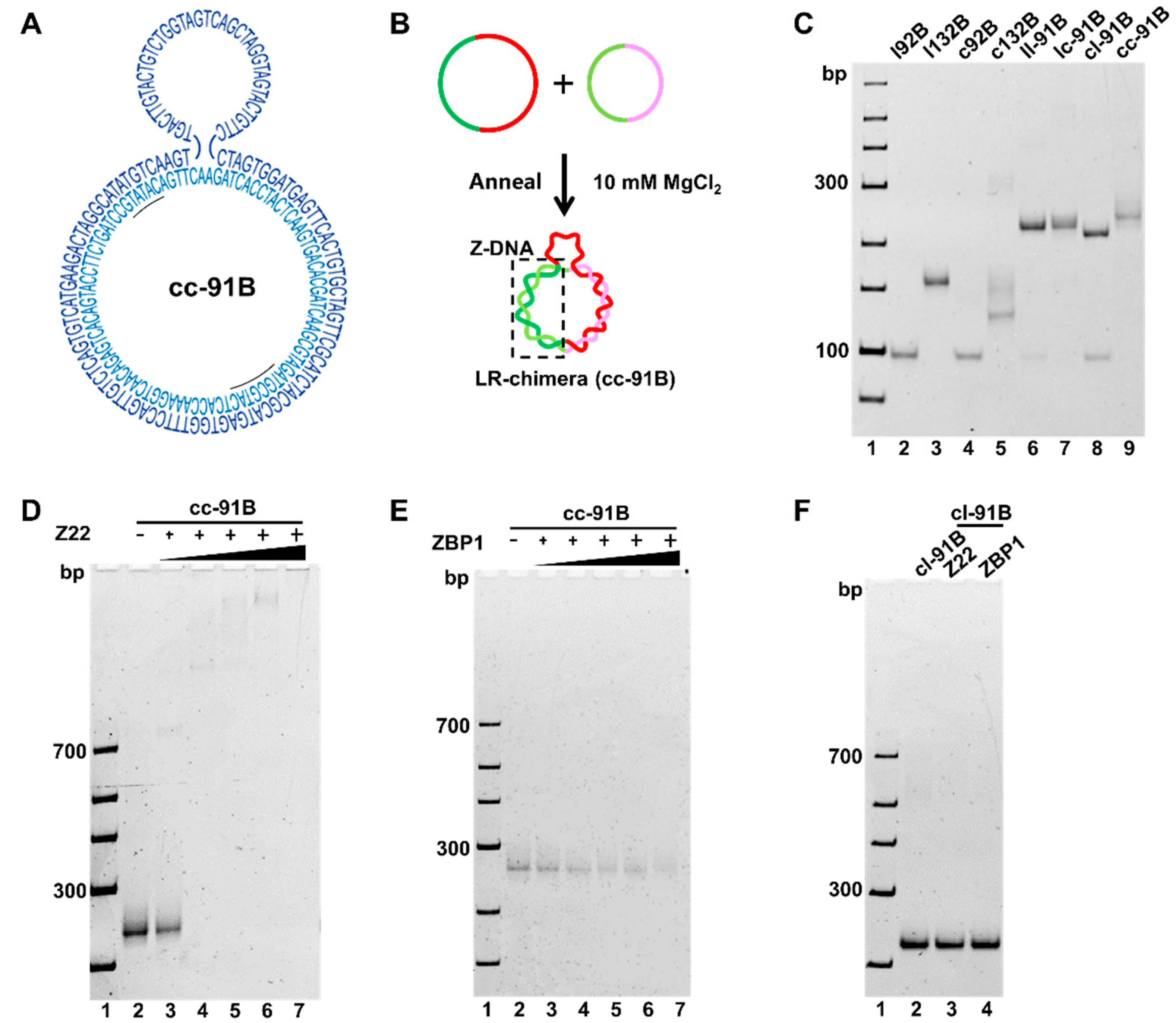
2.3. Confirmation of the Z-DNA Part Present in cc-91A Using Z-DNA-Specific Binding Proteins
2.4. Preparation and Characterization of APP-Poor LR-Chimera with an ssDNA Hook
2.5. Evaluation of ZBP1 Binding Affinity to Non-Modified Z-DNA
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Preparation of Circular ssDNA
4.3. Preparation of LR-Chimera with an ssDNA Part
4.4. Preparation of Bait
4.5. Evaluation of Affinity between ZBP1 and Z-DNA
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Sample Availability
References
- Pohl, F.M.; Jovin, T.M. Salt-induced co-operative conformational change of a synthetic DNA: Equilibrium and kinetic studies with poly (dG-dC). J. Mol. Biol. 1972, 67, 375–396. [Google Scholar] [CrossRef]
- Wang, A.H.J.; Quigley, G.J.; Kolpak, F.J.; Crawford, J.L.; Van Boom, J.H.; van der Marel, G.; Rich, A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 1979, 282, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Nishino, M.; Ikegami, H.; Fujisawa, T.; Kawaguchi, Y.; Kawabata, Y.; Shintani, M.; Ono, M.; Ogihara, T. Functional polymorphism in Z-DNA-forming motif of promoter of SLC11A1 gene and type 1 diabetes in Japanese subjects: Association study and meta-analysis. Metab. Clin. Exp. 2005, 54, 628–633. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Kolimi, N.; Rathinavelan, T. Twisting right to left: A…A mismatch in a CAG trinucleotide repeat overexpansion provokes left-handed Z-DNA conformation. PLoS Comput. Biol. 2015, 11, e1004162. [Google Scholar] [CrossRef]
- Geng, J.; Zhao, C.; Ren, J.; Qu, X. Alzheimer’s disease amyloid beta converting left-handed Z-DNA back to right-handed B-form. Chem. Commun. 2010, 46, 7187–7189. [Google Scholar] [CrossRef]
- Suram, A.; Rao, J.K.; Latha, K.S.; Viswamitra, M.A. First evidence to show the topological change of DNA from B-DNA to Z-DNA conformation in the hippocampus of Alzheimer’s brain. Neuromol. Med. 2002, 2, 289–297. [Google Scholar] [CrossRef]
- Ray, B.K.; Dhar, S.; Henry, C.; Rich, A.; Ray, A. Epigenetic regulation by Z-DNA silencer function controls cancer-associated ADAM-12 expression in breast cancer: Cross-talk between MeCP2 and NF1 transcription factor family. Cancer Res. 2013, 73, 736–744. [Google Scholar] [CrossRef]
- Bacolla, A.; Tainer, J.A.; Vasquez, K.M.; Cooper, D.N. Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic Acids Res. 2016, 44, 5673–5688. [Google Scholar] [CrossRef]
- Ray, B.K.; Dhar, S.; Shakya, A.; Ray, A. Z-DNA-forming silencer in the first exon regulates human ADAM-12 gene expression. Proc. Natl. Acad. Sci. USA 2011, 108, 103–108. [Google Scholar] [CrossRef]
- Oh, D.B.; Kim, Y.G.; Rich, A. Z-DNA-binding proteins can act as potent effectors of gene expression in vivo. Proc. Natl. Acad. Sci. USA 2002, 99, 16666–16671. [Google Scholar] [CrossRef]
- Haniford, D.B.; Pulleyblank, D.E. The in-vivo occurrence of Z DNA. J. Biomol. Struct. Dyn. 1983, 1, 593–609. [Google Scholar] [CrossRef] [PubMed]
- Assil, S.; Paludan, S.R. Live and let die: ZBP1 senses viral and cellular RNAs to trigger necroptosis. EMBO J. 2017, 36, 2470–2472. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Kumari, S.; Kim, C.; Van, T.M.; Wachsmuth, L.; Polykratis, A.; Pasparakis, M. RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nature 2016, 540, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Hu, Y.; Fan, L.; Wang, H.; Sun, Z.; Deng, S.; Liu, Y.; Hu, C. Ctenopharyngodon idella PKZ facilitates cell apoptosis through phosphorylating eIF2α. Mol. Immunol. 2016, 69, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Deigendesch, N.; Koch-Nolte, F.; Rothenburg, S. ZBP1 subcellular localization and association with stress granules is controlled by its Z-DNA binding domains. Nucleic Acids Res. 2006, 34, 5007–5020. [Google Scholar] [CrossRef]
- Kwon, J.A.; Rich, A. Biological function of the vaccinia virus Z-DNA-binding protein E3L: Gene transactivation and antiapoptotic activity in HeLa cells. Proc. Natl. Acad. Sci. USA 2005, 102, 12759–12764. [Google Scholar] [CrossRef]
- Runkel, L.; Nordheim, A. Chemical footprinting of the interaction between left-handed Z-DNA and anti-Z-DNA antibodies by diethylpyrocarbonate carbethoxylation. J. Mol. Biol. 1986, 189, 487–501. [Google Scholar] [CrossRef]
- Herbert, A.; Alfken, J.; Kim, Y.G.; Mian, I.S.; Nishikura, K.; Rich, A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl. Acad. Sci. USA 1997, 94, 8421–8426. [Google Scholar] [CrossRef]
- Van Quyen, D.; Ha, S.C.; Lowenhaupt, K.; Rich, A.; Kim, K.K.; Kim, Y.G. Characterization of DNA-binding activity of Za domains from poxviruses and the importance of the β-wing regions in converting B-DNA to Z-DNA. Nucleic Acids Res. 2007, 35, 7714–7720. [Google Scholar] [CrossRef][Green Version]
- Polymenis, M.; Stollar, B.D. Domain interactions and antigen binding of recombinant anti-Z-DNA antibody variable domains. The role of heavy and light chains measured by surface plasmon resonance. J. Immunol. 1995, 154, 2198–2208. [Google Scholar]
- Schwartz, T.; Behlke, J.; Lowenhaupt, K.; Heinemann, U.; Rich, A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 2001, 8, 761–765. [Google Scholar] [CrossRef] [PubMed]
- Sultana, A.; Lee, J.E. Measuring Protein-protein and protein-nucleic acid interactions by biolayer interferometry. Curr. Protoc. Protein Sci. 2015, 79, 19.25.1–19.25.26. [Google Scholar] [CrossRef] [PubMed]
- Ziu, I.; Laryea, E.T.; Alashkar, F.; Wu, C.G.; Martic, S. A dip-and-read optical aptasensor for detection of tau protein. Anal. Bioanal. Chem. 2020, 412, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Tao, L.; Wang, T.; Zhang, J.; He, A.; Lam, K.H.; Liu, Z.; He, X.; Perry, K.; Dong, M.; et al. Structural basis for recognition of frizzled proteins by Clostridium difficile toxin B. Science 2018, 360, 664–669. [Google Scholar] [CrossRef]
- Tao, L.; Zhang, J.; Meraner, P.; Tovaglieri, A.; Wu, X.; Gerhard, R.; Zhang, X.; Stallcup, W.B.; Miao, J.; He, X.; et al. Frizzled are colonic epithelial receptors for Clostridium difficile toxin B. Nature 2016, 538, 350–355. [Google Scholar] [CrossRef]
- Amin-Wetzel, N.; Saunders, R.A.; Kamphuis, M.J.; Rato, C.; Preissler, S.; Harding, H.P.; Ron, D. A J-protein co-chaperone recruits BiP to monomerize IRE1 and repress the unfolded protein response. Cell 2017, 171, 1625–1637. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Wang, X.; Jin, W.; Yun Tan, Y.; Fang, H.; Chen, S.; Chen, Z.; Wang, K. Genome-wide studies identify a novel interplay between AML1 and AML1/ETO in t (8;21) acute myeloid leukemia. Blood 2016, 127, 233–242. [Google Scholar] [CrossRef]
- Gao, S.; Hu, B.; Zheng, X.; Cao, Y.; Liu, D.; Sun, M.; Jiao, B.; Wang, L. Gonyautoxin 1/4 aptamers with high-affinity and high-specificity: From efficient selection to aptasensor application. Biosens. Bioelectron. 2016, 79, 938–944. [Google Scholar] [CrossRef]
- Gao, S.; Zheng, X.; Hu, B.; Sun, M.; Wu, J.; Jiao, B.; Wang, L. Enzyme-linked, aptamer-based, competitive biolayer interferometry biosensor for palytoxin. Biosens. Bioelectron. 2017, 89, 952–958. [Google Scholar] [CrossRef]
- Zhang, Y.; Cui, Y.; An, R.; Liang, X.; Li, Q.; Wang, H.; Wang, H.; Fan, Y.; Dong, P.; Li, J.; et al. Topologically constrained formation of stable Z-DNA from normal sequence under physiological conditions. J. Am. Chem. Soc. 2019, 141, 7758–7764. [Google Scholar] [CrossRef]
- An, R.; Li, Q.; Fan, Y.; Li, J.; Pan, X.; Komiyama, M.; Liang, X. Highly efficient preparation of single-stranded DNA rings by T4 ligase at abnormally low Mg(II) concentration. Nucleic Acids Res. 2017, 45, e139. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Han, X.; An, R.; Zhang, Y.; Cheng, K.; Liang, X.; Komiyama, M. Terminal hairpin in oligonucleotide dominantly prioritizes intramolecular cyclization by T4 ligase over intermolecular polymerization: An exclusive methodology for producing ssDNA rings. Nucleic Acids Res. 2018, 46, e132. [Google Scholar] [CrossRef] [PubMed]
- Gubu, A.; Wang, J.; Jin, H.; Tang, X. Synthesis and “DNA interlocks” formation of small circular oligodeoxynucleotides. ACS Appl. Mater. Interfaces 2020, 12, 12584–12590. [Google Scholar] [CrossRef] [PubMed]
- Sui, Z.; Liu, M.; Wang, W.; Chen, H.; Wang, G.; An, R.; Liang, X.; Komiyama, M. Efficient preparation of large-sized rings of single-stranded DNA through one-pot ligation of multiple fragments. Chem. Asian J. 2019, 14, 3251–3254. [Google Scholar] [CrossRef]
- Takaoka, A.; Wang, Z.; Choi, M.K.; Yanai, H.; Negishi, H.; Ban, T.; Lu, Y.; Miyagishi, M.; Kodama, T.; Honda, K.; et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 2007, 448, 501–505. [Google Scholar] [CrossRef]
- Kaiser, W.J.; Upton, J.W.; Mocarski, E.S. Receptor-interacting protein homotypic interaction motif-dependent control of NF-kB activation via the DNA-dependent activator of IFN regulatory factors. J. Immunol. 2008, 181, 6427–6434. [Google Scholar] [CrossRef]
- Sridharan, H.; Ragan, K.B.; Guo, H.; Gilley, R.P.; Landsteiner, V.J.; Kaiser, W.J.; Upton, J.W. Murine cytomegalovirus IE3-dependent transcription is required for DAI/ZBP1-mediated necroptosis. EMBO Rep. 2017, 18, 1429–1441. [Google Scholar] [CrossRef]
- Guo, H.; Gilley, R.P.; Fisher, A.; Lane, R.; Landsteiner, V.J.; Ragan, K.B.; Dovey, C.M.; Carette, J.E.; Upton, J.W.; Mocarski, E.S.; et al. Species-independent contribution of ZBP1/DAI/DLM-1-triggered necroptosis in host defense against HSV1. Cell Death Dis. 2018, 9, 816. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Y.P.; Ma, W.Z.; Chen, H.; Liu, M.Q.; An, R.; Cheng, B.X.; Liang, X.G. Nonalternating purine pyrimidine sequences can form stable left-handed DNA duplex by strong topological constraint. Nucleic Acids Res. 2022, 50, 684–696. [Google Scholar] [CrossRef]
- Möller, A.; Gabriels, J.E.; Lafer, E.M.; Nordheim, A.; Rich, A.; Stollar, B.D. Monoclonal antibodies recognize different parts of Z-DNA. J. Biol. Chem. 1982, 257, 12081–12085. [Google Scholar] [CrossRef]
- Lafer, E.M.; Sousa, R.; Rich, A. Anti-Z-DNA antibody binding can stabilize Z-DNA in relaxed and linear plasmids under physiological conditions. EMBO J. 1985, 4, 3655–3660. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ramelot, T.A.; Lee, H.W.; Xiao, R.; Everrtt, J.K.; Montelione, G.T.; Prestegard, J.H.; Kennedy, M.A. Solution structure of the free Zα domain of human DLM-1 (ZBP1/DAI), a Z-DNA binding domain. J. Biomol. NMR 2014, 60, 189–195. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; An, R.; Liang, X. Construction of ssDNA-Attached LR-Chimera Involving Z-DNA for ZBP1 Binding Analysis. Molecules 2022, 27, 3706. https://doi.org/10.3390/molecules27123706
Li L, An R, Liang X. Construction of ssDNA-Attached LR-Chimera Involving Z-DNA for ZBP1 Binding Analysis. Molecules. 2022; 27(12):3706. https://doi.org/10.3390/molecules27123706
Chicago/Turabian StyleLi, Lin, Ran An, and Xingguo Liang. 2022. "Construction of ssDNA-Attached LR-Chimera Involving Z-DNA for ZBP1 Binding Analysis" Molecules 27, no. 12: 3706. https://doi.org/10.3390/molecules27123706
APA StyleLi, L., An, R., & Liang, X. (2022). Construction of ssDNA-Attached LR-Chimera Involving Z-DNA for ZBP1 Binding Analysis. Molecules, 27(12), 3706. https://doi.org/10.3390/molecules27123706







