Notch3-Mediated mTOR Signaling Pathway Is Involved in High Glucose-Induced Autophagy in Bovine Kidney Epithelial Cells
Abstract
:1. Introduction
2. Results
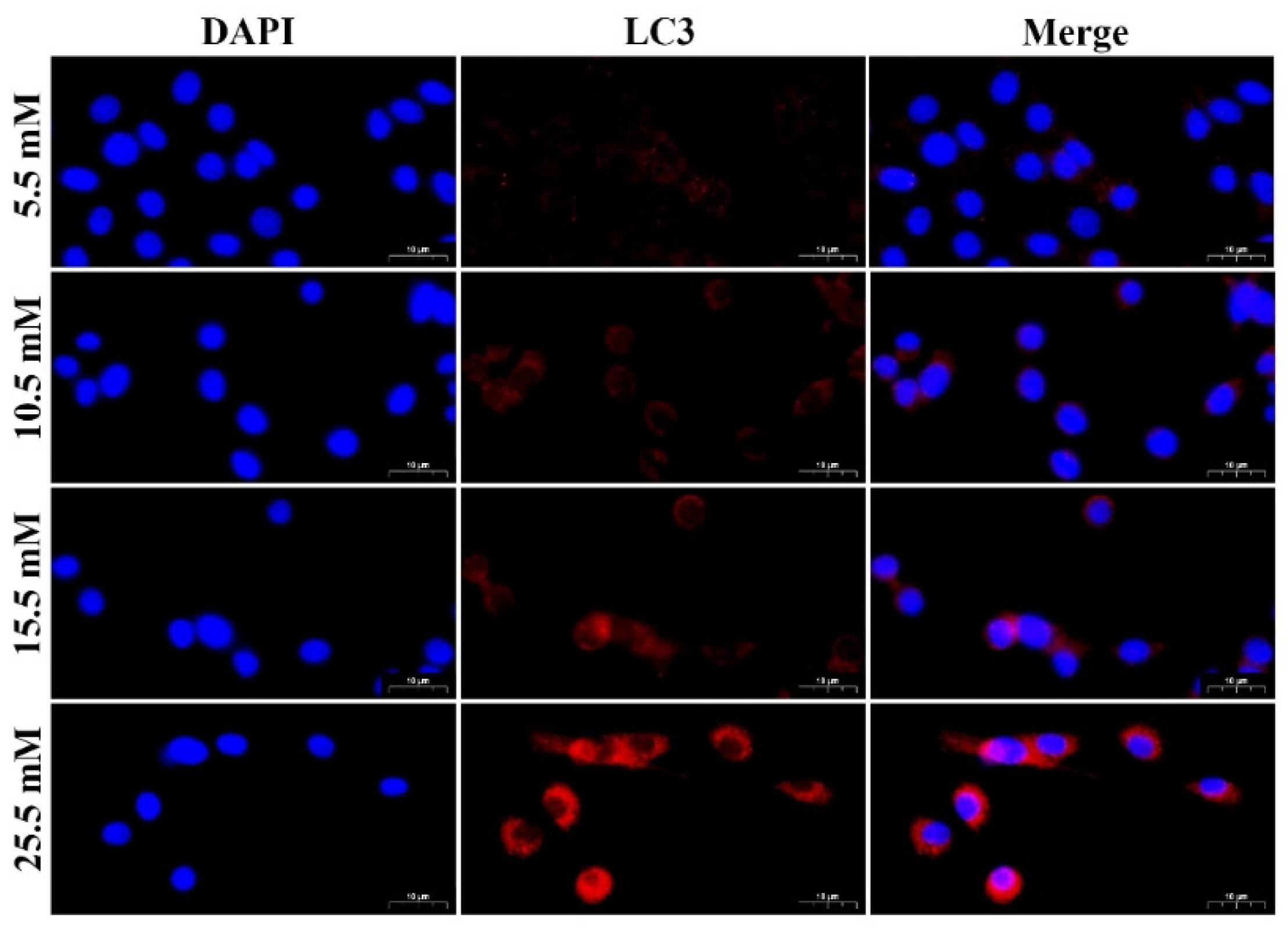
2.1. HG Treatment Induced Autophagy Markers in MDBK Cells
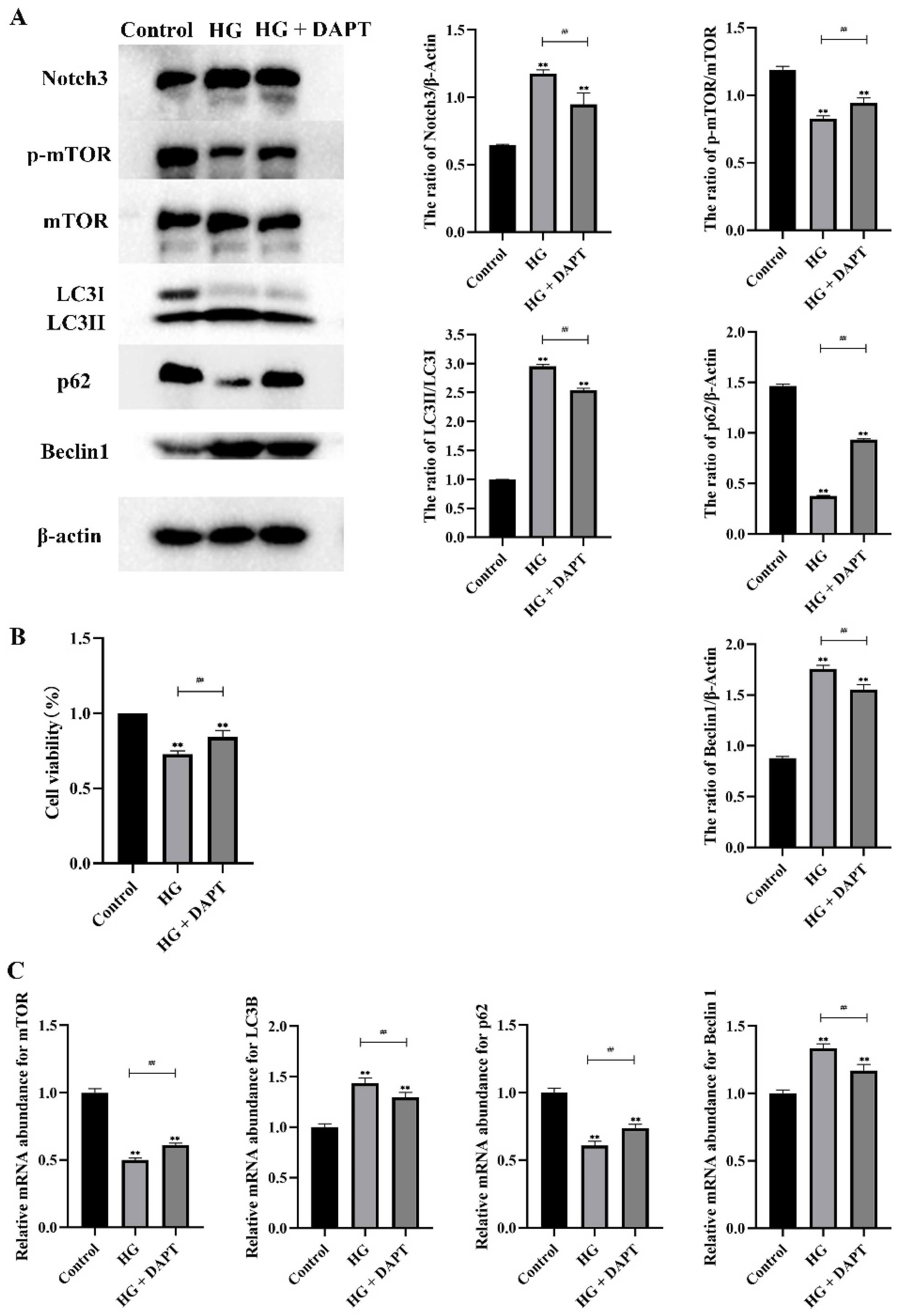
2.2. HG Induced Notch3 and mTOR Signaling Pathways in MDBK Cells
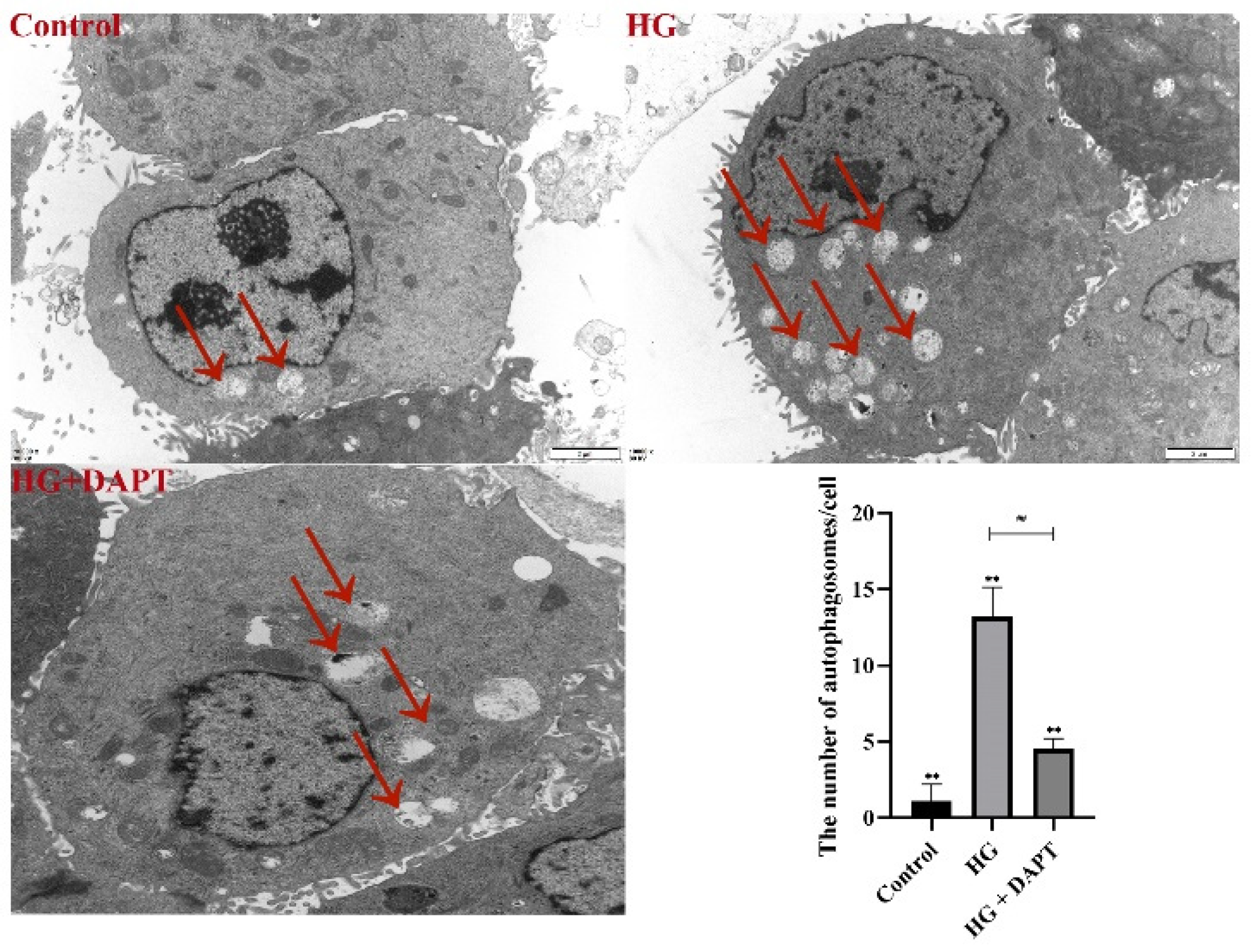
2.3. Notch3-Mediated mTOR Signaling Pathway was Involved in HG-Induced Autophagy in MDBK Cells
3. Discussion
4. Materials and Methods
4.1. Cell, Chemical and Reagents
4.2. Cell Culture
4.3. Cell Counting Kit-8 (CCK-8) Assay for Cell Growth
4.4. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.5. Western Blot Analysis
4.6. Immunofluorescence Analysis
4.7. Transmission Electron Microscopy (TEM) Analysis
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Jiang, Y.; Xie, F.; Lv, X.; Wang, S.; Liao, X.; Yu, Y.; Dai, Q.; Zhang, Y.; Meng, J.; Hu, G.; et al. Mefunidone ameliorates diabetic kidney disease in STZ and db/db mice. FASEB J. 2021, 35, e21198. [Google Scholar] [CrossRef] [PubMed]
- Alicic, R.Z.; Rooney, M.T.; Tuttle, K.R. Diabetic Kidney Disease: Challenges, Progress, and Possibilities. Clin. J. Am. Soc. Nephrol. 2017, 12, 2032–2045. [Google Scholar] [CrossRef] [PubMed]
- Duggan, E.W.; Carlson, K.; Umpierrez, G.E. Perioperative Hyperglycemia Management: An Update. Anesthesiology 2017, 126, 547–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, G.; Hattingh, J.; Ganhao, M. Stress in cattle assessed after handling, after transport and after slaughter. Vet. Rec. 1988, 123, 201–205. [Google Scholar] [CrossRef]
- Hagenmaier, J.A.; Reinhardt, C.D.; Bartle, S.J.; Henningson, J.N.; Ritter, M.J.; Calvo-Lorenzo, M.S.; Vogel, G.J.; Guthrie, C.A.; Siemens, M.G.; Thomson, D.U. Effect of handling intensity at the time of transport for slaughter on physiological response and carcass characteristics in beef cattle fed ractopamine hydrochloride. J. Anim. Sci. 2017, 95, 1963–1976. [Google Scholar]
- Godfrey, R.W.; Smith, S.D.; Guthrie, M.J.; Stanko, R.L.; Neuendorff, D.A.; Randel, R.D. Physiological responses of newborn Bos indicus and Bos indicus x Bos taurus calves after exposure to cold. J. Anim. Sci. 1991, 69, 258–263. [Google Scholar] [CrossRef]
- Gronda, E.; Jessup, M.; Iacoviello, M.; Palazzuoli, A.; Napoli, C. Glucose Metabolism in the Kidney: Neurohormonal Activation and Heart Failure Development. J. Am. Heart Assoc. 2020, 9, e018889. [Google Scholar] [CrossRef]
- Bai, Y.; Li, H.; Dong, J. Up-regulation of miR-20a weakens inflammation and apoptosis in high-glucose-induced renal tubular cell mediating diabetic kidney disease by repressing CXCL8 expression. Arch. Physiol. Biochem. 2020, 1, 1–8. [Google Scholar] [CrossRef]
- Lin, J.; Cheng, A.; Cheng, K.; Deng, Q.; Zhang, S.; Lan, Z.; Wang, W.; Chen, J. New Insights into the Mechanisms of Pyroptosis and Implications for Diabetic Kidney Disease. Int. J. Mol. Sci. 2020, 21, 7057. [Google Scholar] [CrossRef]
- Yang, D.; Livingston, M.J.; Liu, Z.; Dong, G.; Zhang, M.; Chen, J.-K.; Dong, Z. Autophagy in diabetic kidney disease: Regulation, pathological role and therapeutic potential. Cell. Mol. Life Sci. 2018, 75, 669–688. [Google Scholar] [CrossRef]
- Gong, L.; Pan, Q.; Yang, N. Autophagy and Inflammation Regulation in Acute Kidney Injury. Front. Physiol. 2020, 11, 576463. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Li, J.; Mao, L.; Wu, J.; Deng, Z.; He, M.; An, S.; Zeng, Z.; Huang, Q.; Chen, Z. p53 Deacetylation Alleviates Sepsis-Induced Acute Kidney Injury by Promoting Autophagy. Front. Immunol. 2021, 12, 685523. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-F.; Kuo, Y.-T.; Chen, T.-Y.; Chien, C.-T. Quercetin-Rich Guava (Psidium guajava) Juice in Combination with Trehalose Reduces Autophagy, Apoptosis and Pyroptosis Formation in the Kidney and Pancreas of Type II Diabetic Rats. Molecules 2016, 21, 334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, J.; Shi, Y.; Gong, J.; Zhao, L.; Li, Y.; He, Q.; Huang, H. Exosome secreted from adipose-derived stem cells attenuates diabetic nephropathy by promoting autophagy flux and inhibiting apoptosis in podocyte. Stem Cell. Res. Ther. 2019, 10, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siebel, C.; Lendahl, U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef] [Green Version]
- Nowell, C.S.; Radtke, F. Notch as a tumour suppressor. Nat. Rev. Cancer 2017, 17, 145–159. [Google Scholar] [CrossRef]
- Sanchez-Niño, M.D.; Ortiz, A. Notch3 and kidney injury: Never two without three. J. Pathol. 2012, 228, 266–273. [Google Scholar] [CrossRef]
- Kavvadas, P.; Keuylian, Z.; Prakoura, N.; Placier, S.; Dorison, A.; Chadjichristos, C.E.; Dussaule, J.-C.; Chatziantoniou, C. Notch3 orchestrates epithelial and inflammatory responses to promote acute kidney injury. Kidney Int. 2018, 94, 126–138. [Google Scholar] [CrossRef]
- Huang, M.; Zhang, J.; Xu, H.; Ding, T.; Tang, D.; Yuan, Q.; Tao, L.; Ye, Z. The TGFβ-ERK pathway contributes to Notch3 upregulation in the renal tubular epithelial cells of patients with obstructive nephropathy. Cell. Signal. 2018, 51, 139–151. [Google Scholar] [CrossRef]
- Al-Bari, M.A.A.; Xu, P. Molecular regulation of autophagy machinery by mTOR-dependent and -independent pathways. Ann. N. Y. Acad. Sci. 2020, 1467, 3–20. [Google Scholar] [CrossRef]
- Wang, W.; Liu, J.; Ma, A.; Miao, R.; Jin, Y.; Zhang, H.; Xu, K.; Wang, C.; Wang, J. mTORC1 is involved in hypoxia-induced pulmonary hypertension through the activation of Notch3. J. Cell. Physiol. 2014, 229, 2117–2125. [Google Scholar] [CrossRef] [PubMed]
- Ivanovska, J.; Shah, S.; Wong, M.J.; Kantores, C.; Jain, A.; Post, M.; Yeganeh, B.; Jankov, R.P. mTOR-Notch3 signaling mediates pulmonary hypertension in hypoxia-exposed neonatal rats independent of changes in autophagy. Pediatr. Pulmonol. 2017, 52, 1443–1454. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, Y.; Tooze, S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018, 14, 207–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Sui, W.; Xing, Y.; Cheng, J.; Cheng, C.; Xue, F.; Zhang, J.; Wang, X.; Zhang, C.; Hao, P.; et al. Angiotensin IV attenuates diabetic cardiomyopathy suppressing FoxO1-induced excessive autophagy, apoptosis and fibrosis. Theranostics 2021, 11, 8624–8639. [Google Scholar] [CrossRef]
- Lin, X.; Li, S.; Wang, Y.-J.; Wang, Y.; Zhong, J.-Y.; He, J.-Y.; Cui, X.-J.; Zhan, J.-K.; Liu, Y.-S. Exosomal Notch3 from high glucose-stimulated endothelial cells regulates vascular smooth muscle cells calcification/aging. Life Sci. 2019, 232, 116582. [Google Scholar] [CrossRef]
- Ma, Y.; Li, M.; Si, J.; Xiong, Y.; Lu, F.; Zhang, J.; Zhang, L.; Zhang, P.; Yang, Y. Blockade of Notch3 inhibits the stem-like property and is associated with ALDH1A1 and CD44 via autophagy in non-small lung cancer. Int. J. Oncol. 2016, 48, 2349–2358. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.-H.; Shen, W.-L.; Shih, C.-M.; Ho, K.-H.; Cheng, C.-H.; Lin, C.-W.; Lee, C.-C.; Liu, A.-J.; Chen, K.-C. The CHAC1-inhibited Notch3 pathway is involved in temozolomide-induced glioma cytotoxicity. Neuropharmacology 2017, 116, 300–314. [Google Scholar] [CrossRef]
- Wang, W.; Chen, D.; Zhu, K. SOX2OT variant 7 contributes to the synergistic interaction between EGCG and Doxorubicin to kill osteosarcoma via autophagy and stemness inhibition. J. Exp. Clin. Cancer Res. 2018, 37, 37. [Google Scholar] [CrossRef] [Green Version]
- Glatigny, M.; Moriceau, S.; Rivagorda, M.; Ramos-Brossier, M.; Nascimbeni, A.C.; Lante, F.; Shanley, M.R.; Boudarene, N.; Rousseaud, A.; Friedman, A.K.; et al. Autophagy Is Required for Memory Formation and Reverses Age-Related Memory Decline. Curr. Biol. 2019, 29, 435–448. [Google Scholar] [CrossRef] [Green Version]
- Xue, M.; Yang, F.; Le, Y.; Yang, Y.; Wang, B.; Jia, Y.; Zheng, Z.; Xue, Y. Klotho protects against diabetic kidney disease via AMPK- and ERK-mediated autophagy. Acta Diabetol. 2021, 58, 1413–1423. [Google Scholar] [CrossRef]
- Pasquel, F.J.; Lansang, M.C.; Dhatariya, K.; Umpierrez, G.E. Management of diabetes and hyperglycaemia in the hospital. Lancet Diabetes Endocrinol. 2021, 9, 174–188. [Google Scholar] [CrossRef]
- Wang, J.; Yue, X.; Meng, C.; Wang, Z.; Jin, X.; Cui, X.; Yang, J.; Shan, C.; Gao, Z.; Yang, Y.; et al. Acute Hyperglycemia May Induce Renal Tubular Injury Through Mitophagy Inhibition. Front. Endocrinol. 2020, 11, 536213. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Sun, Z.; Zhu, X.; Ma, Y. Astilbin inhibits high glucose-induced autophagy and apoptosis through the PI3K/Akt pathway in human proximal tubular epithelial cells. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 106, 1175–1181. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-C.; Chiu, C.-H.; Chen, J.-B.; Chen, C.-H.; Chang, H.-W. Mitochondrial Fission Increases Apoptosis and Decreases Autophagy in Renal Proximal Tubular Epithelial Cells Treated with High Glucose. DNA Cell Biol. 2016, 35, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satyavarapu, E.M.; Das, R.; Mandal, C.; Mukhopadhyay, A.; Mandal, C. Autophagy-independent induction of LC3B through oxidative stress reveals its non-canonical role in anoikis of ovarian cancer cells. Cell Death Dis. 2018, 9, 934. [Google Scholar] [CrossRef] [Green Version]
- Zaffagnini, G.; Savova, A.; Danieli, A.; Romanov, J.; Tremel, S.; Ebner, M.; Peterbauer, T.; Sztacho, M.; Trapannone, R.; Tarafder, A.K.; et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018, 37, e98308. [Google Scholar] [CrossRef]
- Parzych, K.R.; Klionsky, D.J. An Overview of Autophagy: Morphology, Mechanism, and Regulation. Antioxid. Redox Signal. 2014, 20, 460–473. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.-G.; Li, P.-L.; Dai, Y.; Deng, J.-L.; Shan, M.-Y.; Chen, B.; Zhang, K.-B.; Guo, S.-D.; Xu, Z.-H. Mibefradil Alleviates High-Glucose-induced Cardiac Hypertrophy by Inhibiting PI3K/Akt/mTOR-mediated Autophagy. J. Cardiovasc. Pharmacol. 2020, 76, 246–254. [Google Scholar] [CrossRef]
- Brandt, S.; Ballhause, T.M.; Bernhardt, A.; Becker, A.; Salaru, D.; Le-Deffge, H.M.; Fehr, A.; Fu, Y.; Philipsen, L.; Djudjaj, S.; et al. Fibrosis and Immune Cell Infiltration Are Separate Events Regulated by Cell-Specific Receptor Notch3 Expression. J. Am. Soc. Nephrol. 2020, 31, 2589–2608. [Google Scholar] [CrossRef]
- Wimmer, R.A.; Leopoldi, A.; Aichinger, M.; Wick, N.; Hantusch, B.; Novatchkova, M.; Taubenschmid, J.; Hämmerle, M.; Esk, C.; Bagley, J.A.; et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature 2019, 565, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Ni, L.; Zhang, C.; Wu, X. The Role of Notch3 Signaling in Kidney Disease. Oxid. Med. Cell. Longev. 2020, 2020, 1809408. [Google Scholar] [CrossRef] [PubMed]






| Gene Name | Primer | Sequence (5′ and 3′) | Product Length (bp) | Annealing Temperature (°C) | Sequence Number |
|---|---|---|---|---|---|
| Notch3 | Forward | CAGACACCAATGCCCAGGAC | 110 | 60 | XM_003586246.3 |
| Reverse | CAGGTCTGTAGAGCGGTTCC | ||||
| mTOR | Forward | GCTGGCACTTGCTCACAAAA | 148 | 60 | XM_002694043.6 |
| Reverse | GAAGGCATCAATCTTGCGGG | ||||
| LC3B | Forward | TGCCGTCCGAGAAAACCTTCAAAC | 89 | 60 | XM_027513856.1 |
| Reverse | CGGGATTTTGGTAGGATGCTGCTC | ||||
| p62 | Forward | CTGGGAGATGGGCACACC | 107 | 60 | XM_024993877.1 |
| Reverse | TGGGATCTTCCGATGGACCA | ||||
| Beclin1 | Forward | TCCATTACTTGCCACAGCC | 184 | 60 | AM051355.1 |
| Reverse | GCCATCAGATGCCTCCC | ||||
| β-actin | Forward | CGTCCGTGACATCAAGGAGAAGC | 143 | 60 | BT030480.1 |
| Reverse | GGAACCGCTCATTGCCGATGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Y.; Fang, J.; Guo, H.; Cui, H.; Deng, J.; Yu, S.; Gou, L.; Wang, F.; Ma, X.; Ren, Z.; et al. Notch3-Mediated mTOR Signaling Pathway Is Involved in High Glucose-Induced Autophagy in Bovine Kidney Epithelial Cells. Molecules 2022, 27, 3121. https://doi.org/10.3390/molecules27103121
Cui Y, Fang J, Guo H, Cui H, Deng J, Yu S, Gou L, Wang F, Ma X, Ren Z, et al. Notch3-Mediated mTOR Signaling Pathway Is Involved in High Glucose-Induced Autophagy in Bovine Kidney Epithelial Cells. Molecules. 2022; 27(10):3121. https://doi.org/10.3390/molecules27103121
Chicago/Turabian StyleCui, Yaocheng, Jing Fang, Hongrui Guo, Hengmin Cui, Junliang Deng, Shumin Yu, Liping Gou, Fengyuan Wang, Xiaoping Ma, Zhihua Ren, and et al. 2022. "Notch3-Mediated mTOR Signaling Pathway Is Involved in High Glucose-Induced Autophagy in Bovine Kidney Epithelial Cells" Molecules 27, no. 10: 3121. https://doi.org/10.3390/molecules27103121
APA StyleCui, Y., Fang, J., Guo, H., Cui, H., Deng, J., Yu, S., Gou, L., Wang, F., Ma, X., Ren, Z., Xie, Y., Geng, Y., Wang, Y., & Zuo, Z. (2022). Notch3-Mediated mTOR Signaling Pathway Is Involved in High Glucose-Induced Autophagy in Bovine Kidney Epithelial Cells. Molecules, 27(10), 3121. https://doi.org/10.3390/molecules27103121







