Protein Engineering of an Artificial P450BM3 Peroxygenase System Enables Highly Selective O-Demethylation of Lignin Monomers
Abstract
:1. Introduction
2. Results and Discussions
2.1. O-demethylation of Guaiacol Catalyzed by the P450BM3 Peroxygenase System
2.2. O-demethylation of Syringol Catalyzed by the P450BM3 Peroxidase System
2.3. O-demethylation of Anisole Catalyzed by the P450BM3 Peroxidase System
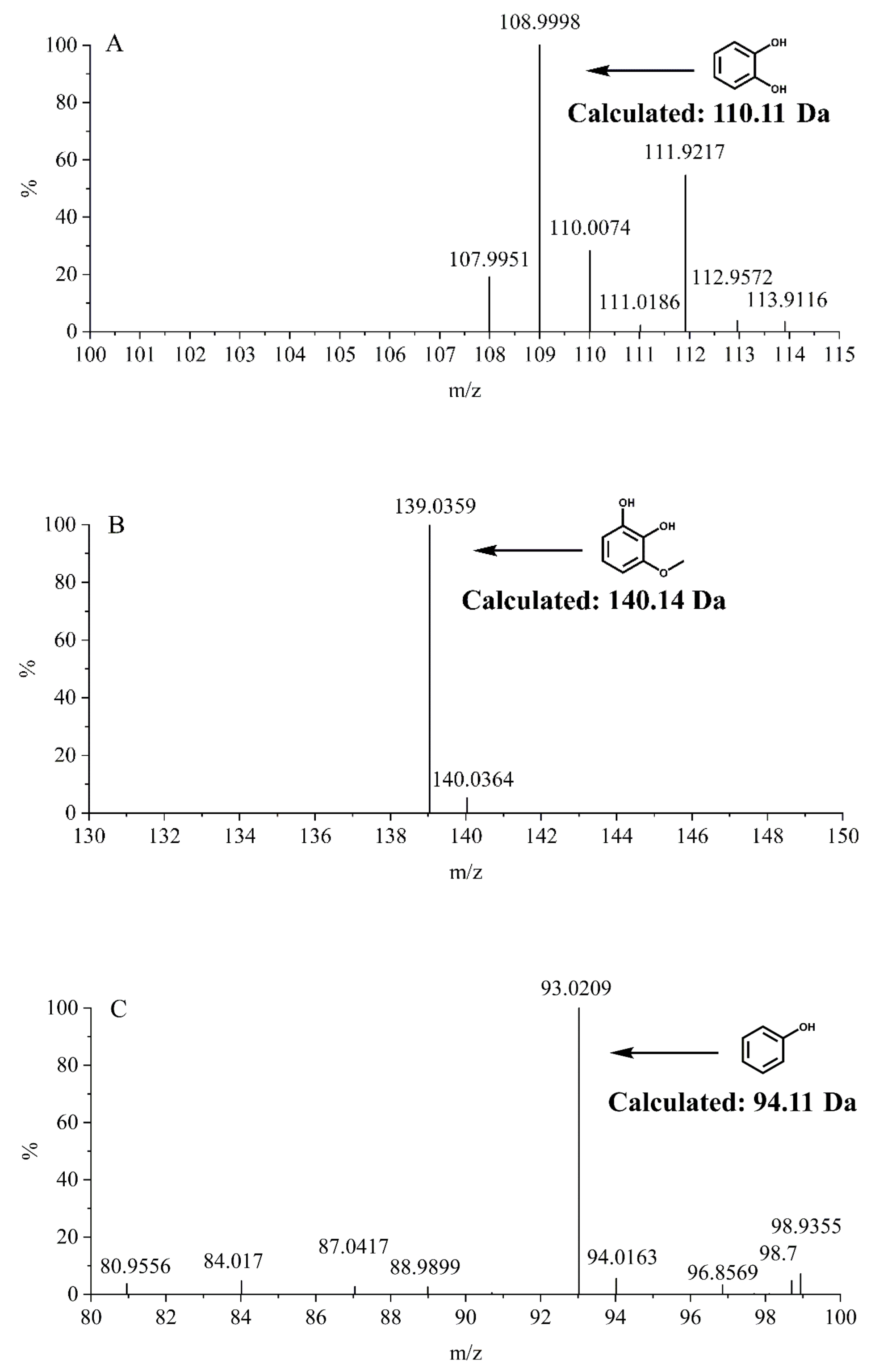
2.4. MS Assay of Reaction Products
2.5. 1 H NMR Assay of Reaction Products
3. Materials and Methods
3.1. Materials
3.2. Methods
3.2.1. Expression and Purification of P450BM3
3.2.2. Mutagenesis
3.2.3. General Procedure for O-demethylation of Aromatic Ethers
3.2.4. Product Analysis by HPLC
3.2.5. UPLC-MS Assay of Reaction Products
3.2.6. 1 H NMR Spectroscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gazi, S. Valorization of wood biomass-lignin via selective bond scission: A minireview. Appl. Catal. B Environ. 2019, 257, 117936. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.P.; Chow, M.; Liu, C.C.; Lau, A.; Liu, J.; Eltis, L.D. Vanillin catabolism in Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 2012, 78, 586–588. [Google Scholar] [CrossRef] [Green Version]
- Kamimura, N.; Takahashi, K.; Mori, K.; Araki, T.; Fujita, M.; Higuchi, Y.; Masai, E. Bacterial catabolism of lignin-derived aromatics: New findings in a recent decade: Update on bacterial lignin catabolism. Environ. Microbiol. Rep. 2017, 9, 679–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasai, D.; Fujinami, T.; Abe, T.; Mase, K.; Katayama, Y.; Fukuda, M.; Masai, E. Uncovering the protocatechuate 2,3-cleavage pathway genes. J. Bacteriol. 2009, 191, 6758–6768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nogales, J.; Canales, A.; Jimenez-Barbero, J.; Serra, B.; Pingarron, J.M.; Garcia, J.L.; Diaz, E. Unravelling the gallic acid degradation pathway in bacteria: The gal cluster from Pseudomonas putida. Mol. Microbiol. 2011, 79, 359–374. [Google Scholar] [CrossRef]
- Richter, N.; Zepeck, F.; Kroutil, W. Cobalamin-dependent enzymatic O-, N-, and S-demethylation. Trends Biotechnol. 2015, 33, 371–373. [Google Scholar] [CrossRef]
- Venkatesagowda, B. Enzymatic demethylation of lignin for potential biobased polymer applications. Fungal Biol. Rev. 2019, 33, 190–224. [Google Scholar] [CrossRef]
- Abe, T.; Masai, E.; Miyauchi, K.; Katayama, Y.; Fukuda, M. A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 2005, 187, 2030–2037. [Google Scholar] [CrossRef] [Green Version]
- Bell, S.G.; Yang, W.; Tan, A.B.; Zhou, R.; Johnson, E.O.; Zhang, A.; Zhou, W.; Rao, Z.; Wong, L.L. The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: Structural changes on substrate binding and the identification of an anion binding site. Dalton Trans. 2012, 41, 8703–8714. [Google Scholar] [CrossRef]
- Bell, S.G.; Zhou, R.; Yang, W.; Tan, A.B.; Gentleman, A.S.; Wong, L.L.; Zhou, W. Investigation of the substrate range of CYP199A4: Modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering. Chemistry 2012, 18, 16677–16688. [Google Scholar] [CrossRef] [PubMed]
- Harada, A.; Kamimura, N.; Takeuchi, K.; Yu, H.Y.; Masai, E.; Senda, T. The crystal structure of a new O-demethylase from Sphingobium sp. strain SYK-6. FEBS J. 2017, 284, 1855–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohler, A.C.; Mills, M.J.L.; Adams, P.D.; Simmons, B.A.; Sale, K.L. Structure of aryl O-demethylase offers molecular insight into a catalytic tyrosine-dependent mechanism. Proc. Natl. Acad. Sci. USA 2017, 114, E3205–E3214. [Google Scholar] [CrossRef] [Green Version]
- Machovina, M.M.; Mallinson, S.J.B.; Knott, B.C.; Meyers, A.W.; Garcia-Borras, M.; Bu, L.; Gado, J.E.; Oliver, A.; Schmidt, G.P.; Hinchen, D.J.; et al. Enabling microbial syringol conversion through structure-guided protein engineering. Proc. Natl. Acad. Sci. USA 2019, 116, 13970–13976. [Google Scholar] [CrossRef] [Green Version]
- Yoshikata, T.; Suzuki, K.; Kamimura, N.; Namiki, M.; Hishiyama, S.; Araki, T.; Kasai, D.; Otsuka, Y.; Nakamura, M.; Fukuda, M.; et al. Three-component O-demethylase system essential for catabolism of a lignin-derived biphenyl compound in Sphingobium sp. strain SYK-6. Appl. Environ. Microbiol. 2014, 80, 7142–7153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellis, E.S.; Hinchen, D.J.; Bleem, A.; Bu, L.; Mallinson, S.J.B.; Allen, M.D.; Streit, B.R.; Machovina, M.M.; Doolin, Q.V.; Michener, W.E.; et al. Engineering a cytochrome P450 for demethylation of lignin-derived aromatic aldehydes. JACS Au 2021, 1, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Notonier, S.; Werner, A.Z.; Kuatsjah, E.; Dumalo, L.; Abraham, P.E.; Hatmaker, E.A.; Hoyt, C.B.; Amore, A.; Ramirez, K.J.; Woodworth, S.P.; et al. Metabolism of syringyl lignin-derived compounds in Pseudomonas putida enables convergent production of 2-pyrone-4,6-dicarboxylic acid. Metab. Eng. 2021, 65, 111–122. [Google Scholar] [CrossRef]
- Chen, J.; Kong, F.; Ma, N.; Zhao, P.; Liu, C.; Wang, X.; Cong, Z. Peroxide-driven hydroxylation of small alkanes catalyzed by an artificial P450BM3 peroxygenase system. ACS Catal. 2019, 9, 7350–7355. [Google Scholar] [CrossRef]
- Ma, N.; Chen, Z.; Chen, J.; Chen, J.; Wang, C.; Zhou, H.; Yao, L.; Shoji, O.; Watanabe, Y.; Cong, Z. Dual-functional small molecules for generating an efficient cytochrome P450BM3 peroxygenase. Angew Chem. Int. Ed. Engl. 2018, 57, 7628–7633. [Google Scholar] [CrossRef]
- Xu, J.; Wang, C.; Cong, Z. Strategies for substrate-regulated P450 catalysis: From substrate engineering to co-catalysis. Chemistry 2019, 25, 6853–6863. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, C.; Ma, N.; Chen, J.; Liu, C.; Wang, F.; Xu, J.; Cong, Z. Regioselective aromatic O-demethylation with an artificial P450BM3 peroxygenase system. Catal. Sci. Technol. 2020, 10, 1219–1223. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, Y.; Chen, Q.; Dong, S.; Feng, Y.; Cong, Z.; Shaik, S.; Wang, B. H-bonding networks dictate the molecular mechanism of H2O2 activation by P450. ACS Catal. 2021, 11, 8774–8785. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: Palo Alto, CA, USA, 2002; Available online: www.pymol.org (accessed on 1 April 2022).
- Omura, T.; Sato, R. The carbon monoxide-binding pigment of liver microsomes. J. Biol. Chem. 1964, 239, 2370–2378. [Google Scholar] [CrossRef]
- Wang, Z.J.; Renata, H.; Peck, N.E.; Farwell, C.C.; Coelho, P.S.; Arnold, F.H. Improved cyclopropanation activity of histidine-ligated cytochrome P450 enables the enantioselective formal synthesis of levomilnacipran. Angew. Chem. Int. Ed. 2014, 53, 6810–6813. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Limon, A.; Aguila, S.; Ayala, M.; Batista, C.V.; Vazquez-Duhalt, R. Peroxidase activity stabilization of cytochrome P450(BM3) by rational analysis of intramolecular electron transfer. J. Inorg. Biochem. 2013, 122, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Fernandez-Fueyo, E.; Ni, Y.; van Schie, M.; Gacs, J.; Renirie, R.; Wever, R.; Mutti, F.G.; Rother, D.; Alcalde, M.; et al. Selective aerobic oxidation reactions using a combination of photocatalytic water oxidation and enzymatic oxyfunctionalisations. Nat. Catal. 2018, 1, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Lan, D.; Tan, X.; Hollmann, F.; Bornscheuer, U.T.; Yang, B.; Wang, Y. How to break the Janus effect of H2O2 in biocatalysis? Understanding inactivation mechanisms to generate more robust enzymes. ACS Catal. 2019, 9, 2916–2921. [Google Scholar] [CrossRef] [Green Version]

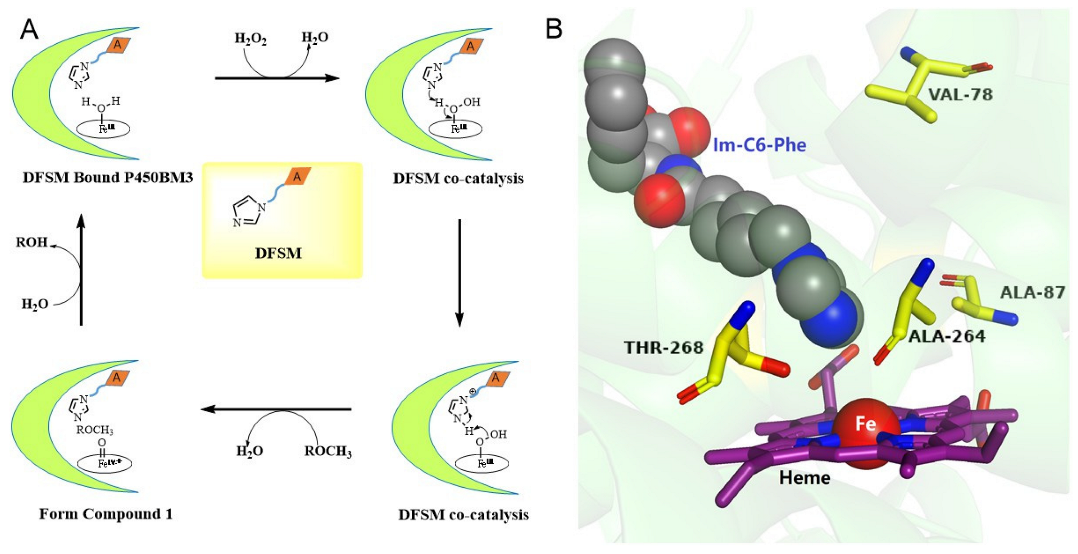
| Enzyme | DFSM | TON b | Catechol Selectivity % | |||
|---|---|---|---|---|---|---|
| 1a | 1b | 1c | 1d | |||
| F87A [21] | Im-C6-Phe c | 539 ± 2 | nd d | nd | 344 ± 11 | 61 |
| F87G [21] | Im-C6-Phe | 495 ± 9 | nd | nd | 236 ± 12 | 68 |
| F87A/T268L | Im-C6-Phe | 10 ± 1 | nd | nd | nd | 100 |
| F87A/T268P | Im-C6-Phe | 321 ± 2 | nd | nd | 50 ± 2 | 86 |
| F87A/T268I [21] | Im-C6-Phe | 96 ± 1 | nd | nd | 16 ± 0.1 | 85 |
| F87A/V78A | Im-C6-Phe | 448 ± 18 | nd | nd | 37 ±3 | 92 |
| F87G/V78A | Im-C6-Phe | 285 ± 12 | nd | nd | 46 ± 3 | 86 |
| F87A/V78A/A264G | Im-C6-Phe | 333 ± 14 | nd | nd | 15 ± 1 | 96 |
| F87G/V78A/A264G | Im-C6-Phe | 67 ± 2 | nd | nd | 2 ± 1 | 97 |
| F87A/V78A/T268I | Im-C6-Phe | 428 ± 4 | 800 ± 6 | 18 ± 1 | 26 ± 1 | 34 |
| F87A/V78G/T268I | Im-C6-Phe | 188 ± 5 | 521 ± 15 | 12 ± 1 | 17 ± 1 | 25 |
| F87A/V78A/T268P | Im-C6-Phe | 174 ± 3 | 26 ± 1 | 10 ± 0.6 | 70 ± 1 | 62 |
| F87A/V78G/T268P | Im-C6-Phe | 353 ± 2 | nd | 9 ± 1 | 79 ± 1 | 80 |
| F87A/T268P/A264G | Im-C6-Phe | 269 ± 9 | nd | nd | nd | 100 |
| F87A/V78A/T268I/A264G | Im-C6-Phe | 839 ± 7 | nd | nd | nd | 100 |
| F87A/V78A/A264G/T268A | Im-C6-Phe | 154 ± 5 | nd | nd | nd | 100 |
| Enzyme | DFSM | TON b | 3-Methoxycatechol Selectivity % | |
|---|---|---|---|---|
| 2a | 2b | |||
| F87A/T268P | Im-C6-Phe c | 238 ± 2 | nd d | 100 |
| F87A/V78A | Im-C6-Phe | nd | nd | — |
| F87A/V78G | Im-C6-Phe | 30 ± 1 | nd | 100 |
| F87A/V78A/T268I | Im-C6-Phe | 200 ± 1 | nd | 100 |
| F87A/V78G/T268I | Im-C6-Phe | 208 ± 1 | nd | 100 |
| F87A/T268P/V78A | Im-C6-Phe | 236 ± 5 | nd | 100 |
| F87A/T268P/A264G | Im-C6-Phe | 97 ± 12 | nd | 100 |
| F87A/V78A/T268I/A264G | Im-C6-Phe | 67 ± 1 | nd | 100 |
| Enzyme | DFSM | TON b | Phenol Selectivity % | ||
|---|---|---|---|---|---|
| 3a | 3b | 3c | |||
| F87A [21] | Im-C6-Phe c | 145 ± 3 | 12 ± 1 | 393 ± 9 | 26 |
| F87A/T268I [21] | Im-C6-Phe | 262 ± 2 | nd d | nd | 100 |
| F87A/T268P | Im-C6-Phe | 128 ± 3 | 28 ± 2 | 333 ± 5 | 26 |
| F87A/V78A/T268I | Im-C6-Phe | 265 ± 6 | nd | nd | 100 |
| F87A/V78G/T268I | Im-C6-Phe | 108 ± 2 | nd | nd | 100 |
| F87A/V78A/T268I/A264G | Im-C6-Phe | 67 ± 3 | nd | nd | 100 |
| F87A/V78A/T268I/A264S | Im-C6-Phe | 132 ± 6 | nd | nd | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Miao, H.; Li, Y.; Wang, F.; Xu, J. Protein Engineering of an Artificial P450BM3 Peroxygenase System Enables Highly Selective O-Demethylation of Lignin Monomers. Molecules 2022, 27, 3120. https://doi.org/10.3390/molecules27103120
Li M, Miao H, Li Y, Wang F, Xu J. Protein Engineering of an Artificial P450BM3 Peroxygenase System Enables Highly Selective O-Demethylation of Lignin Monomers. Molecules. 2022; 27(10):3120. https://doi.org/10.3390/molecules27103120
Chicago/Turabian StyleLi, Maosheng, Hengmin Miao, Yanqing Li, Fang Wang, and Jiakun Xu. 2022. "Protein Engineering of an Artificial P450BM3 Peroxygenase System Enables Highly Selective O-Demethylation of Lignin Monomers" Molecules 27, no. 10: 3120. https://doi.org/10.3390/molecules27103120
APA StyleLi, M., Miao, H., Li, Y., Wang, F., & Xu, J. (2022). Protein Engineering of an Artificial P450BM3 Peroxygenase System Enables Highly Selective O-Demethylation of Lignin Monomers. Molecules, 27(10), 3120. https://doi.org/10.3390/molecules27103120






