Abstract
Antimicrobial resistance (AMR) poses a serious threat to our society from both the medical and economic point of view, while the antibiotic discovery pipeline has been dwindling over the last decades. Targeting non-essential bacterial pathways, such as those leading to antibiotic persistence, a bacterial bet-hedging strategy, will lead to new molecular entities displaying low selective pressure, thereby reducing the insurgence of AMR. Here, we describe a way to target (p)ppGpp (guanosine tetra- or penta-phosphate) signaling, a non-essential pathway involved in the formation of persisters, with a structure-based approach. A superfamily of enzymes called RSH (RelA/SpoT Homolog) regulates the intracellular levels of this alarmone. We virtually screened several fragment libraries against the (p)ppGpp synthetase domain of our RSH chosen model RelSeq, selected three main chemotypes, and measured their interaction with RelSeq by thermal shift assay and STD-NMR. Most of the tested fragments are selective for the synthetase domain, allowing us to select the aminobenzoic acid scaffold as a hit for lead development.
1. Introduction
The rising of bacteria resistant to the currently available antibiotic arsenal is posing a serious threat to the way of life we have become accustomed to over the past century since Fleming isolated and characterized penicillin [1,2]. If left unchecked, antimicrobial resistance (AMR), paired with the decreasing number of new antibiotics progressing through the clinical pipeline, will lead to millions of deaths each year in a few decades [3].
AMR manifests itself in stable, heritable genetic forms, as well as in lesser-known transient phenotypes that are more elusive and difficult to recognize and tackle [4]. On the other hand, antimicrobial compounds currently in the clinical phase consist mostly of derivatives of established classes, with an urgent need for drugs that address multidrug-resistant Gram-negative bacteria [3].
The search for new chemical entities, targeting non-essential pathways that play a role in both the infection process (e.g., bacterial adhesion, quorum sensing, virulence, and biofilm formation) and the insurgence of genetic resistance, is an attractive strategy for attaining new antimicrobial drugs exerting minimal selective pressure.
We posit (p)ppGpp-signalling molecules (guanosine tetra- or penta-phosphate) [5] as key players in both processes [6]. Indeed, (p)ppGpp is a ubiquitous alarmone that directs bacterial adaptation to environmental changes (e.g., nutrient starvation, oxidative stress, etc.) by binding various targets involved, e.g., in nucleotide metabolism, DNA replication and repair [7], transcription [8], and translation [9,10]. This alarmone thus has pleiotropic effects on bacterial cell physiology [11], regulating cell size, virulence, quorum sensing, and biofilm formation [12,13,14,15]. In particular, its ability to downregulate cell metabolism and cell growth hints at its role in the insurgence of the bacterial non-heritable dormant phenotype called persister [16]. Persisters are transiently tolerant to antibiotic treatment (i.e., they are a form of phenotypic AMR) until they revert to the awake state and resume growth, constituting an infection reservoir that sustains chronic and recurrent infections and paves the way to the acquisition of genetic resistance [17,18]. Over the last two decades, extensive research has revealed different molecular mechanisms leading to their formation [19], and one of them is the accumulation of (p)ppGpp together with its downstream effects.
Intracellular (p)ppGpp levels are regulated by enzymes belonging to the RSH (RelA/SpoT Homolog) superfamily [20]. These enzymes catalyze (p)ppGpp synthesis via a Mg2+-dependent pyrophosphate transfer from ATP to the 3′-OH group of either GDP or GTP. They also catalyze (p)ppGpp hydrolysis leading to the release of pyrophosphate (PPi) using distinct active sites in different protein domains (Figure 1a). “Short” RSH proteins harbor either only the synthetase (SAS, small alarmone synthetases) or hydrolase (SAH, small alarmone hydrolases) domain, respectively. “Long” RSH proteins contain both catalytic domains and a C-terminal regulatory domain (CTD) that activates alarmone synthesis by promoting Rel oligomerization [21] and/or upon binding to stalled ribosomes (i.e., ribosomes bound to uncharged tRNAs during aminoacid starvation) [22,23] or favors alarmone hydrolysis by directly inhibiting the synthetase site [24]. In addition, reciprocal regulation of the two catalytic domains has been postulated with mechanisms that vary among species [25,26,27,28].
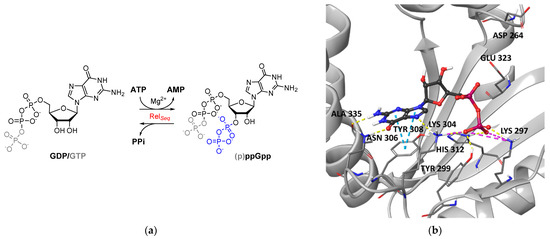
Figure 1.
(a) Enzymatic reaction that regulates intracellular (p)ppGpp levels catalyzed by RSH enzymes such as RelSeq; (b) X-ray structure of GDP in the SYNTH site of RelSeq (1VJ7, chainA). GDP is represented in ball and stick, and the key interacting amino acids are labeled.
The search for inhibitors of Rel enzymes synthetase activity dates back to the early 2010s, when Relacin [29] and a few other (p)ppGpp analogs [30,31] were described by Wexselblatt and co-workers. In all cases, IC50 values measured against RelA (E. coli) and Rel from D. radiodurans were estimated to be between 1 and 5 mM, with relatively low ligand efficiency and no subsequent further optimization reported. More recently, extensive efforts, including high-throughput screening (RelBs, B. subtilis) and an expanded library of (p)ppGpp analogs (RelA), have identified some low µM, non-specific Rel inhibitors [32,33], while screening of a large pharmaceutical library (GSK, >2 M compounds) for inhibitors of RelMtb (M. tuberculosis) identified only one compound (X9) as a potential lead for a combination TB therapy with isoniazid [34].
In this framework, we optimized the synthesis of a fluorescent (p)ppGpp selective chemosensor [35], and here, we report the identification of three novel chemical scaffolds for the design of selective RSH inhibitors through fragments virtual screening campaigns followed by experimental validation of representative fragments on the synthetase site of the “long” RSH protein RelSeq (S. equisimilis).
2. Results and Discussion
2.1. Fragment Libraries Virtual Screening in RelSeq Synthetase Site
We chose as a protein model the X-ray crystal structure reported for RelSeq in a so-called synthetase-ON conformation, i.e., the chain A of the pdb structure 1VJ7 (residues 1–385), which carries the GDP substrate in the synthetase catalytic site [25]. This is a truncated form of the protein, lacking the C-terminal regulatory domain.
By analyzing the interactions of GDP within the catalytic site (Figure 1b), we could observe that it binds to the G-loop (Tyr299-Ser310), forming a π–π stacking interaction with the side chain of Tyr308 through its guanine ring. H-bonds with the side chains of Lys304 and Asn306 and with the backbone of Ala335 stabilize this core interaction. In addition, the GDP pyrophosphate moiety forms salt bridges with Lys304 and Lys297 side chains and H-bonds with Tyr299 and His312 side chains.
Our analysis revealed that the RelSeq synthetase-ON catalytic site conformation could not be catalytically competent, as it lacks the space necessary to accommodate the pyrophosphate donor ATP, and the reported catalytic residues Asp264 and Glu323 are unfavorably oriented to promote the reaction (Figure 1b). On this basis, we constructed and reported a catalytically competent RelSeq chimera model based on the structure of the SAS RelP from S. aureus, where the resulting catalytic site is considerably more extended [36]. However, we performed an initial virtual screening on the RelSeq X-ray crystal structure in order to focus our binding site exploration on the region occupied by the enzyme substrate GDP.
During protein preparation (1VJ7, chain A, see Section 3), we took a closer look at the protonation state of His312, interacting with the β-phosphate group of GDP in the crystal. Although the neutral form should be more plausible, given the pH working conditions of the enzyme (activity usually tested at pH 7–9), we decided to generate a model with the residue in the protonated form as well (Hip312) in order to assess the validity of this assumption. We set up and validated a docking protocol within the synthetase binding site for both models by re-docking the co-crystallized GDP molecule using GLIDE v8.0 (Supplementary Figures S1 and S2). [37]
Several chemical vendors currently make available virtual structure datasets of fragment libraries, often organized according to specific experimental properties (e.g., solubility). In order to maximize the chemical space explored in our screening [38,39], we selected seven different libraries of commercially available fragments, Maybridge Rule of 3, Asinex Fragments, Life Chemicals Fragment Library with Experimental Solubility Data I and II, OTAVA Solubility fragment library, Chembridge Fragment library, and SPECS fragment library, amounting to a total of 58,321 2D entries (see Section 3 and Supplementary Table S1). We implemented the validated docking protocol for the virtual screening (VS) of the selected fragment libraries following the workflow shown in Figure 2. For each library, Ligprep [40] converted 2D entries into 3D structures considering stereoisomers, tautomers, and protonation states, leading to an increase in the total number of structures up to 114,966.
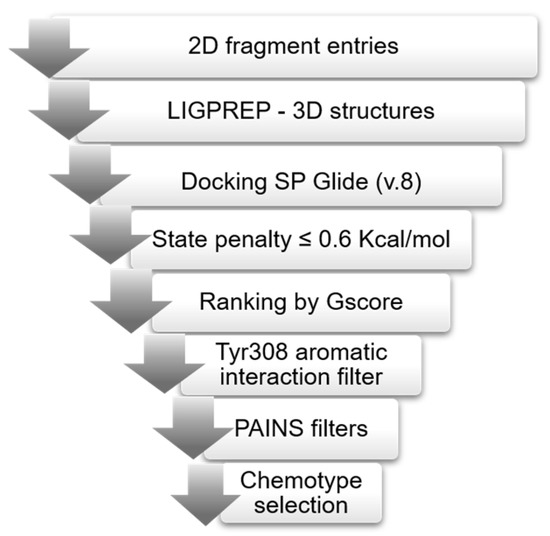
Figure 2.
Virtual screening workflow. Ligprep generated 3D structure from 2D fragments; Glide docked the 3D structures into both RelSeq models (His312 and Hip312). A state penalty filter excluded unfavorable states, and the resulting docking poses were ranked according to Gscore. Only fragments forming an aromatic interaction with Tyr308 were retained, and after removing PAINS, we evaluated the top 1% ranked poses identifying the most representative chemotypes.
2.2. Post-Docking Analysis and Chemotype Selection
We removed from the docking outputs (one pose saved for each fragment) the less stable tautomeric and ionization forms, as determined by Epik [41] (state penalty value ≤ 0.6 kcal/mol). We removed duplicates from the merged outputs and ranked them by Gscore. Finally, we applied an aromatic interaction filter with Tyr308 (i.e., an aromatic ligand atom must be within 5 Å from any heavy atom of the Tyr308 side chain), a key residue that, when mutated into asparagine or serine, inhibits the synthetic activity of the enzyme, and removed PAINS (pan-assay interference compounds) using the filters provided by Canvas [42,43] in order to exclude frequent hitters [44]. This work resulted in the selection of 30,126 and 30,960 fragments for the His312 and Hip312 grids, respectively. We visually inspected and assessed the top 1% of docked poses, identifying three recurrent chemotypes, i.e., benzimidazole, aminobenzoic acid, and indole (for the calculated enrichment factors, see Supplementary Table S2) and three singletons (the best pose of representative fragments is shown in Figure 3 and Supplementary Figure S3). Aminobenzoic acids emerged mainly from the Hip312 model and benzimidazoles mainly from the His312 model, while indoles emerged from both to a lesser extent.
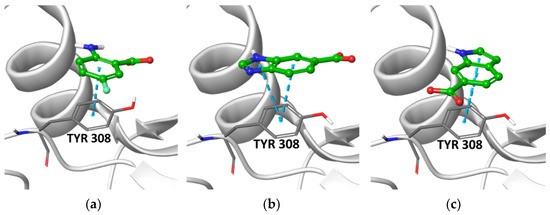
Figure 3.
Best poses of representative structures of the three chemotypes selected for experimental validation. (a) aminobenzoic acid A3 docked in the Hip312 grid; (b) Benzimidazole B2 docked in the His312 grid; (c) indole I1 docked in the His312 grid. Fragments are shown as balls and sticks with green carbon atoms. The protein is shown in grey, with the side chain of Tyr308 highlighted.
In order to maximize the chemical space explored, we performed a similarity search (Tanimoto index ≥ 90%) using the PubChem database [45] and the representative structures of each chemotype as input. We applied the same screening workflow (see Supplementary Information) to the expanded set of fragments leading to the final selection of eighteen fragments for experimental validation (Figure 4).
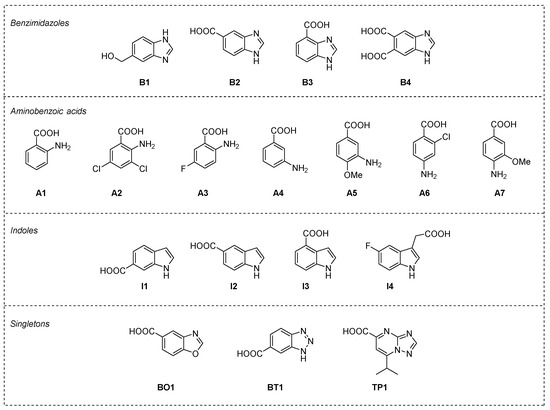
Figure 4.
Fragments selected for experimental validation (i.e., thermal shift assay) grouped by chemotype: benzimidazoles (B1–B4), aminobenzoic acids (A1–A7), indoles (I1–I4), and the three singletons (BO1, BT1, and TP1).
Prior to conducting biochemical assays, we assessed the docking poses stability by running molecular dynamics (MD) simulations. Starting from the best pose for each fragment we ran 100 ns simulations using Desmond [46] (NPT, T = 300 K, p = 1 atm, TIP3P [47], OPLS3e [48], dt = 2 fs). We considered the aromatic interaction with Tyr308 described above as the key feature to be maintained and monitored during the simulations. All the examined fragments form stable interactions with the residues involved in the binding of the GDP guanine ring in the X-ray structure, retaining, in particular, stable contact with the side chain of Tyr308, with the exception of fragments B1 and I2 that exit the binding pocket (Supplementary Figure S4).
2.3. Thermal Shift Assay on Selected Fragments vs. RelSeq Protein Constructs
With the aim of studying protein–ligand interactions between RelSeq constructs and selected fragments, we evaluated the use of different techniques, such as tryptophan assay, microscale thermophoresis (MST), isothermal calorimetry (ITC) or thermal shift assay (TSA). The tryptophan assay is based on ligand-induced conformational changes in the local environment surrounding tryptophan residues in the target protein. Irradiation at 280 nm is followed by the detection of tryptophan fluorescence emission at different wavelengths. We used this technique to measure the Kd values of the natural substrates GDP and ATP (see below), but it is not applicable in the case of fragments due to their high absorbance at 280 nm. On the other hand, the currently available protein-labeling reagents for MST were not compatible with our protein constructs. We chose TSA over ITC due to its higher potential throughput and lower amount of protein required.
Thermal shift is an experimental technique in which protein thermal denaturation is monitored following the increase in fluorescence reported by a protein-bound dye [49]. In particular, an environment-sensitive hydrophobic dye (e.g., SYPRO Orange) binds to hydrophobic regions that become progressively exposed during thermal denaturation, resulting in an increase in its fluorescence emission. Since the binding of small molecules (e.g., fragments) to the protein can cause conformational changes that affect its melting temperature, this technique allows the screening of several compounds in a range of concentrations without consuming sizeable amounts of protein.
We preliminary determined by TSA the dissociation constants (Kd) of complexes engaged by RelSeq with its natural substrates, ATP (0.49 ± 0.09 mM) and GDP (0.26 ± 0.06 mM), finding values comparable to those obtained by tryptophan assay (KdATP = 0.39 ± 0.04 mM, KdGDP = 0.15 ± 0.01 mM) and confirming the robustness of this technique (Table 1, entries 1 and 2).

Table 1.
Thermal shift assay. Kd (mM) of protein–ligand complexes engaged by RelSeq constructs with the tested fragments.
We therefore evaluated the affinity of the 18 fragments selected from our in silico screening for the bifunctional protein RelSeq by titration of the protein in a thermal shift assay (Table 1). We initially used RelSeq 1–385, a truncated construct lacking the abovementioned C-terminal regulatory domain with a catalytic activity intrinsically shifted towards (p)ppGpp synthesis (12-fold higher than the full-length protein) [50].
Interestingly, all but four of the tested fragments showed a dose-dependent interaction with the bifunctional protein with Kd values in the low millimolar range.
In particular, only one of the four tested benzimidazoles showed a measurable affinity for RelSeq (1–385) (B3, Table 1, entry 5), whereas fragment B2 yielded a biphasic curve that requires further investigation (Table 1, entry 4). All of the selected aminobenzoic acids showed good affinities for the protein, with the exception of A2 (Table 1, entries 7–13), while among the four indoles tested, all interacted in a dose-dependent manner with RelSeq (1–385) (Table 1, entries 14–17). Finally, two of the three singletons tested (BO1 and TP1) showed a low mM affinity for the protein.
We assessed the fragments’ selectivity for RelSeq synthetase domain by performing TSA experiments on two mono-functional truncated protein constructs: RelSeq SYNTH (residues 79–385) and RelSeq HYD (residues 1–224), presenting only the synthetase or hydrolase protein domain, respectively. Both constructs retain part of the central 3-helix bundle to ensure proper folding, especially in the case of the less stable SYNTH domain [50].
The four fragments that failed to show binding to the bifunctional protein (B1, B4, A2, and BT1) also failed to show dose-dependent effects on the two truncated constructs, while the biphasic curve initially observed for B2 was determined to be a selective interaction with the HYD domain, with no affinity for the SYNTH domain.
With the exception of B3, which shows comparable affinities for both catalytic domains, all the fragments binding to RelSeq (1–385) showed a remarkable selectivity for the SYNTH domain, even with a generally increased absolute value for the measured Kd. This is probably due to the lower overall stability of the SYNTH domain compared to the full protein, as previously reported by Mechold et al. [50]. Indeed, RelSeq SYNTH (79–385), despite being catalytically functional (data not shown), requires the use of a non-ionic surfactant in the purification steps in order to avoid precipitation.
2.4. STD-NMR Protein–Fragment Interaction Experiments
The relatively weak affinity of the fragments measured by TSA directed us towards NMR methods to assess the specificity of the fragments-protein interactions. Indeed, ligand-based NMR methods [51] can be applied to weak and transient protein–ligand complexes that are difficult to study with other structural techniques and do not require protein labeling (since only NMR signals of the small molecule are detected), and only a small amount of protein is required. In particular, STD (Saturation Transfer Difference) exploits NOE effects between the protein and the ligand to map target–ligand interactions and to characterize biologically relevant complexes [52,53].
Considering the promising selectivity profile of the aminobenzoic acids for the RelSeq synthetase domain, we performed STD-NMR experiments with fragment A1 as the representative chemotype. The results confirmed the binding event and showed a good interaction for the aromatic protons of A1 with RelSeq (1–385) (Figure 5b). Comparable overall STD intensities, suggesting a similar binding mode, were observed for RelSeq SYNTH (79–385), confirming the specificity of the interaction (Figure 5c). On the other hand, the RelSeq HYD (1–224) construct did not produce any magnetization transfer (Figure 5d). STD experiments performed on RelSeq (1–385) with the two non-binding fragments B1 and A2 did not show any significant interaction, refuting artifacts or non-specific binding (Supplementary Figures S5 and S6, respectively).
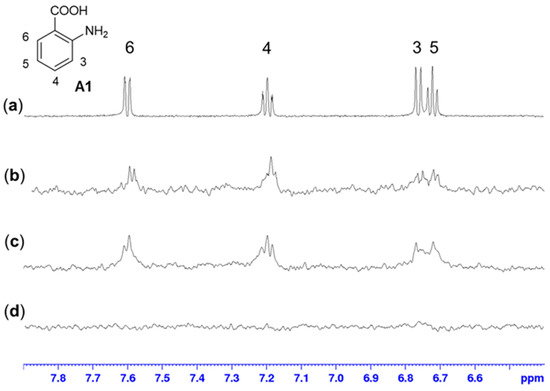
Figure 5.
STD-NMR experiments (fragment:protein ratio 1000:1; fragment concentration 3 mM). (a) 1H-NMR of fragment A1 in phosphate buffer at 298 K. (b) STD-NMR experiment of A1 with RelSeq (1–385). (c,d) STD-NMR experiment of A1 with RelSeq SYNTH (79–385) and with RelSeq HYD (1–224), respectively. The same binding mode can be observed for full-length RelSeq and RelSeq SYNTH, while no binding can be detected with RelSeq HYD.
3. Materials and Methods
3.1. Computational Methods
Protein Preparation. RelSeq three-dimensional structure (PDB 1VJ7, chain A, residues 5–341) was prepared for docking calculations using the ‘Protein Preparation Wizard’ of Schrödinger® suite [40] and OPLS_2005 force field [54]. All water molecules were deleted, and the gaps of the HD domain (K110-N123 and K153-D158) were built using Prime [55]. The residues’ protonation states were determined according to PROPKA at pH 7. Two models were built considering the two possible protonation states of His312, i.e., the neutral (His312) and the protonated (Hip312) form. According to Epik [41] results at pH 7± 2, the GDP molecule is fully deprotonated (total formal charge of −3). Hydrogen bonds were optimized according to the exhaustive sampling option, and the entire complexes were optimized by using a restrained minimization (root-mean-square deviation on heavy atoms < 0.30 Å). The K110-N123 and K153-D158 gaps of the HD domain were built, and the former was further refined using the ‘refine loops’ tool of Prime (OPLS3e [48], VSGB [56]) with default parameters. Five structures were generated, and the model with the lowest Prime energy was selected for the docking calculation.
Docking protocol. Grid-Based Ligand Docking With Energetics (Glide) [37] v.8.0 software was used with the OPLS_2005 force field. Receptor grids for HIS312 and HIP312 systems were generated in a cubic region (24.5 Å) centered on GDP molecules with an inner cubic box of 10 Å. The receptor was considered a rigid body, while the ligands were considered flexible. The standard precision (SP) method was applied with default parameters. No Epik state penalty was added to the Glide score. The docking protocol was validated for the X-ray ligand by saving five poses after a post-minimization of the first 10 poses.
The top-ranked poses succeeded in reproducing the experimental binding mode of GDP (RMSD on heavy atoms of 0.78 Å and 1.23 Å in the HIS312 and HIP312 models, respectively) (Supplementary Figures S1 and S2).
Fragment libraries preparation. Seven fragment libraries were downloaded:
- Maybridge Ro3 Diversity Set (2500 fragments) (www.maybridge.com, accessed on 9 June 2015);
- Asinex-Fragments-21872 (21,872 fragments) (www.asinex.com, accessed on 18 January 2019);
- ‘Fragment Libraries with Experimental Solubility Data’, two datasets from Life Chemicals (11,667 and 2921 fragments, respectively) (www.lifechemicals.com, accessed on 1 February 2019);
- OTAVA Solubility Fragment Library (1021 fragments) (www.otavachemicals.com, accessed on 4 February 2019);
- FragmentLibrary_sdf_13808 (13,808 fragments) from CHEMBRIDGE (www.chembridge.com, accessed on 18 April 2019)
- Preplated Fragment-Based Library (4532 fragments) from SPECS (www.specs.net, accessed on 11 February 2019).
For each fragment, we generated 3D structures, tautomers, stereoisomers (at most 32 per ligand), and protonation states (Epik at pH = 7 ± 2) using the Ligprep tool [40]. Their energy was minimized using ‘MacroModel’ [40], implemented with truncated Newton conjugated gradient method [57], and the resulting structures were used as input for docking calculations (see Supplementary Table S1).
Molecular Dynamics simulations. Molecular dynamics (MD) simulations (100 ns, NPT, OPLS3e [48], T = 300 K, Langevin thermostat [58] relaxation time = 1.0 ps; p = 1 atm; barostat relaxation time = 2.0 ps [59]) were carried out using Desmond [46] starting from the best pose of the eighteen fragments selected for the TSA (Hip312 best pose for aminobenzoic acids, His312 best pose for all the other fragments). Atomic coordinates were saved every 100 ps for a total of about 1000 frames. The systems were solvated into a (10 Å side) cubic box of TIP3P [47] water molecules and neutralized by adding Cl− and Na+ ions at a physiological concentration of 0.15 M NaCl. The systems were equilibrated by applying the ‘desmond_npt_relax.msj’ protocol available in Desmond with default parameters.
3.2. Experimental Methods
Cloning, Expression, and Purification ofRelSeqconstructs. A pET21 expression vector containing the DNA sequence coding for RelSeq 1–385 fused with a C-terminal His-tag was purchased from Giotto Biotech. Two truncated constructs of the bifunctional enzyme, RelSeq 79–385 (RelSeq SYNTH) and RelSeq 1–224 (RelSeq HYD) [50], presenting only the synthetase or hydrolase domain, respectively, were obtained with the Q5 Site-directed mutagenesis kit (New England Biolabs). Each protein construct was overproduced in BL21(DE3) Escherichia coli cells (Merck), grown in LB medium. Protein expression was induced by the addition of 0.5 mM IPTG and prolonged overnight at 25 °C for RelSeq 1–385 and RelSeq HYD and at 20 °C for RelSeq SYNTH.
In a typical purification, bacterial cells harvested by centrifugation were resuspended in lysis buffer (50 mM Tris-HCl pH 8.0, 250 mM NaCl, 10 mM imidazole, 0.5 mM TCEP), supplemented with 1 mM phenylmethanesulfonylfluoride, 20 μg/mL DNAse I (Merck) and, only in the case of RelSeq SYNTH, 0.1% Triton X-100. Cell disruption was performed by sonication, and, after high-speed centrifugation and microfiltration, the resulting bacterial soluble extract was loaded on two 1 mL Ni Sepharose HisTrap columns (GE Healthcare), connected in series and equilibrated with lysis buffer. Elution of RelSeq constructs was achieved by applying a linear gradient of elution buffer (50 mM Tris-HCl pH 8.0, 250 mM NaCl, 500 mM imidazole, 0.5 mM TCEP) over 15 column volumes. After a size exclusion chromatography step on a HiPrep 16/60 Sephacryl S-200 HR (GE Healthcare), RelSeq constructs were stored at −80 °C in 50 mM Tris-HCl pH 8, 200 mM NaCl, 5% glycerol.
Thermal shift assays on selected fragments vs. RelSeq constructs. The binding of the selected fragments to RelSeq constructs was assessed by titration of the protein in thermal shift assays, performed using a Step One Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA, USA). Assays were performed in 20 mM Tris-HCl, pH 8, containing 150 mM NaCl. The final protein concentration was kept at 0.5 mg/mL for all RelSeq constructs, and the fluorescent Protein Thermal Shift Dye (Thermo Fisher Scientific, Waltham, MA, USA) was used to monitor protein unfolding within the excitation/emission ranges 470–505/540–700 nm. Each fragment was dissolved in DMSO at a stock concentration of 200 mM, and two-fold dilution series were prepared to have final compound concentrations ranging from 0.3 mM to 10 mM; 2.5% DMSO was added in place of the fragments for control samples. Assays were performed in triplicate at a final volume of 15 µL in MicroAmp™ Fast Optical 48-well reaction plates (Thermo Fisher Scientific, Waltham, MA, USA) sealed with adhesive films. Plates were heated from 25 to 90 °C with a heating rate of 0.5 °C/min. The Kd of protein–ligand complexes engaged by RelSeq constructs with the tested fragments was calculated from the plot of protein melting temperature variations as a function of fragment concentrations with the equation for Ligand Binding (1 site) provided with the software GraFit 5.0 (Erithacus Software, Staines, UK).
STD-NMR experiments on selected fragments vs. RelSeq constructs. Experiments were performed on a 600 MHz Bruker Avance spectrometer. All experiments were acquired at 298 and 283 K on the free ligands in deuterated phosphate buffer pH 7.4. A DMSO-d6 percentage of about 5% was added to dissolve the fragments. In 1D spectra, water suppression was achieved by excitation sculpting sequence. STD NMR experiments were performed using WATERGATE 3-9-19 pulse sequence for water suppression. On-resonance irradiation of the protein was performed at a chemical shift of −0.05 ppm and 10 ppm; off-resonance irradiation was applied at 200 ppm, where no protein signals were visible. Selective pre-saturation of the protein was achieved by a train of Gauss-shaped pulses of 49 ms in length each. STD spectra were acquired with a saturation time of 2.94 s for all compounds. Blank experiments were conducted in the absence of protein in order to avoid artifacts. We tested several protein/fragment ratios (i.e., 1:100, 1:200, and 1:1000) and found that a 1:1000 ratio with a protein concentration of 3 µM (500 μL final volume) afforded the best signal-to-noise ratio.
4. Conclusions
In conclusion, we targeted the intracellular accumulation of (p)ppGpp, a bacterial stringent-response-signaling molecule involved in the insurgence of persisters, a form of phenotypic AMR, and in bacterial virulence. New chemical entities with antimicrobial activity targeting non-essential pathways, such as (p)ppGpp signaling, are urgently needed to fight and prevent antimicrobial resistance.
We performed an extensive structure-based in silico fragment screening on the synthetase site of the bifunctional enzyme RelSeq, selecting three main chemotypes. Protein–fragment interaction experiments evidenced several low mM affinity binders. In particular, the aminobenzoic acid scaffold showed a marked synthetase domain selectivity and was therefore selected for the rational design of enzyme inhibitors that will be described in due course. Potent and selective Rel inhibitors will enable to shed light on the role of (p)ppGpp signaling in persisters’ formation and pave the way to low-selective-pressure antimicrobial therapeutic approaches.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/molecules27103097/s1, Figure S1: Superposition of X-ray vs. Hip312 best pose GDP, Figure S2: Superposition of X-ray vs. His312 best pose GDP, Figure S3: Best poses of the three singletons selected from the His312 grid, Figure S4: Tyr308-fragment centroids distances monitored during MD simulations. Figure S5: STD-NMR of B1 with RelSeq (1–385), Figure S6: STD-NMR of A2 with RelSeq (1–385), Table S1: Databases used for the virtual screening campaign, Table S2: 3D Datasets used in the VS workflow and enrichment factors calculated for each chemotype, Table S3: Fragment datasets obtained by using PubChem database, Table S4: Docking results for the fragment sets into HIP312 and HIS312 models.
Author Contributions
Conceptualization, S.S.; methodology, S.S., L.S. and M.C.; validation, S.S., M.C. and L.S.; investigation, C.C., L.S., M.M. and F.V.; writing—original draft preparation, S.S., C.C., M.C., L.S. and F.V.; writing—review and editing, S.S. and M.C.; visualization, C.C. and F.V.; supervision, S.S.; project administration, S.S. and M.C.; funding acquisition, S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This project received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme (ERACHRON project, grant agreement no. 758108). The APC was funded by the ERC (grant no. 758108).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request to the corresponding author.
Acknowledgments
The authors would like to thank Stefania Cimbari for the administrative support.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Fleming, A. On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to their Use in the Isolation of B. influenzæ. Br. J. Exp. Pathol. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations in the Review on Antimicrobial Resistance. 2016, pp. 1–81. Available online: https://apo.org.au/node/63983 (accessed on 19 April 2022).
- Theuretzbacher, U.; Gottwalt, S.; Beyer, P.; Butler, M.; Czaplewski, L.; Lienhardt, C.; Moja, L.; Paul, M.; Paulin, S.; Rex, J.H.; et al. Analysis of the clinical antibacterial and antituberculosis pipeline. Lancet Infect. Dis. 2019, 19, e40–e50. [Google Scholar] [CrossRef]
- Schrader, S.M.; Vaubourgeix, J.; Nathan, C. Biology of antimicrobial resistance and approaches to combat it. Sci. Transl. Med. 2020, 12, eaaz6992. [Google Scholar] [CrossRef] [PubMed]
- Cashel, M.; Gallant, J. Two Compounds implicated in the Function of the RC Gene of Escherichia coli. Nature 1969, 221, 838–841. [Google Scholar] [CrossRef]
- Pulschen, A.A.; Fernandes, A.Z.N.; Cunha, A.F.; Sastre, D.E.; Matsuguma, B.E.; Gueiros-Filho, F.J. Many birds with one stone: Targeting the (p)ppGpp signaling pathway of bacteria to improve antimicrobial therapy. Biophys. Rev. 2021, 13, 1039–1051. [Google Scholar] [CrossRef]
- Kamarthapu, V.; Epshtein, V.; Benjamin, B.; Proshkin, S.; Mironov, A.; Cashel, M.; Nudler, E. ppGpp couples transcription to DNA repair in E. coli. Science 2016, 352, 993–996. [Google Scholar] [CrossRef]
- Molodtsov, V.; Sineva, E.; Zhang, L.; Huang, X.; Cashel, M.; Ades, S.E.; Murakami, K.S. Allosteric Effector ppGpp Potentiates the Inhibition of Transcript Initiation by DksA. Mol. Cell 2018, 69, 828–839.e5. [Google Scholar] [CrossRef]
- Diez, S.; Ryu, J.; Caban, K.; Gonzalez, R.L., Jr.; Dworkin, J. The alarmones (p)ppGpp directly regulate translation initiation during entry into quiescence. Proc. Natl. Acad. Sci. USA 2020, 117, 15565–15572. [Google Scholar] [CrossRef]
- Steinchen, W.; Zegarra, V.; Bange, G. (p)ppGpp: Magic Modulators of Bacterial Physiology and Metabolism. Front. Microbiol. 2020, 11, 2072. [Google Scholar] [CrossRef]
- Irving, S.E.; Choudhury, N.R.; Corrigan, R.M. The stringent response and physiological roles of (pp)pGpp in bacteria. Nat. Rev. Microbiol. 2021, 19, 256–271. [Google Scholar] [CrossRef]
- Buke, F.; Grilli, J.; Cosentino Lagomarsino, M.; Bokinsky, G.; Tans, S.J. ppGpp is a bacterial cell size regulator. Curr. Biol. 2022, 32, 870–877.e5. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Islam, M.; Jung, H.W.; Lim, D.; Kim, K.; Lee, S.G.; Park, C.; Lee, J.C.; Shin, M. ppGpp signaling plays a critical role in virulence of Acinetobacter baumannii. Virulence 2021, 12, 2122–2132. [Google Scholar] [CrossRef] [PubMed]
- Kalia, D.; Merey, G.; Nakayama, S.; Zheng, Y.; Zhou, J.; Luo, Y.; Guo, M.; Roembke, B.T.; Sintim, H.O. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 2013, 42, 305–341. [Google Scholar] [CrossRef] [PubMed]
- Hauryliuk, V.; Atkinson, G.C.; Murakami, K.S.; Tenson, T.; Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 2015, 13, 298–309. [Google Scholar] [CrossRef]
- Bigger, J.W. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 1944, 244, 497–500. [Google Scholar] [CrossRef]
- Cohen, N.R.; Lobritz, M.A.; Collins, J.J. Microbial persistence and the road to drug resistance. Cell Host Microbe 2013, 13, 632–642. [Google Scholar] [CrossRef]
- Huemer, M.; Mairpady Shambat, S.; Brugger, S.D.; Zinkernagel, A.S. Antibiotic resistance and persistence—Implications for human health and treatment perspectives. EMBO Rep. 2020, 21, e51034. [Google Scholar] [CrossRef]
- Wilmaerts, D.; Windels, E.M.; Verstraeten, N.; Michiels, J. General Mechanisms Leading to Persister Formation and Awakening. Trends Genet 2019, 35, 401–411. [Google Scholar] [CrossRef]
- Atkinson, G.C.; Tenson, T.; Hauryliuk, V. The RelA/SpoT homolog (RSH) superfamily: Distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS ONE 2011, 6, e23479. [Google Scholar] [CrossRef]
- Kaspy, I.; Glaser, G. Escherichia coli RelA Regulation via Its C-Terminal Domain. Front. Microbiol. 2020, 11, 572419. [Google Scholar] [CrossRef]
- Winther, K.S.; Roghanian, M.; Gerdes, K. Activation of the Stringent Response by Loading of RelA-tRNA Complexes at the Ribosomal A-Site. Mol. Cell. 2018, 70, 95–105.e4. [Google Scholar] [CrossRef] [PubMed]
- Takada, H.; Roghanian, M.; Caballero-Montes, J.; Van Nerom, K.; Jimmy, S.; Kudrin, P.; Trebini, F.; Murayama, R.; Akanuma, G.; Garcia-Pino, A.; et al. Ribosome association primes the stringent factor Rel for tRNA-dependent locking in the A-site and activation of (p)ppGpp synthesis. Nucleic Acids Res. 2021, 49, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Pausch, P.; Abdelshahid, M.; Steinchen, W.; Schafer, H.; Gratani, F.L.; Freibert, S.A.; Wolz, C.; Turgay, K.; Wilson, D.N.; Bange, G. Structural Basis for Regulation of the Opposing (p)ppGpp Synthetase and Hydrolase within the Stringent Response Orchestrator Rel. Cell Rep. 2020, 32, 108157. [Google Scholar] [CrossRef] [PubMed]
- Hogg, T.; Mechold, U.; Malke, H.; Cashel, M.; Hilgenfeld, R. Conformational Antagonism between Opposing Active Sites in a Bifunctional RelA/SpoT Homolog Modulates (p)ppGpp Metabolism during the Stringent Response. Cell 2004, 117, 57–68. [Google Scholar] [CrossRef]
- Avarbock, A.; Avarbock, D.; Teh, J.-S.; Buckstein, M.; Wang, Z.-m.; Rubin, H. Functional Regulation of the Opposing (p)ppGpp Synthetase/Hydrolase Activities of RelMtb from Mycobacterium tuberculosis †. Biochemistry 2005, 44, 9913–9923. [Google Scholar] [CrossRef] [PubMed]
- Tamman, H.; Van Nerom, K.; Takada, H.; Vandenberk, N.; Scholl, D.; Polikanov, Y.; Hofkens, J.; Talavera, A.; Hauryliuk, V.; Hendrix, J.; et al. A nucleotide-switch mechanism mediates opposing catalytic activities of Rel enzymes. Nat. Chem. Biol. 2020, 16, 834–840. [Google Scholar] [CrossRef]
- Sinha, A.K.; Winther, K.S. The RelA hydrolase domain acts as a molecular switch for (p)ppGpp synthesis. Commun. Biol. 2021, 4, 434. [Google Scholar] [CrossRef]
- Wexselblatt, E.; Oppenheimer-Shaanan, Y.; Kaspy, I.; London, N.; Schueler-Furman, O.; Yavin, E.; Glaser, G.; Katzhendler, J.; Ben-Yehuda, S. Relacin, a novel antibacterial agent targeting the Stringent Response. PLoS Pathog. 2012, 8, e1002925. [Google Scholar] [CrossRef]
- Wexselblatt, E.; Katzhendler, J.; Saleem-Batcha, R.; Hansen, G.; Hilgenfeld, R.; Glaser, G.; Vidavski, R.R. ppGpp analogues inhibit synthetase activity of Rel proteins from Gram-negative and Gram-positive bacteria. Bioorg. Med. Chem. 2010, 18, 4485–4497. [Google Scholar] [CrossRef]
- Wexselblatt, E.; Kaspy, I.; Glaser, G.; Katzhendler, J.; Yavin, E. Design, synthesis and structure-activity relationship of novel Relacin analogs as inhibitors of Rel proteins. Eur. J. Med. Chem. 2013, 70, 497–504. [Google Scholar] [CrossRef]
- Andresen, L.; Varik, V.; Tozawa, Y.; Jimmy, S.; Lindberg, S.; Tenson, T.; Hauryliuk, V. Auxotrophy-based High Throughput Screening assay for the identification of Bacillus subtilis stringent response inhibitors. Sci. Rep. 2016, 6, 35824. [Google Scholar] [CrossRef] [PubMed]
- Beljantseva, J.; Kudrin, P.; Jimmy, S.; Ehn, M.; Pohl, R.; Varik, V.; Tozawa, Y.; Shingler, V.; Tenson, T.; Rejman, D.; et al. Molecular mutagenesis of ppGpp: Turning a RelA activator into an inhibitor. Sci. Rep. 2017, 7, 41839. [Google Scholar] [CrossRef] [PubMed]
- Dutta, N.K.; Klinkenberg, L.G.; Vazquez, M.J.; Segura-Carro, D.; Colmenarejo, G.; Ramon, F.; Rodriguez-Miquel, B.; Mata-Cantero, L.; Porras-De Francisco, E.; Chuang, Y.M.; et al. Inhibiting the stringent response blocks Mycobacterium tuberculosis entry into quiescence and reduces persistence. Sci. Adv. 2019, 5, eaav2104. [Google Scholar] [CrossRef] [PubMed]
- Conti, G.; Minneci, M.; Sattin, S. Optimised Synthesis of the Bacterial Magic Spot (p)ppGpp Chemosensor PyDPA. ChemBioChem 2019, 20, 1717–1721. [Google Scholar] [CrossRef]
- Civera, M.; Sattin, S. Homology Model of a Catalytically Competent Bifunctional Rel Protein. Front. Mol. Biosci. 2021, 8, 628596. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Erlanson, D.A.; Fesik, S.W.; Hubbard, R.E.; Jahnke, W.; Jhoti, H. Twenty years on: The impact of fragments on drug discovery. Nat. Rev. Drug Discov. 2016, 15, 605–619. [Google Scholar] [CrossRef]
- Lamoree, B.; Hubbard, R.E. Using Fragment-Based Approaches to Discover New Antibiotics. SLAS Discov. 2018, 23, 495–510. [Google Scholar] [CrossRef]
- Schrödinger. Maestro, Schrödinger Release 2018-1; Schrödinger, LLC: New York, NY, USA, 2018. [Google Scholar]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pK(a) prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef]
- Duan, J.; Dixon, S.L.; Lowrie, J.F.; Sherman, W. Analysis and comparison of 2D fingerprints: Insights into database screening performance using eight fingerprint methods. J. Mol. Graph. Model. 2010, 29, 157–170. [Google Scholar] [CrossRef]
- Sastry, M.; Lowrie, J.F.; Dixon, S.L.; Sherman, W. Large-Scale Systematic Analysis of 2D Fingerprint Methods and Parameters to Improve Virtual Screening Enrichments. J. Chem. Inf. Model. 2010, 50, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B.; Holloway, G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2020, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossváry, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the ACM/IEEE Conference on Supercomputing (SC06), Tampa, FL, USA, 11–17 November 2006. [Google Scholar]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Roos, K.; Wu, C.; Damm, W.; Reboul, M.; Stevenson, J.M.; Lu, C.; Dahlgren, M.K.; Mondal, S.; Chen, W.; Wang, L.; et al. OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. J. Chem. Theory Comput. 2019, 15, 1863–1874. [Google Scholar] [CrossRef] [PubMed]
- Huynh, K.; Partch, C.L. Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein. Sci. 2015, 79, 28.9.1–28.9.14. [Google Scholar] [CrossRef]
- Mechold, U.; Murphy, H.; Brown, L.; Cashel, M. Intramolecular Regulation of the Opposing (p)ppGpp Catalytic Activities of RelSeq, the Rel/Spo Enzyme from Streptococcus equisimilis. J. Bacteriol. 2002, 184, 2878–2888. [Google Scholar] [CrossRef][Green Version]
- Meyer, B.; Peters, T. NMR Spectroscopy Techniques for Screening and Identifying Ligand Binding to Protein Receptors. Angew. Chem. Int. Ed. 2003, 42, 864–890. [Google Scholar] [CrossRef]
- Guzzetti, I.; Civera, M.; Vasile, F.; Araldi, E.M.; Belvisi, L.; Gennari, C.; Potenza, D.; Fanelli, R.; Piarulli, U. Determination of the binding epitope of RGD-peptidomimetics to αvβ3 and αIIbβ3 integrin-rich intact cells by NMR and computational studies. Org. Biomol. Chem. 2013, 11, 3886–3893. [Google Scholar] [CrossRef]
- Sattin, S.; Panza, M.; Vasile, F.; Berni, F.; Goti, G.; Tao, J.H.; Moroni, E.; Agard, D.; Colombo, G.; Bernardi, A. Synthesis of Functionalized 2-(4-Hydroxyphenyl)-3-methylbenzofuran Allosteric Modulators of Hsp90 Activity. Eur. J. Org. Chem. 2016, 2016, 3349–3364. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.; Honig, B.; Shaw, D.E.; Friesner, R.A. A hierarchical approach to all-atom protein loop prediction. Proteins 2004, 55, 351–367. [Google Scholar] [CrossRef]
- Li, J.; Abel, R.; Zhu, K.; Cao, Y.; Zhao, S.; Friesner, R.A. The VSGB 2.0 model: A next generation energy model for high resolution protein structure modeling. Proteins 2011, 79, 2794–2812. [Google Scholar] [CrossRef] [PubMed]
- Ponder, J.W.; Richards, F.M. An efficient newton-like method for molecular mechanics energy minimization of large molecules. J. Comput. Chem. 1987, 8, 1016–1024. [Google Scholar] [CrossRef]
- Grest, G.S.; Kremer, K. Molecular dynamics simulation for polymers in the presence of a heat bath. Phys. Rev. A 1986, 33, 3628–3631. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Vangunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular-Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).