Bioactive Phenolic Compounds from Peperomia obtusifolia
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Methods
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Biological activities
3.4.1. Anthelmintic Activity
3.4.2. Antifungal Activity
3.4.3. Antibacterial Activity
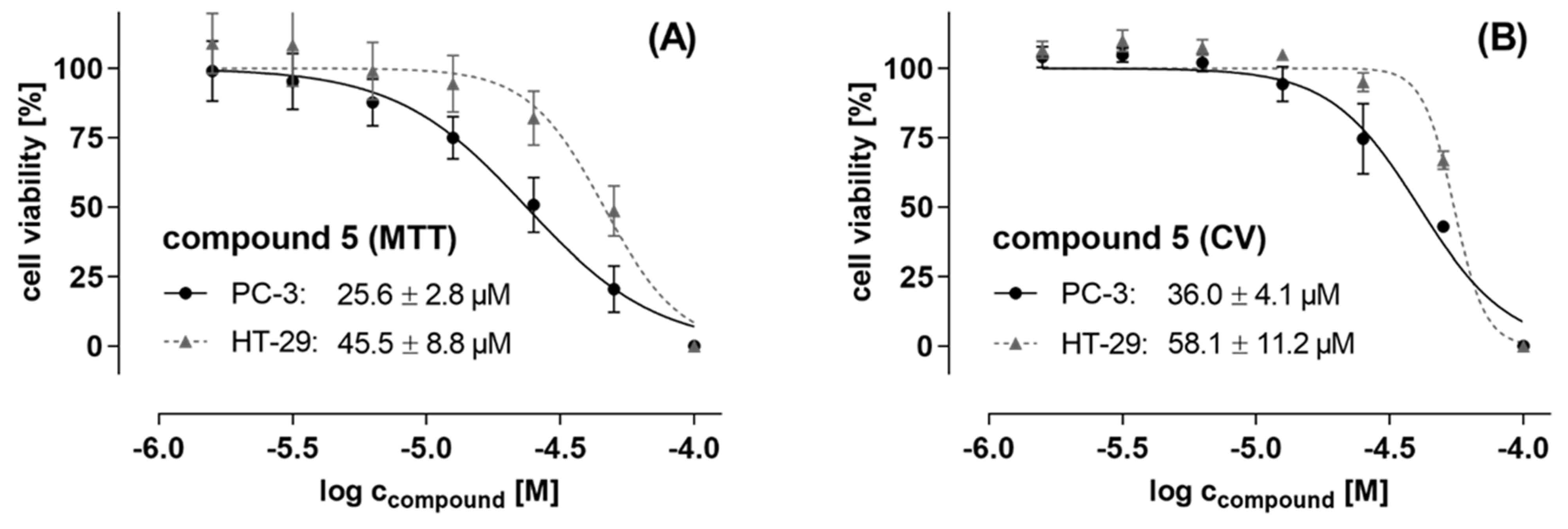
3.4.4. Cytotoxic Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Felippe, L.G.; Baldoqui, D.C.; Kato, M.J.; Bolzani, V.d.S.; Guimarães, E.F.; Cicarelli, R.M.B.; Furlan, M. Trypanocidal tetrahydrofuran lignans from Peperomia blanda. Phytochemistry 2008, 69, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Al-Madhagi, W.M.; Mohd Hashim, N.; Awad Ali, N.A.; Alhadi, A.A.; Abdul Halim, S.N.; Othman, R. Chemical profiling and biological activity of Peperomia blanda (Jacq.) Kunth. PeerJ 2018, 6, e4839. [Google Scholar] [CrossRef] [PubMed]
- Malquichagua Salazar, K.J.; Delgado Paredes, G.E.; Lluncor, L.R.; Young, M.C.M.; Kato, M.J. Chromenes of polyketide origin from Peperomia villipetiola. Phytochemistry 2005, 66, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Seeram, N.P.; Lewis, A.W.; Jacobs, H.; Nair, M.G.; McLean, S.; Reynolds, W.F. Proctoriones A−C: 2-Acylcyclohexane-1,3-dione derivatives from Peperomia proctorii. J. Nat. Prod. 2000, 63, 399–402. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, N.; Hasegawa, T.; Sakai, J.; Kakuta, S.; Tang, W.; Oka, S.; Kiuchi, M.; Ogura, H.; Kataoka, T.; et al. Bioactive tetrahydrofuran lignans from Peperomia dindygulensis. J. Nat. Prod. 2005, 68, 1656–1660. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Li, N.; Ning, M.M.; Zhou, C.H.; Yang, Q.R.; Wang, M.W. Bioactive compounds from Peperomia pellucida. J. Nat. Prod. 2006, 69, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wu, J.; Hasegawa, T.; Sakai, J.; Bai, L.; Wang, L.; Kakuta, S.; Furuya, Y.; Ogura, H.; Kataoka, T.; et al. Bioactive polyketides from Peperomia duclouxii. J. Nat. Prod. 2007, 70, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cheng, Y.; Wu, H.; Li, N.; Liu, R.; Yang, X.; Qiu, Y.Y.; Wen, H.; Liang, J. The natural secolignan peperomin E induces apoptosis of human gastric carcinoma cells via the mitochondrial and PI3K/Akt signaling pathways in vitro and in vivo. Phytomedicine 2016, 23, 818–827. [Google Scholar] [CrossRef] [PubMed]
- Batista, A.N.L.; Santos-Pinto, J.; Batista, J.M., Jr.; Souza-Moreira, T.M.; Santoni, M.M.; Zanelli, C.F.; Kato, M.J.; López, S.N.; Palma, M.S.; Furlan, M. The combined use of proteomics and transcriptomics reveals a complex secondary metabolite network in Peperomia obtusifolia. J. Nat. Prod. 2017, 80, 1275–1286. [Google Scholar] [CrossRef]
- Tanaka, T.; Asai, F.; Iinuma, M. Phenolic compounds from Peperomia obtusifolia. Phytochemistry 1998, 49, 229–232. [Google Scholar] [CrossRef]
- da Silva Mota, J.; Leite, A.C.; Batista Junior, J.M.; Noelí López, S.; Luz Ambrósio, D.; Duó Passerini, G.; Kato, M.J.; da Silva Bolzani, V.; Barretto Cicarelli, R.M.; Furlan, M. In vitro trypanocidal activity of phenolic derivatives from Peperomia obtusifolia. Planta Med. 2009, 75, 620–623. [Google Scholar] [CrossRef]
- Mota, J.d.S.; Leite, A.C.; Kato, M.J.; Young, M.C.M.; Bolzani, V.d.S.; Furlan, M. Isoswertisin flavones and other constituents from Peperomia obtusifolia. Nat. Prod. Res. 2011, 25, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Batista, J.M.; Batista, A.N.L.; Kato, M.J.; Bolzani, V.S.; López, S.N.; Nafie, L.A.; Furlan, M. Further monoterpene chromane esters from Peperomia obtusifolia: VCD determination of the absolute configuration of a new diastereomeric mixture. Tetrahedron Lett. 2012, 53, 6051–6054. [Google Scholar] [CrossRef]
- Ruiz Mostacero, N.; Castelli, M.V.; Cutró, A.C.; Hollmann, A.; Batista, J.M., Jr.; Furlan, M.; Valles, J.; Fulgueira, C.L.; López, S.N. Antibacterial activity of prenylated benzopyrans from Peperomia obtusifolia (Piperaceae). Nat. Prod. Res. 2021, 35, 1706–1710. [Google Scholar] [CrossRef] [PubMed]
- Tamayose, C.I.; Romoff, P.; Toyama, D.O.; Gaeta, H.H.; Costa, C.R.C.; Belchor, M.N.; Ortolan, B.D.; Velozo, L.S.M.; Kaplan, M.A.C.; Ferreira, M.J.P.; et al. Non-clinical studies for evaluation of 8-c-rhamnosyl apigenin purified from Peperomia obtusifolia against acute edema. Int. J. Mol. Sci. 2017, 18, 1972. [Google Scholar] [CrossRef]
- Wu, W.; Zhao, F.; Ding, R.; Bao, L.; Gao, H.; Lu, J.C.; Yao, X.S.; Zhang, X.Q.; Liu, H.W. Four new cryptoporic acid derivatives from the fruiting bodies of Cryptoporus sinensis, and their inhibitory effects on nitric oxide production. Chem. Biodivers. 2011, 8, 1529–1538. [Google Scholar] [CrossRef]
- Zhou, L.Y.; Yu, X.H.; Lu, B.; Hua, Y. Bioassay-guided isolation of cytotoxic isocryptoporic acids from Cryptoporus volvatus. Molecules 2016, 21, 1692. [Google Scholar] [CrossRef] [PubMed]
- Pham, H.T.; Lee, K.H.; Jeong, E.; Woo, S.; Yu, J.; Kim, W.Y.; Lim, Y.W.; Kim, K.H.; Kang, K.B. Species prioritization based on spectral dissimilarity: A case study of polyporoid fungal species. J. Nat. Prod. 2021, 84, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Chepkirui, C.; Yuyama, K.T.; Wanga, L.A.; Decock, C.; Matasyoh, J.C.; Abraham, W.-R.; Stadler, M. Microporenic acids A–G, biofilm inhibitors, and antimicrobial agents from the Basidiomycete Microporus species. J. Nat. Prod. 2018, 81, 778–784. [Google Scholar] [CrossRef]
- Liao, Y.; Ye, Y.; Li, S.; Zhuang, Y.; Chen, L.; Chen, J.; Cui, Z.; Huo, L.; Liu, S.; Song, G. Synthesis and SARs of dopamine derivatives as potential inhibitors of influenza virus PAN endonuclease. Eur. J. Med. Chem. 2020, 189, 112048. [Google Scholar] [CrossRef]
- Park, J.B. Protective effects of veskamide, enferamide, becatamide, and oretamide on H2O2-induced apoptosis of PC-12 cells. Phytomedicine 2011, 18, 843–847. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-M.; Jan, F.Y.; Chen, M.-T.; Lee, T.-J. Peperomins A, B, and C, novel secolignans from Peperomia japonica. Heterocycles 1989, 29, 411–414. [Google Scholar] [CrossRef]
- Li, H.X.; Yang, S.Y.; Kim, Y.S.; Jang, H.D.; Kim, Y.H.; Li, W. Nitro derivatives and other compounds from sugar apple (Annona squamosa L.) leaves exhibit soluble epoxide hydrolase inhibitory activity. Med. Chem. Res. 2019, 28, 1939–1944. [Google Scholar] [CrossRef]
- Li, Y.Z.; Tong, A.P.; Huang, J. Two new norlignans and a new lignanamide from Peperomia tetraphylla. Chem. Biodivers. 2012, 9, 769–776. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Yamaguchi, S.; Murakami, T.; Matsuda, H.; Yamahara, J.; Murakami, N. Absolute stereostructures of trifoliones A, B, C, and D, new biologically active diterpenes from the tuber of Sagittaria trifolia L. Chem. Pharm. Bull. 1993, 41, 1677–1679. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, Z.; Luo, J.; Pan, K.; Kong, L. A new alkaloid glycoside from the rhizomes of Aristolochia fordiana. Nat. Prod. Res. 2014, 28, 1065–1069. [Google Scholar] [CrossRef]
- Alves, N.S.F.; Setzer, W.N.; da Silva, J.K.R. The chemistry and biological activities of Peperomia pellucida (Piperaceae): A critical review. J. Ethnopharmacol. 2019, 232, 90–102. [Google Scholar] [CrossRef]
- Ng, Z.X.; Than, M.J.Y.; Yong, P.H. Peperomia pellucida (L.) Kunth herbal tea: Effect of fermentation and drying methods on the consumer acceptance, antioxidant and anti-inflammatory activities. Food Chem. 2021, 344, 128738. [Google Scholar] [CrossRef]
- Gutierrez, Y.V.; Yamaguchi, L.F.; de Moraes, M.M.; Jeffrey, C.S.; Kato, M.J. Natural products from Peperomia: Occurrence, biogenesis and bioactivity. Phytochem Rev. 2016, 15, 1009–1033. [Google Scholar] [CrossRef]
- Batista, J.M., Jr.; Lopez, S.N.; Mota Jda, S.; Vanzolini, K.L.; Cass, Q.B.; Rinaldo, D.; Vilegas, W.; Bolzani Vda, S.; Kato, M.J.; Furlan, M. Resolution and absolute configuration assignment of a natural racemic chromane from Peperomia obtusifolia (Piperaceae). Chirality 2009, 21, 799–801. [Google Scholar] [CrossRef]
- Shi, L.; Qin, H.; Jin, X.; Yang, X.; Lu, X.; Wang, H.; Wang, R.; Yu, D.; Feng, B. The natural phenolic peperobtusin A induces apoptosis of lymphoma U937 cells via the Caspase dependent and p38 MAPK signaling pathways. Biomed. Pharmacother. 2018, 102, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Seeram, N.P.; Jacobs, H.; McLean, S.; Reynolds, W.F. A prenylated benzopyran derivative from Peperomia clusiifolia. Phytochemistry 1998, 49, 1389–1391. [Google Scholar] [CrossRef]
- Thomsen, H.; Reider, K.; Franke, K.; Wessjohann, L.A.; Keiser, J.; Dagne, E.; Arnold, N. Characterization of constituents and anthelmintic properties of Hagenia abyssinica. Sci. Pharm. 2012, 80, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Otto, A.; Porzel, A.; Schmidt, J.; Brandt, W.; Wessjohann, L.; Arnold, N. Structure and absolute configuration of pseudohygrophorones A12 and B12, alkyl cyclohexenone derivatives from Hygrophorus abieticola (Basidiomycetes). J. Nat. Prod. 2016, 79, 74–80. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, C.H.C.; de Carvalho, M.G.; Franke, K.; Wessjohann, L. Dammarane-type triterpenoids from the stem of Ziziphus glaziovii Warm. (Rhamnaceae). Phytochemistry 2019, 162, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.F.; Nasr, F.A.; Noman, O.M.; Alyhya, N.A.; Ali, I.; Saoud, M.; Rennert, R.; Dube, M.; Hussain, W.; Green, I.R.; et al. Cichorins D-F: Three new compounds from Cichorium intybus and their biological effects. Molecules 2020, 25, 4160. [Google Scholar] [CrossRef]
- Krammer, G.; Ley, J.P.; Geißler, K.; Geißler, T.; Gomoll, F.; Welters, P.; Jach, G.; Wessjohann, L. Method for Biotechnological Production of Methylized Cinnamic Acids and Cinnamic Acid Esters, Methylized Phenethylamines and the Coupling Products Thereof, Particularly of Cinnamic Acid Amides. U.S. Patent US10227620B2, 12 March 2019. [Google Scholar]
- Greger, H, Alkamides: A critical reconsideration of a multifunctional class of unsaturated fatty acid amides. Phytochem. Rev. 2016, 15, 729–770. [CrossRef]
- Taha, H.; Looi, C.Y.; Arya, A.; Wong, W.F.; Yap, L.F.; Hasanpourghadi, M.; Mohd, M.A.; Paterson, I.C.; Ali, H.A. (6E,10E) Isopolycerasoidol and (6E,10E) isopolycerasoidol methyl ester, prenylated benzopyran derivatives from Pseuduvaria monticola induce mitochondrial-mediated apoptosis in human breast adenocarcinoma cells. PLoS ONE 2015, 10, e0126126. [Google Scholar] [CrossRef] [PubMed]
- Zafra-Polo, M.C.; González, M.C.; Tormo, J.R.; Estornell, E.; Cortes, D. Polyalthidin: New prenylated benzopyran inhibitor of the mammalian mitochondrial respiratory chain. J. Nat. Prod. 1996, 10, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Corés Martinez, D.M.; Sanz Ferrando, M.J.; Bermejo del Castillo, A.; Piqueras Ruiz, L.; Collado Sánchez, A.; Gomes Marques, P.; Vila Dasí, L. Prenylated Benzopyranes as PPAR. Agonists Patent WO2019129909, 4 July 2019. [Google Scholar]
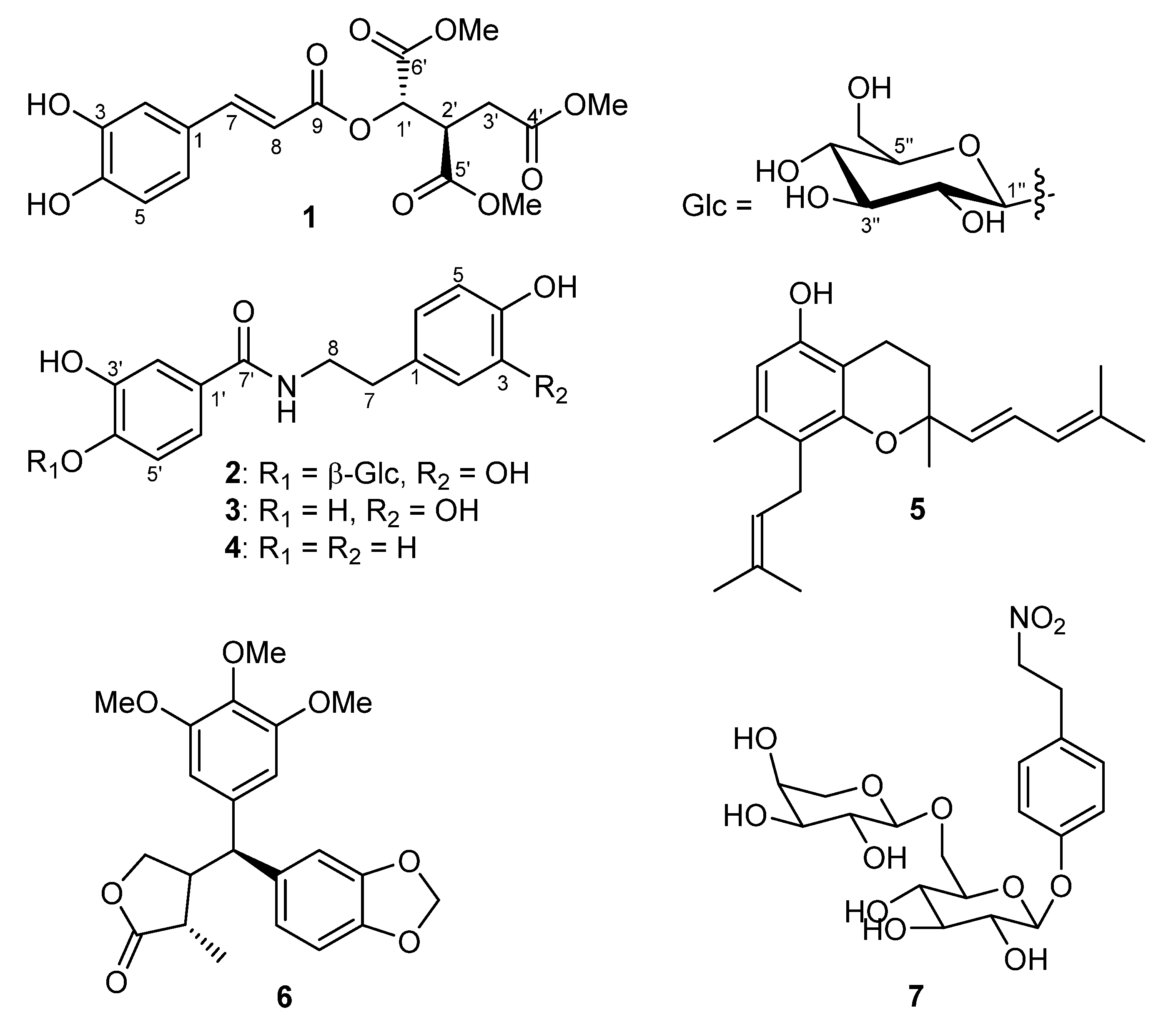
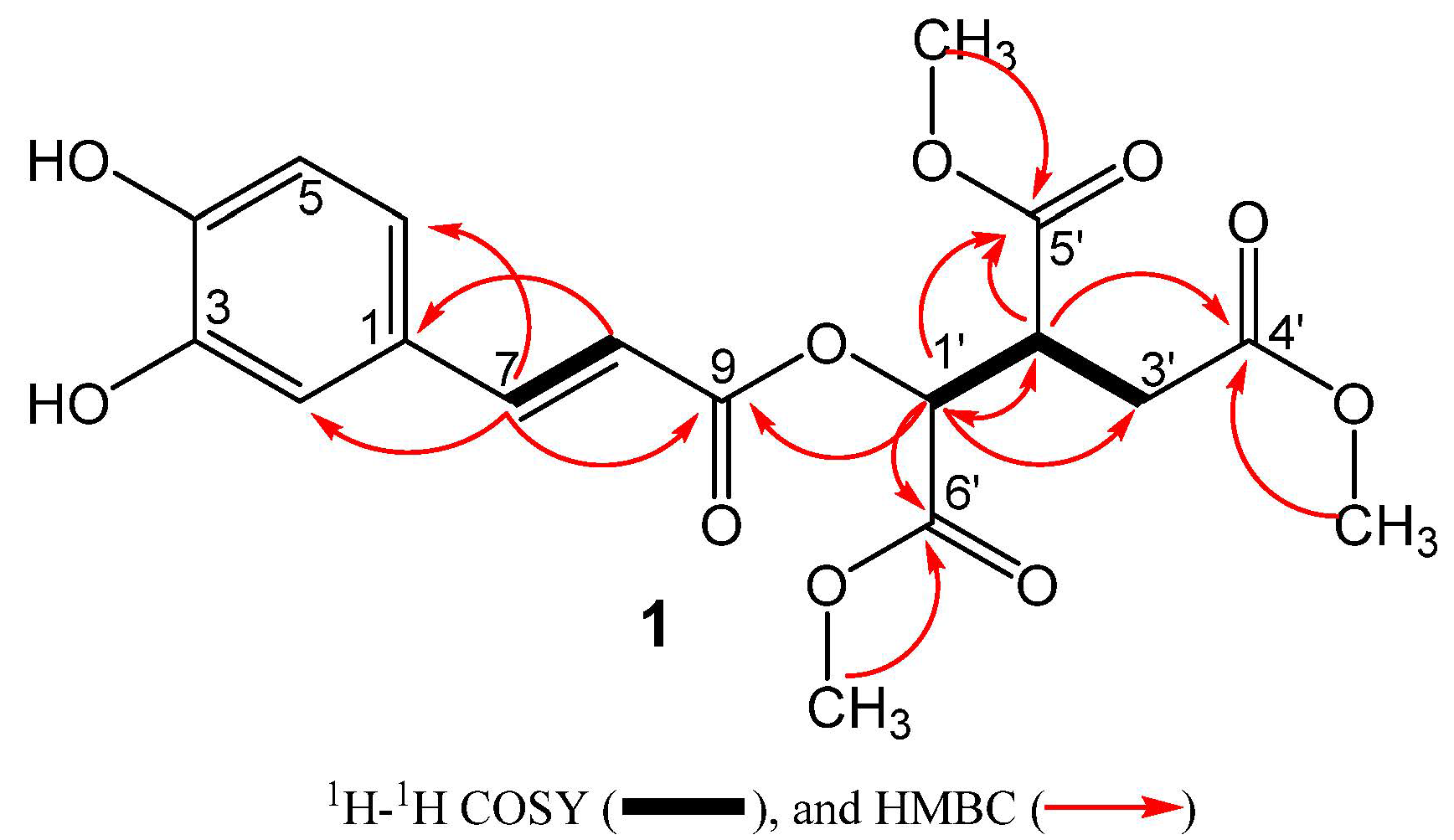
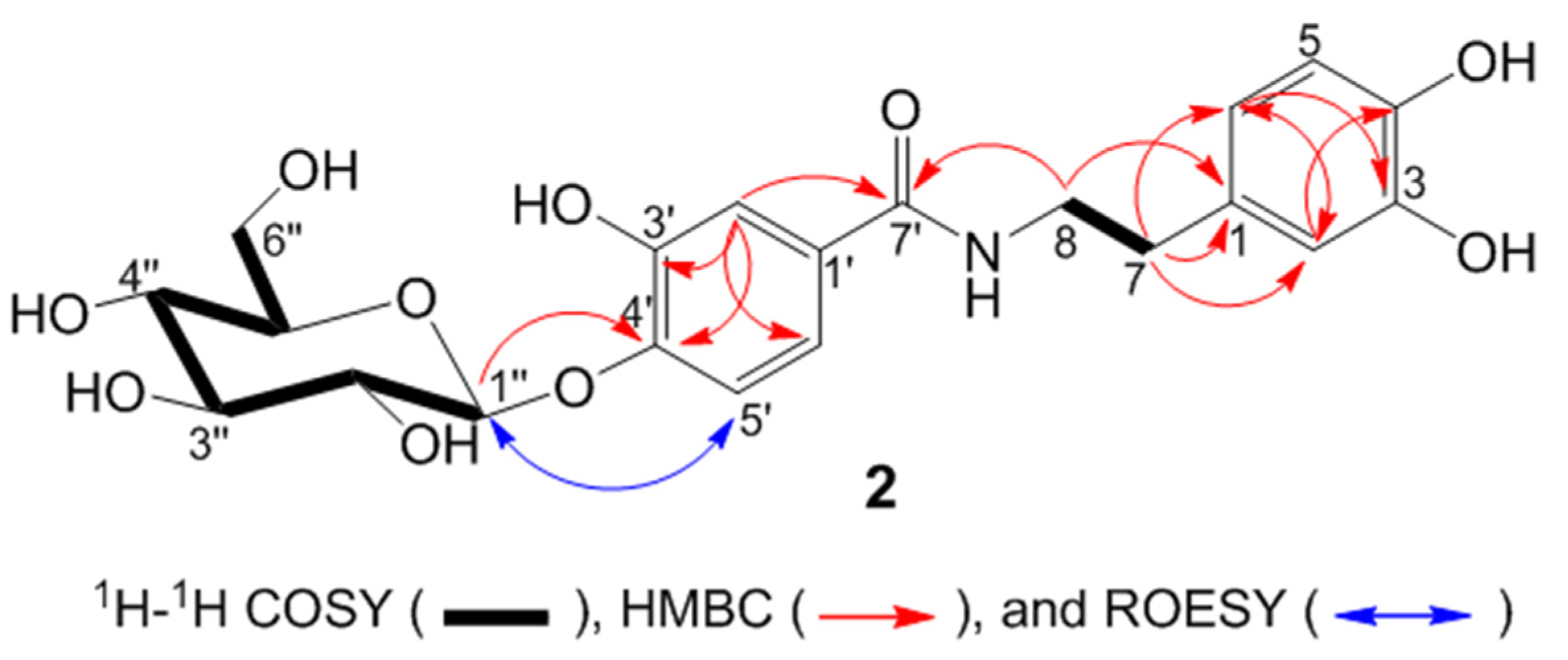
| No. | δ (H) a | δ (C) b | HMBC |
|---|---|---|---|
| 1 | - | 127.4 | |
| 2 | 7.06 (d, J = 2.1) | 115.2 | |
| 3 | - | 146.8 | |
| 4 | - | 150.0 | |
| 5 | 6.79 (d, J = 8.2) | 116.5 | |
| 6 | 6.97 (dd, J = 8.2, 2.1) | 123.3 | |
| 7 | 7.59 (d, J = 15.8) | 148.6 | C1, C2, C6, C8, C9 |
| 8 9 | 6.29 (d, J = 15.8) - | 113.4 167.5 | C1, C7, C9 |
| 1’ | 5.46 (d, J = 3.9) | 72.6 | C2’, C3’, C5’, C6’, C9’ |
| 2’ | 3.62 (ddd, J = 9.0, 5.5, 3.9) | 44.2 | C1’, C3’, C4’, C5’, C6’ |
| 3’a | 2.85 (d, J = 17.2, 9.0) | 32.8 | C2’, C1’, C4’, C6’ |
| 3’b | 2.66 (d, J = 17.2, 5.5) | 32.8 | C2’, C1’, C4’, C6’ |
| 4’ | - | 173.2 | |
| 5’ | - | 170.2 | |
| 6’ | - | 172.1 | |
| OMe | 3.73 | 52.9 | C6’ |
| OMe OMe | 3.77 3.68 | 53.1 53.4 | C5’ C4’ |
| No. | δ (H) a | δ (C) b | HMBC |
|---|---|---|---|
| 1 | - | 132.2 | |
| 2 | 6.70 (d, J = 2.1) | 117.0 | C1, C3, C4, C6, C7 |
| 3 | - | 144.7 | |
| 4 | - | 146.2 | |
| 5 | 6.69 (d, J = 8.0) | 116.5 | C1, C3, C4, C6, C7 |
| 6 | 6.59 (dd, J = 8.0, 2.1) | 121.1 | C2, C3, C7 |
| 7 | 2.75 (t, J = 7.6) | 35.9 | C1, C2, C6, C8 |
| 8a | 3.57 (dt, J = 13.2, 7.6) | 42.8 | C1, C7, C7’ |
| 8b | 3.50 (dt, J = 13.2, 7.6) | 42.8 | C1, C7, C7’ |
| 1’ | - | 126.3 | |
| 2’ | 7.25 (d, J = 3.1) | 120.7 | C1’, C3’, C4’, C5’, C6’, C7’ |
| 3’ | - | 154,3 | |
| 4’ | - | 150.1 | |
| 5’ | 7.24 (d, J = 8.8) | 117.3 | C1’, C3’, C4’, C6’ |
| 6’ | 6.86 (dd, J = 8.8, 3.1) | 120.3 | C1’, C3’, C4’, C5’ |
| 7’ | - | 167.8 | |
| 1’’ | 4.79 (d, J = 7.5) | 105.1 | C4’, C3’’, C5’’ |
| 2’’ | 3.38 (m) | 74.9 | |
| 3’’ | 3.41 (m) | 78.1 | |
| 4’’ | 3.38 (m) | 78.5 | |
| 5’’ | 3.39 (m) | 71.3 | |
| 6a’’ | 3.70 (dd, J = 12.0, 5.0) | 62.5 | |
| 6b’’ | 3.89 (d, J = 12.0) | 62.5 |
| Anthelmintic Assay | Antifungal Assays | Antibacterial Assays | ||||
|---|---|---|---|---|---|---|
| Mortality [%] | Growth Inhibition [%] a | Growth Inhibition [%] a | ||||
| C. elegans | B. cinerea | S. tritici | P. infestans | B. subtilis | A. fischeri | |
| Compound | 200 µM | 125 µM | 125 µM | 125 µM | 100 µM b | 100 µM b |
| 1 | 1.9 ± 0.8 | −13.6 ± 17.3 | −12.5 ± 17.9 | 10.1 ± 2.3 | 8.0 ± 19.0 | −5.6 ± 2.60 |
| 2 | 0.0 ± 0.0 | −27.6 ± 7.6 | 21.8 ± 8.9 | −51.5 ± 16.0 | 7.0 ± 15.0 | −3.1 ± 1.80 |
| 3 | 0.0 ± 0.0 | −18.5 ± 3.1 | −40.0 ± 35.2 | 56.2 ± 10.3 | 7.0 ± 2.8 | −211.6 ± 27.6 |
| 4 | 2.9 ± 1.4 | −32.3 ± 28.7 | -9.8 ± 8.3 | −66.4 ± 30.4 | −21.0 ± 28.0 | −35.9 ± 19.4 |
| 5 | 4.8 ± 1.4 | 27.0 ± 25.8 | 46.9 ± 8.3 | −6.0 ± 64.4 | 90.0 ± 2.0 | −32.7 ± 12.7 |
| 6 | 2.0 ± 1.7 | 51.4 ± 22.3 | −12.9 ± 2.3 | −9.4 ± 27.4 | −5.0 ± 51.0 | 30.4 ± 8.4 |
| 7 | 1.7 ± 2.4 | 2.8 ± 19.5 | −7.2 ± 1.9 | 17.1 ± 12.1 | 5.0 ± 17.0 | 1.6 ± 3.0 |
| Pos. control | 10 μg/mL ivermectin | 125 µM epoxiconazole | 125 µM terbinafine | 100 µM chloramphenicol | ||
| 98.7 ± 1.9 | 92.0 ± 1.4 | 96.8 ± 1.2 | 99.0 ± 0 | 100.0 ± 0 | ||
| Metabolic cell Viability (MTT)/Cytotoxicity (CV) Assays | ||||
|---|---|---|---|---|
| Cell Viability/Survivals [%] | ||||
| PC-3 (MTT Assay) | PC-3 (CV Assay) | HT-29 (MTT Assay) | HT-29 (CV Assay) | |
| Compound | 10 µM | 10 µM | 10 µM | 10 µM |
| 1 | 101.4 ± 5.8 | 97.5 ± 4.1 | 100.3 ± 4.4 | 91.0 ± 5.1 |
| 2 | 84.1 ± 4.9 | 84.0 ± 6.9 | 74.9 ± 6.5 | 78.2 ± 4.4 |
| 3 | 90.6 ± 3.3 | 98.3 ± 1.3 | 82.6 ± 4.8 | 100.3 ± 3.7 |
| 4 | 114.4 ± 1.2 | 92.9 ± 5.0 | 111.9 ± 1.8 | 89.0 ± 3.0 |
| 5 | 2.4 ± 6.3 | −3.4 ± 24.7 | 59.7 ± 20.5 | 82.5 ± 4.3 |
| 6 | 120.9 ± 4.3 | 93.7 ± 2.6 | 117.1 ± 3.0 | 88.2 ± 3.9 |
| 7 | 113.7 ± 4.0 | 112.2 ± 4.7 | 115.3 ± 3.9 | 107.5 ± 2.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ware, I.; Franke, K.; Hussain, H.; Morgan, I.; Rennert, R.; Wessjohann, L.A. Bioactive Phenolic Compounds from Peperomia obtusifolia. Molecules 2022, 27, 4363. https://doi.org/10.3390/molecules27144363
Ware I, Franke K, Hussain H, Morgan I, Rennert R, Wessjohann LA. Bioactive Phenolic Compounds from Peperomia obtusifolia. Molecules. 2022; 27(14):4363. https://doi.org/10.3390/molecules27144363
Chicago/Turabian StyleWare, Ismail, Katrin Franke, Hidayat Hussain, Ibrahim Morgan, Robert Rennert, and Ludger A. Wessjohann. 2022. "Bioactive Phenolic Compounds from Peperomia obtusifolia" Molecules 27, no. 14: 4363. https://doi.org/10.3390/molecules27144363
APA StyleWare, I., Franke, K., Hussain, H., Morgan, I., Rennert, R., & Wessjohann, L. A. (2022). Bioactive Phenolic Compounds from Peperomia obtusifolia. Molecules, 27(14), 4363. https://doi.org/10.3390/molecules27144363








