Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis
Abstract
:1. Introduction
2. Results
2.1. PyuS Contains the Signature CXPXR Sulfatase Motif
2.2. Production and Purification of Recombinant PyuS
2.3. PyuS Crystal Structure
2.4. PyuS Oligomerization
2.5. PyuS Activity on Sulfatase and Phosphatase Substrates
2.5.1. PyuS Activity on Aromatic Sulfate Esters and Steroid Sulfates
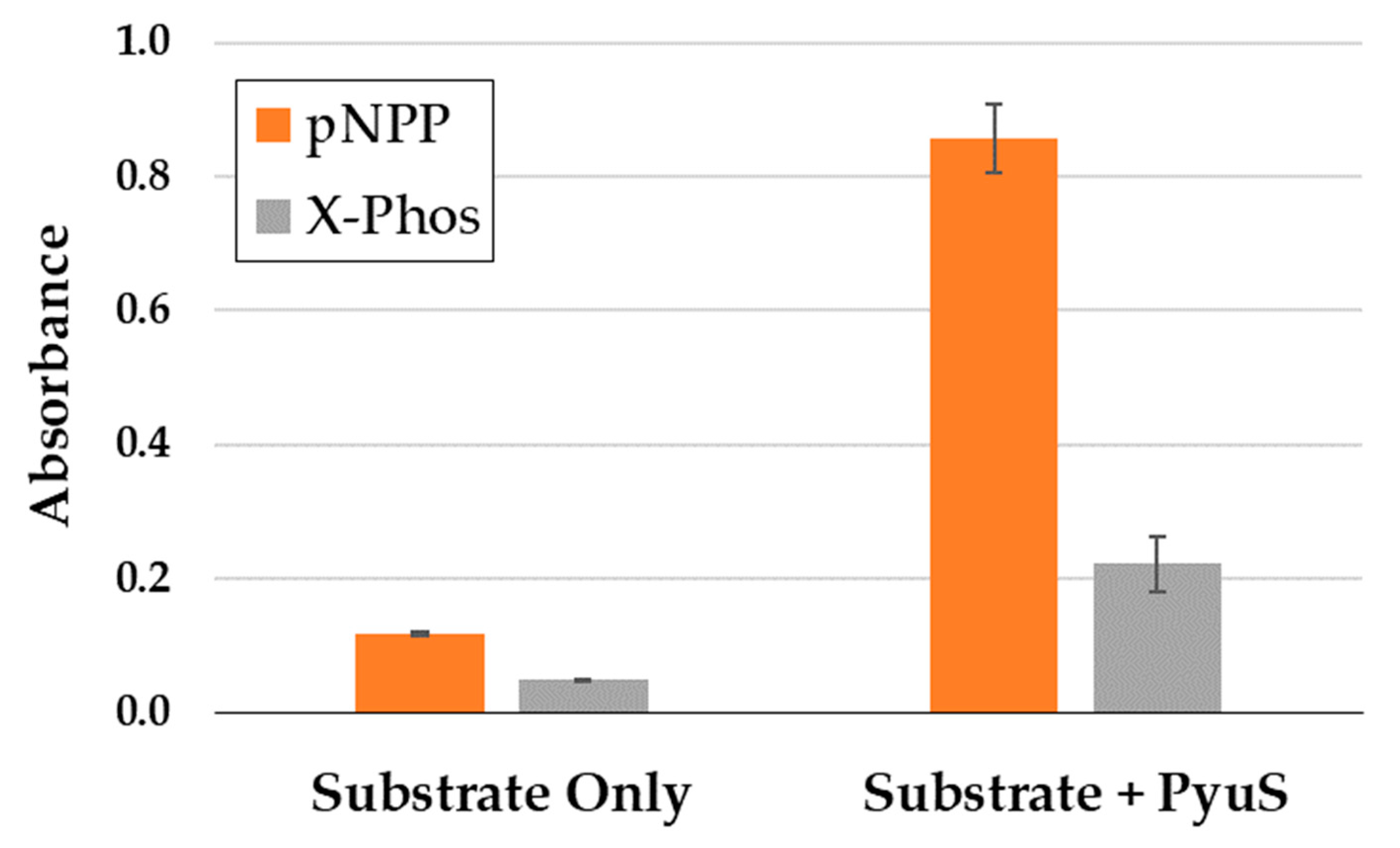
2.5.2. PyuS Activity on Phosphatase Substrates
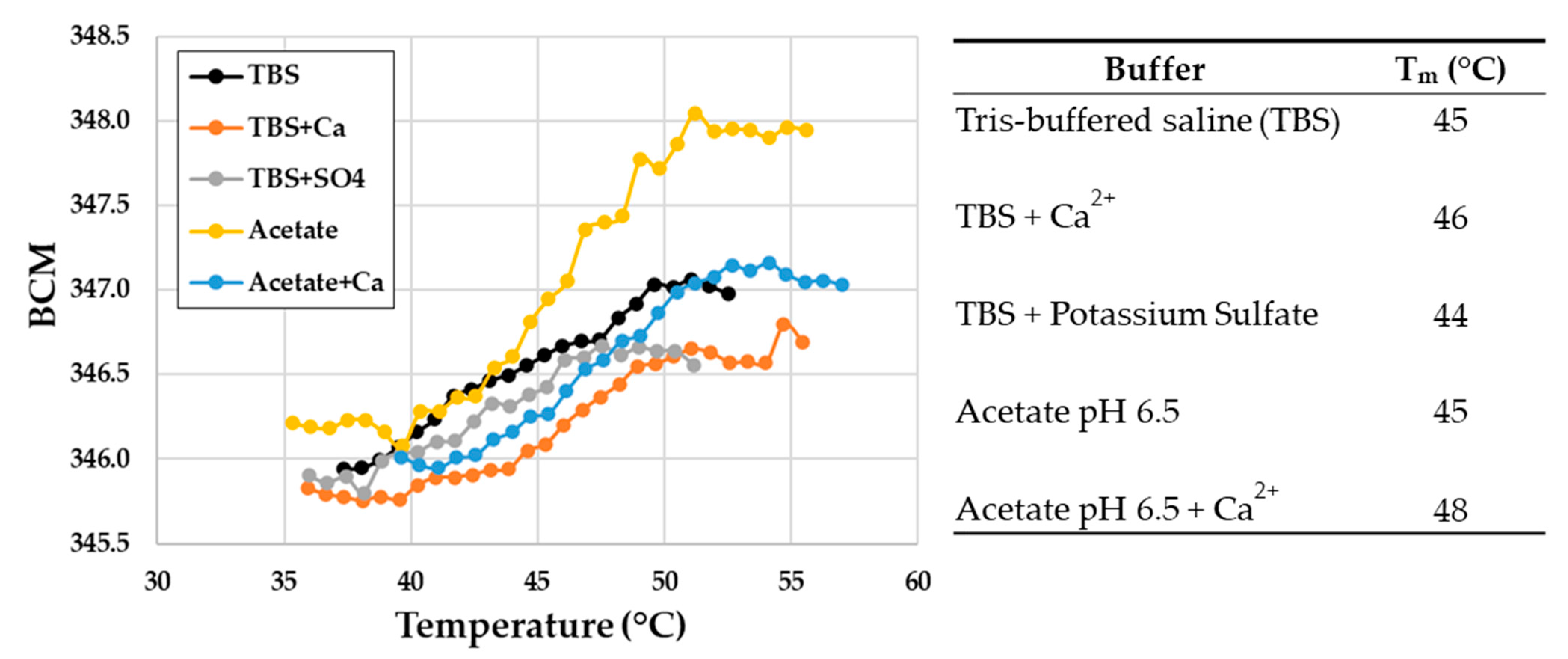
2.6. PyuS Stability
3. Discussion
4. Materials and Methods
4.1. Gene Synthesis, Protein Expression, and Purification
4.2. Metal Analysis
4.3. Protein Crystallization, Data Collection, and Refinement
4.4. Structure Determination, Refinement, and Validation
4.5. Protein Modeling and Structure Analysis
4.6. Hydrolysis Assays with 4MUS, pNCS, and X-Sulf
4.7. HPLC with Steroid Sulfates
4.8. Hydrolysis Assays with pNPP and X-Phos
4.9. Melting Temperature of PyuS
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Varin, L.; DeLuca, V.; Ibrahim, R.K.; Brisson, N. Molecular Characterization of Two Plant Flavonol Sulfotransferases. Proc. Natl. Acad. Sci. USA 1992, 89, 1286–1290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Sreevidya, V.S.; Udvadia, A.J.; Gyaneshwar, P. Dopamine-Induced Sulfatase and Its Regulator Are Required for Salmonella Enterica Serovar Typhimurium Pathogenesis. Microbiology 2019, 165, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Reed, M.J.; Purohit, A.; Woo, L.W.L.; Newman, S.P.; Potter, B.V.L. Steroid Sulfatase: Molecular Biology, Regulation, and Inhibition. Endocr. Rev. 2005, 26, 171–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevenson, B.J.; Waller, C.C.; Ma, P.; Li, K.; Cawley, A.T.; Ollis, D.L.; McLeod, M.D. Pseudomonas Aeruginosa Arylsulfatase: A Purified Enzyme for the Mild Hydrolysis of Steroid Sulfates: Steroid Sulfate Hydrolysis by Pseudomonas Aeruginosa Arylsulfatase. Drug Test. Anal. 2015, 7, 903–911. [Google Scholar] [CrossRef]
- Iannone, M.; Botrè, F.; Martinez-Brito, D.; Matteucci, R.; de la Torre, X. Development and Application of Analytical Procedures for the GC–MS/MS Analysis of the Sulfates Metabolites of Anabolic Androgenic Steroids: The Pivotal Role of Chemical Hydrolysis. J. Chromatogr. B 2020, 1155, 122280. [Google Scholar] [CrossRef]
- Sasisekharan, R.; Raman, R.; Prabhakar, V. Glycomics Approach to Structure-Function Relationships of Glycosaminoglycans. Annu. Rev. Biomed. Eng. 2006, 8, 181–231. [Google Scholar] [CrossRef]
- Diez-Roux, G.; Ballabio, A. Sulfatases and human disease. Annu. Rev. Genom. Hum. Genet. 2005, 6, 355–379. [Google Scholar] [CrossRef]
- Dierks, T.; Schmidt, B.; Borissenko, L.V.; Peng, J.; Preusser, A.; Mariappan, M.; von Figura, K. Multiple Sulfatase Deficiency Is Caused by Mutations in the Gene Encoding the Human Cα-Formylglycine Generating Enzyme. Cell 2003, 113, 435–444. [Google Scholar] [CrossRef] [Green Version]
- Barbeyron, T.; Brillet-Guéguen, L.; Carré, W.; Carrière, C.; Caron, C.; Czjzek, M.; Hoebeke, M.; Michel, G. Matching the Diversity of Sulfated Biomolecules: Creation of a Classification Database for Sulfatases Reflecting Their Substrate Specificity. PLoS ONE 2016, 11, e0164846. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef] [Green Version]
- Knaust, A.; Schmidt, B.; Dierks, T.; von Bülow, R.; von Figura, K. Residues Critical for Formylglycine Formation and/or Catalytic Activity of Arylsulfatase, A. Biochemistry 1998, 37, 13941–13946. [Google Scholar] [CrossRef]
- Dierks, T. Sequence Determinants Directing Conversion of Cysteine to Formylglycine in Eukaryotic Sulfatases. EMBO J. 1999, 18, 2084–2091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berteau, O.; Guillot, A.; Benjdia, A.; Rabot, S. A New Type of Bacterial Sulfatase Reveals a Novel Maturation Pathway in Prokaryotes. J. Biol. Chem. 2006, 281, 22464–22470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dierks, T.; Schmidt, B.; von Figura, K. Conversion of Cysteine to Formylglycine: A Protein Modification in the Endoplasmic Reticulum. Proc. Natl. Acad. Sci. USA 1997, 94, 11963–11968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, B.L.; Ballister, E.R.; Skordalakes, E.; King, D.S.; Breidenbach, M.A.; Gilmore, S.A.; Berger, J.M.; Bertozzi, C.R. Function and Structure of a Prokaryotic Formylglycine-Generating Enzyme. J. Biol. Chem. 2008, 283, 20117–20125. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Wang, B.; Zhang, L.; Zhang, J.; Chen, S. Pedobacter Yulinensis Sp. Nov., Isolated from Sandy Soil, and Emended Description of the Genus Pedobacter. Int. J. Syst. Evol. Microbiol. 2018, 68, 2523–2529. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Nielsen, H. Predicting Secretory Proteins with SignalP. In Protein Function Prediction; Kihara, D., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; Volume 1611, pp. 59–73. ISBN 978-1-4939-7013-1. [Google Scholar]
- Nielsen, H.; Tsirigos, K.D.; Brunak, S.; von Heijne, G. A Brief History of Protein Sorting Prediction. Protein J. 2019, 38, 200–216. [Google Scholar] [CrossRef] [Green Version]
- Rabuka, D.; Rush, J.S.; deHart, G.W.; Wu, P.; Bertozzi, C.R. Site-Specific Chemical Protein Conjugation Using Genetically Encoded Aldehyde Tags. Nat. Protoc. 2012, 7, 1052–1067. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E. ExPASy: The Proteomics Server for in-Depth Protein Knowledge and Analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Cowtan, K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Schmidt, B.; Selmer, T.; Ingendoh, A.; von Figurat, K. A Novel Amino Acid Modification in Sulfatases That Is Defective in Multiple Sulfatase Deficiency. Cell 1995, 82, 271–278. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Cooper, D.R.; Porebski, P.J.; Shabalin, I.G.; Handing, K.B.; Minor, W. CheckMyMetal: A Macromolecular Metal-Binding Validation Tool. Acta Crystallogr. Sect. Struct. Biol. 2017, 73, 223–233. [Google Scholar] [CrossRef] [Green Version]
- Ponstingl, H.; Henrick, K.; Thornton, J.M. Discriminating between Homodimeric and Monomeric Proteins in the Crystalline State. Proteins Struct. Funct. Bioinform. 2000, 41, 47–57. [Google Scholar] [CrossRef]
- van Loo, B.; Heberlein, M.; Mair, P.; Zinchenko, A.; Schüürmann, J.; Eenink, B.D.G.; Holstein, J.M.; Dilkaute, C.; Jose, J.; Hollfelder, F.; et al. High-Throughput, Lysis-Free Screening for Sulfatase Activity Using Escherichia coli Autodisplay in Microdroplets. ACS Synth. Biol. 2019, 8, 2690–2700. [Google Scholar] [CrossRef]
- Pabis, A.; Duarte, F.; Kamerlin, S.C.L. Promiscuity in the Enzymatic Catalysis of Phosphate and Sulfate Transfer. Biochemistry 2016, 55, 3061–3081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Filutowicz, M. Hexahistidine (His6)-Tag Dependent Protein Dimerization: A Cautionary Tale. Acta Biochim. Pol. 1999, 46, 591–599. [Google Scholar] [CrossRef] [Green Version]
- Booth, W.T.; Schlachter, C.R.; Pote, S.; Ussin, N.; Mank, N.J.; Klapper, V.; Offermann, L.R.; Tang, C.; Hurlburt, B.K.; Chruszcz, M. Impact of an N-Terminal Polyhistidine Tag on Protein Thermal Stability. ACS Omega 2018, 3, 760–768. [Google Scholar] [CrossRef]
- PyMOL. The PyMOL Molecular Graphics System, Version 2.0; Schrödinger, LLC: New York, NY, USA, 2016.
- Krissinel, E.; Henrick, K. Secondary-Structure Matching (SSM), a New Tool for Fast Protein Structure Alignment in Three Dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2256–2268. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Multiple Alignment of Protein Structures in Three Dimensions. In Computational Life Sciences; Berthold, M.R., Glen, R.C., Diederichs, K., Kohlbacher, O., Fischer, I., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2005; Volume 3695, pp. 67–78. ISBN 978-3-540-29104-6. [Google Scholar]
- Holm, L. Using Dali for Protein Structure Comparison. In Structural Bioinformatics; Gáspári, Z., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; Volume 2112, pp. 29–42. ISBN 978-1-07-160269-0. [Google Scholar]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural Summaries of PDB Entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef]
- Sillitoe, I.; Bordin, N.; Dawson, N.; Waman, V.P.; Ashford, P.; Scholes, H.M.; Pang, C.S.M.; Woodridge, L.; Rauer, C.; Sen, N.; et al. CATH: Increased Structural Coverage of Functional Space. Nucleic Acids Res. 2021, 49, D266–D273. [Google Scholar] [CrossRef] [PubMed]
- Lewis, T.E.; Sillitoe, I.; Dawson, N.; Lam, S.D.; Clarke, T.; Lee, D.; Orengo, C.; Lees, J. Gene3D: Extensive Prediction of Globular Domains in Proteins. Nucleic Acids Res. 2018, 46, D435–D439. [Google Scholar] [CrossRef] [PubMed]
- Golovin, A.; Henrick, K. MSDmotif: Exploring Protein Sites and Motifs. BMC Bioinform. 2008, 9, 312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Sogi, K.M.; Gartner, Z.J.; Breidenbach, M.A.; Appel, M.J.; Schelle, M.W.; Bertozzi, C.R. Mycobacterium Tuberculosis Rv3406 Is a Type II Alkyl Sulfatase Capable of Sulfate Scavenging. PLoS ONE 2013, 8, e65080. [Google Scholar] [CrossRef]
- Jorge, A.M.; Schneider, J.; Unsleber, S.; Xia, G.; Mayer, C.; Peschel, A. Staphylococcus Aureus Counters Phosphate Limitation by Scavenging Wall Teichoic Acids from Other Staphylococci via the Teichoicase GlpQ. J. Biol. Chem. 2018, 293, 14916–14924. [Google Scholar] [CrossRef] [Green Version]
- Holder, P.G.; Jones, L.C.; Drake, P.M.; Barfield, R.M.; Bañas, S.; de Hart, G.W.; Baker, J.; Rabuka, D. Reconstitution of Formylglycine-Generating Enzyme with Copper(II) for Aldehyde Tag Conversion. J. Biol. Chem. 2015, 290, 15730–15745. [Google Scholar] [CrossRef] [Green Version]
- Woo, L.W.L.; Ganeshapillai, D.; Thomas, M.P.; Sutcliffe, O.B.; Malini, B.; Mahon, M.F.; Purohit, A.; Potter, B.V.L. Structure-Activity Relationship for the First-in-Class Clinical Steroid Sulfatase Inhibitor Irosustat (STX64, BN83495). ChemMedChem 2011, 6, 2019–2034. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Jiménez, M.M.; Monje-Moreno, J.M.; Brokate-Llanos, A.M.; Venegas-Calerón, M.; Sánchez-García, A.; Sansigre, P.; Valladares, A.; Esteban-García, S.; Suárez-Pereira, I.; Vitorica, J.; et al. Steroid Hormones Sulfatase Inactivation Extends Lifespan and Ameliorates Age-Related Diseases. Nat. Commun. 2021, 12, 49. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-Ray Diffraction Data Collected in Oscillation Mode. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1997; Volume 276, pp. 307–326. ISBN 978-0-12-182177-7. [Google Scholar]
- Vagin, A.; Teplyakov, A. MOLREP: An Automated Program for Molecular Replacement. J. Appl. Crystallogr. 1997, 30, 1022–1025. [Google Scholar] [CrossRef]
- Minor, W.; Cymborowski, M.; Otwinowski, Z.; Chruszcz, M. HKL-3000: The Integration of Data Reduction and Structure Solution—from Diffraction Images to an Initial Model in Minutes. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 859–866. [Google Scholar] [CrossRef] [Green Version]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository—New Features and Functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated Comparative Protein Structure Modeling with SWISS-MODEL and Swiss-PdbViewer: A Historical Perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC 5 for the Refinement of Macromolecular Crystal Structures. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Painter, J.; Merritt, E.A. TLSMD Web Server for the Generation of Multi-Group TLS Models. J. Appl. Crystallogr. 2006, 39, 109–111. [Google Scholar] [CrossRef] [Green Version]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-Atom Structure Validation for Macromolecular Crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Handing, K.B.; Niedzialkowska, E.; Shabalin, I.G.; Kuhn, M.L.; Zheng, H.; Minor, W. Characterizing Metal-Binding Sites in Proteins with X-Ray Crystallography. Nat. Protoc. 2018, 13, 1062–1090. [Google Scholar] [CrossRef]
- Zheng, H.; Chordia, M.D.; Cooper, D.R.; Chruszcz, M.; Müller, P.; Sheldrick, G.M.; Minor, W. Validation of Metal-Binding Sites in Macromolecular Structures with the CheckMyMetal Web Server. Nat. Protoc. 2014, 9, 156–170. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and Development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS Biomolecular Solvation Software Suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 Suite and Current Developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellis, K.J.; Morrison, J.F. [23] Buffers of Constant Ionic Strength for Studying PH-Dependent Processes. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1982; Volume 87, pp. 405–426. ISBN 978-0-12-181987-3. [Google Scholar]
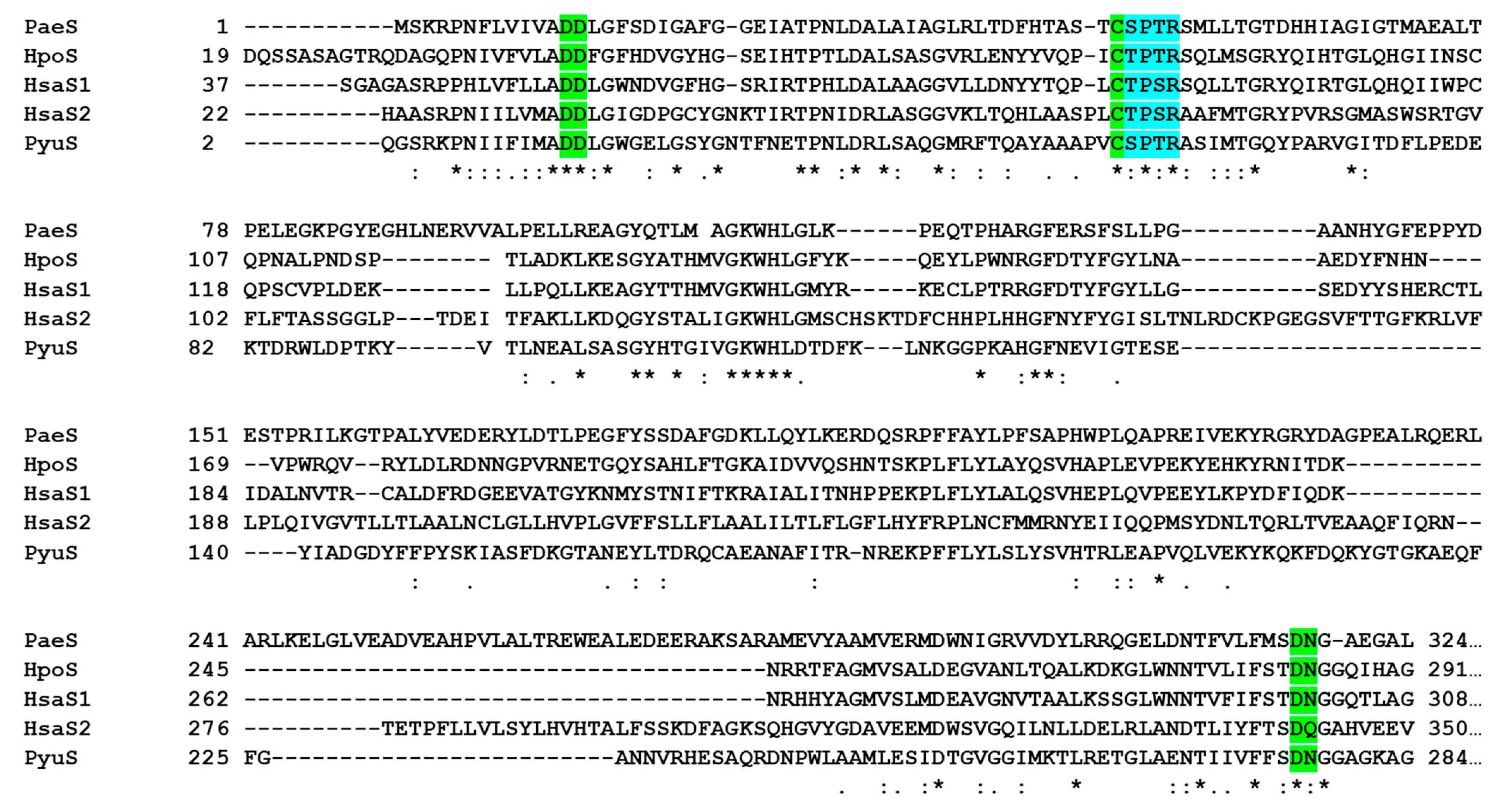
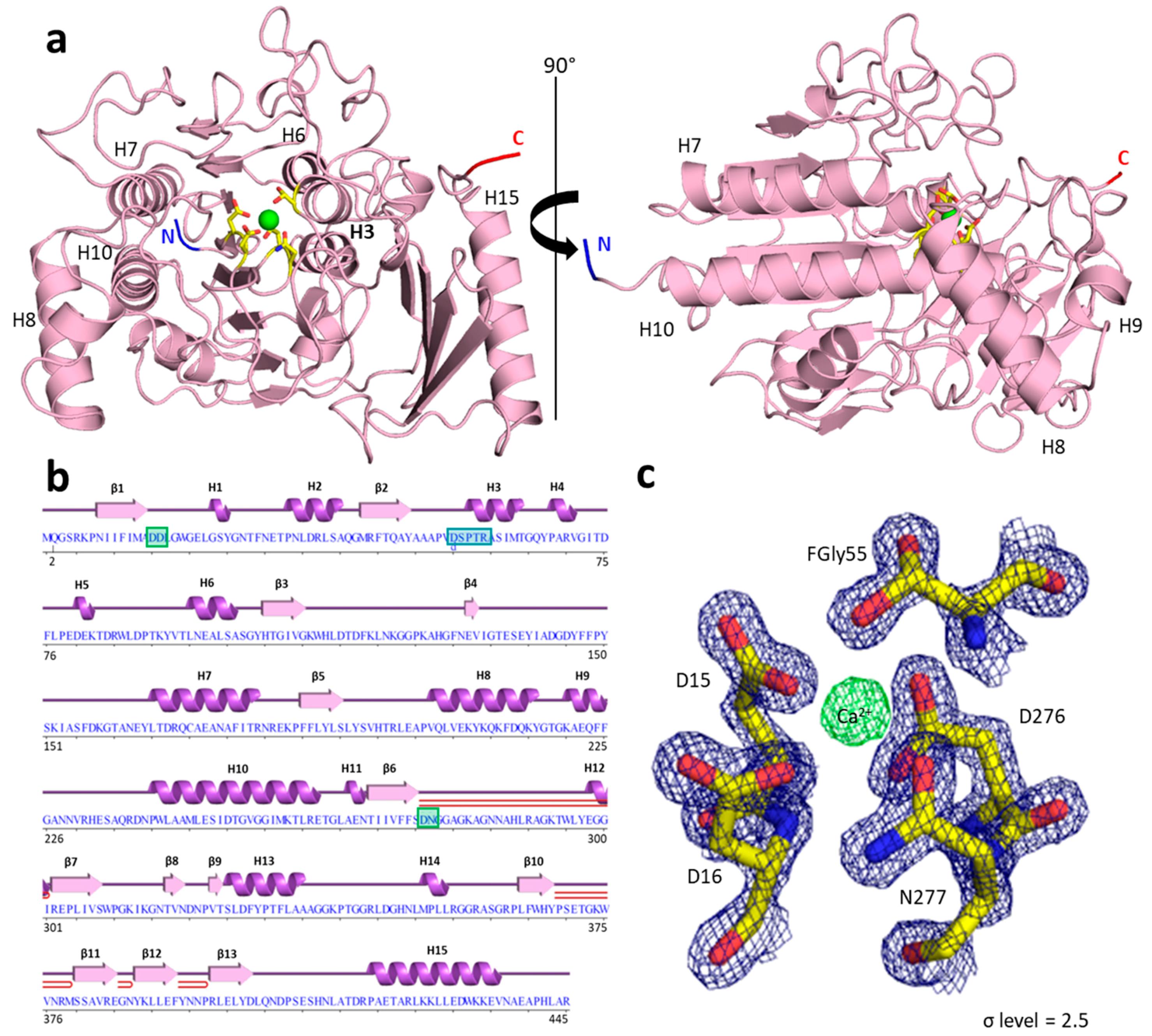
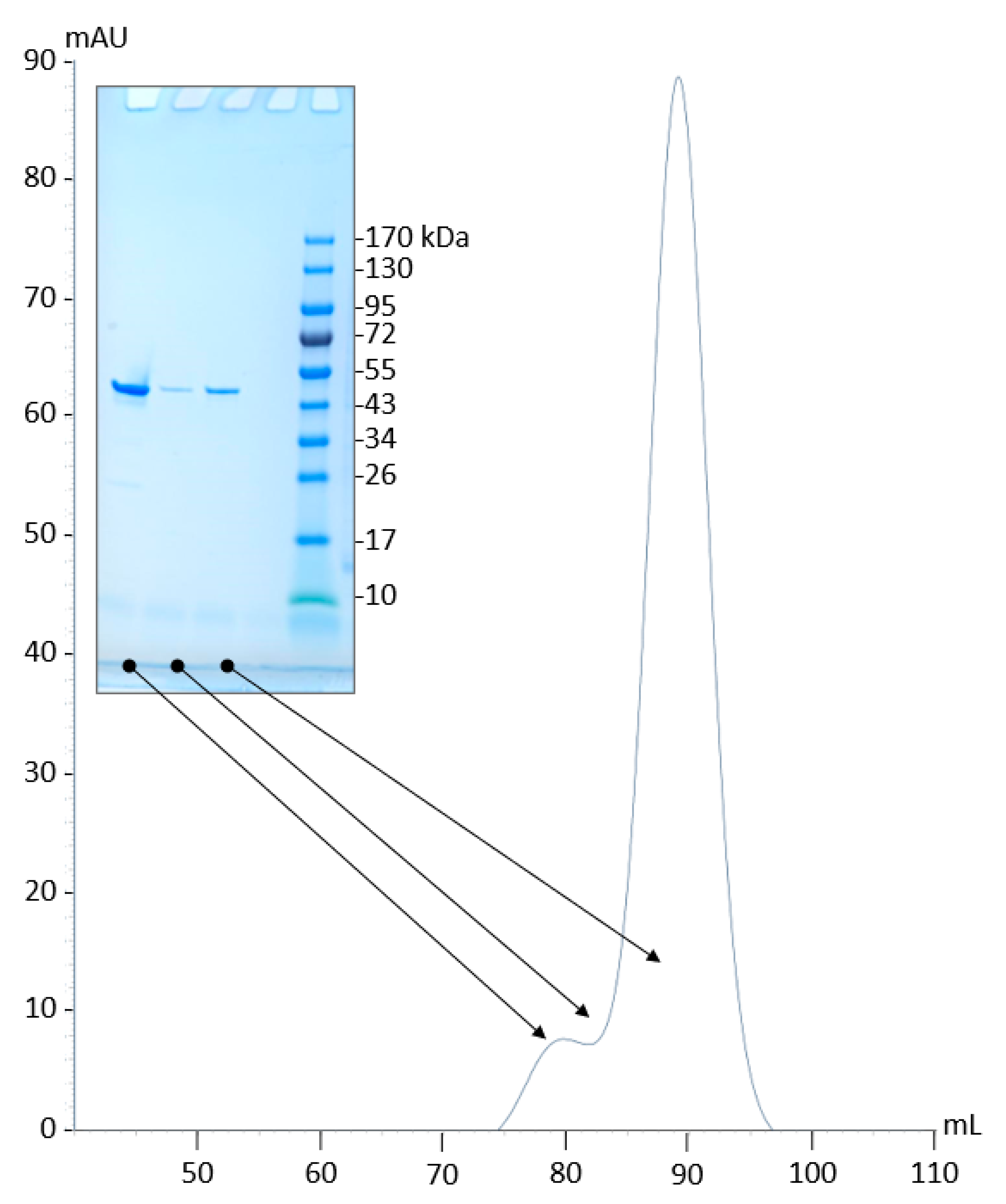


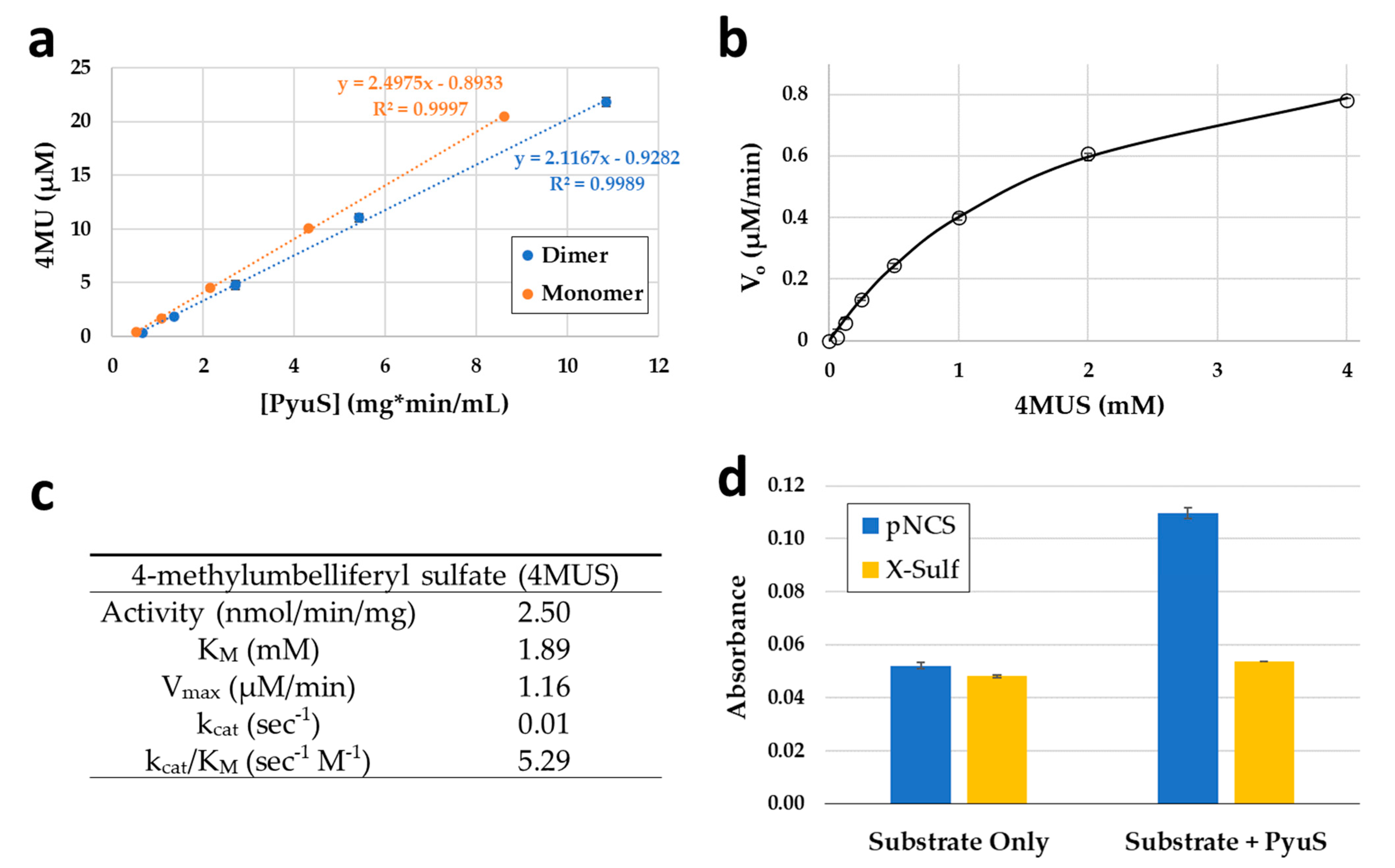
| PDB Code | Protein | RMSD (Å) | Cα Aligned | Sequence Identity to PyuS (%) |
|---|---|---|---|---|
| 1FSU | N-acetylgalactosamine-4-sulfatase | 1.0 | 265 | 18.4 |
| 1HDH | Arylsulfatase | 1.7 | 260 | 15.3 |
| 1P49 | Sterylsulfatase | 0.9 | 251 | 20.0 |
| 6USS | Sulfatase (not classified) | 0.8 | 275 | 24.9 |
| 6UST | N-acetylglucosamine-6-sulfatase | 0.8 | 317 | 29.8 |
| 5G2V | N-acetylglucosamine-6-sulfatase | 1.7 | 251 | 19.8 |
| 4FDJ | N-acetylglucosamine-6-sulfatase | 0.9 | 249 | 23.3 |
| 6B0J | Iota-carrageenan sulfatase | 0.9 | 277 | 23.1 |
| Structure | PyuS (Malonate) | PyuS (Bromine) | PyuS (Citrate) |
|---|---|---|---|
| PDB accession code | 7STT | 7STU | 7STV |
| Data collection | |||
| Diffraction source | APS, 22ID | APS, 22ID | APS, 22ID |
| Wavelength (Å) | 1.0 | 1.0 | 1.0 |
| Space group | P3221 | P3221 | P3221 |
| a, b, c (Å) | 83.6, 83.6, 116.1 | 83.6, 83.6, 115.8 | 82.7, 82.7, 115.3 |
| Resolution range (Å) | 40.0–1.60 (1.63–1.60) | 40.0–2.23 (2.27–2.23) | 40.0–2.34 (2.38–2.34) |
| No. of unique reflections | 59,449 (2069) | 23,149 (1144) | 19,275 (936) |
| Completeness (%) | 95.7 (67.5) | 99.2 (99.6) | 98.8 (98.7) |
| Redundancy | 10.6 (5.8) | 9.1 (6.2) | 9.2 (6.7) |
| <I/σ(I)> | 35.3 (2) | 15.1 (2) | 18.5 (2) |
| Rmeas | 0.079 (0.487) | 0.172 (0.858) | 0.148 (0.525) |
| Rp.i.m. | 0.024 (0.186) | 0.057 (0.339) | 0.048 (0.175) |
| CC1/2 | 0.996 (0.859) | 0.997 (0.738) | 0.998 (0.948) |
| Refinement | |||
| Resolution range (Å) | 39.37–1.60 (1.63–1.60) | 39.22–2.23 (2.29–2.23) | 34.23–2.35 (2.41–2.35) |
| Completeness (%) | 95.5 (68.4) | 99.2 (99.3) | 98.4 (93.3) |
| No. of reflections, working set | 56312 | 21815 | 18285 |
| No. of reflections, test set | 3033 | 1205 | 943 |
| Final Rcryst | 0.148 (0.276) | 0.174 (0.282) | 0.172 (0.286) |
| Final Rfree | 0.178 (0.270) | 0.225 (0.319) | 0.230 (0.301) |
| RMSD bonds (Å) | 0.014 | 0.007 | 0.005 |
| RMSD angles (°) | 1.8 | 1.4 | 1.3 |
| Ramachandran plot | |||
| Allowed regions (%) | 100 | 100 | 100 |
| Favored regions (%) | 97.1 | 97.1 | 97.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schlachter, C.R.; O’Malley, A.; Grimes, L.L.; Tomashek, J.J.; Chruszcz, M.; Lee, L.A. Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis. Molecules 2022, 27, 87. https://doi.org/10.3390/molecules27010087
Schlachter CR, O’Malley A, Grimes LL, Tomashek JJ, Chruszcz M, Lee LA. Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis. Molecules. 2022; 27(1):87. https://doi.org/10.3390/molecules27010087
Chicago/Turabian StyleSchlachter, Caleb R., Andrea O’Malley, Linda L. Grimes, John J. Tomashek, Maksymilian Chruszcz, and L. Andrew Lee. 2022. "Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis" Molecules 27, no. 1: 87. https://doi.org/10.3390/molecules27010087
APA StyleSchlachter, C. R., O’Malley, A., Grimes, L. L., Tomashek, J. J., Chruszcz, M., & Lee, L. A. (2022). Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis. Molecules, 27(1), 87. https://doi.org/10.3390/molecules27010087







