Engineered Bacteriorhodopsin May Induce Lung Cancer Cell Cycle Arrest and Suppress Their Proliferation and Migration
Abstract
:1. Introduction
2. Results
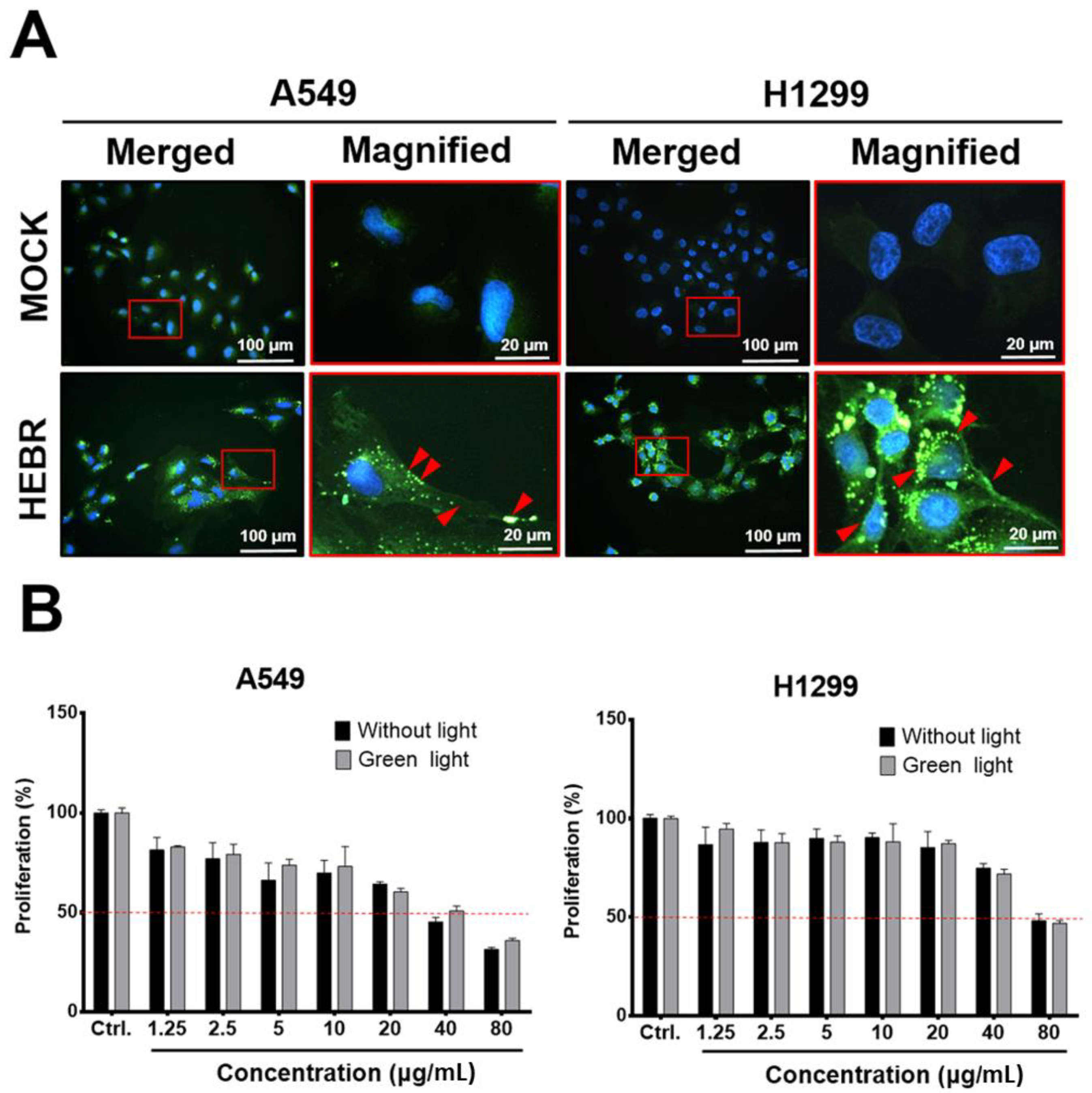
2.1. HEBR Protein Suppresses the Cell Proliferation of A549 and H1299 Cells
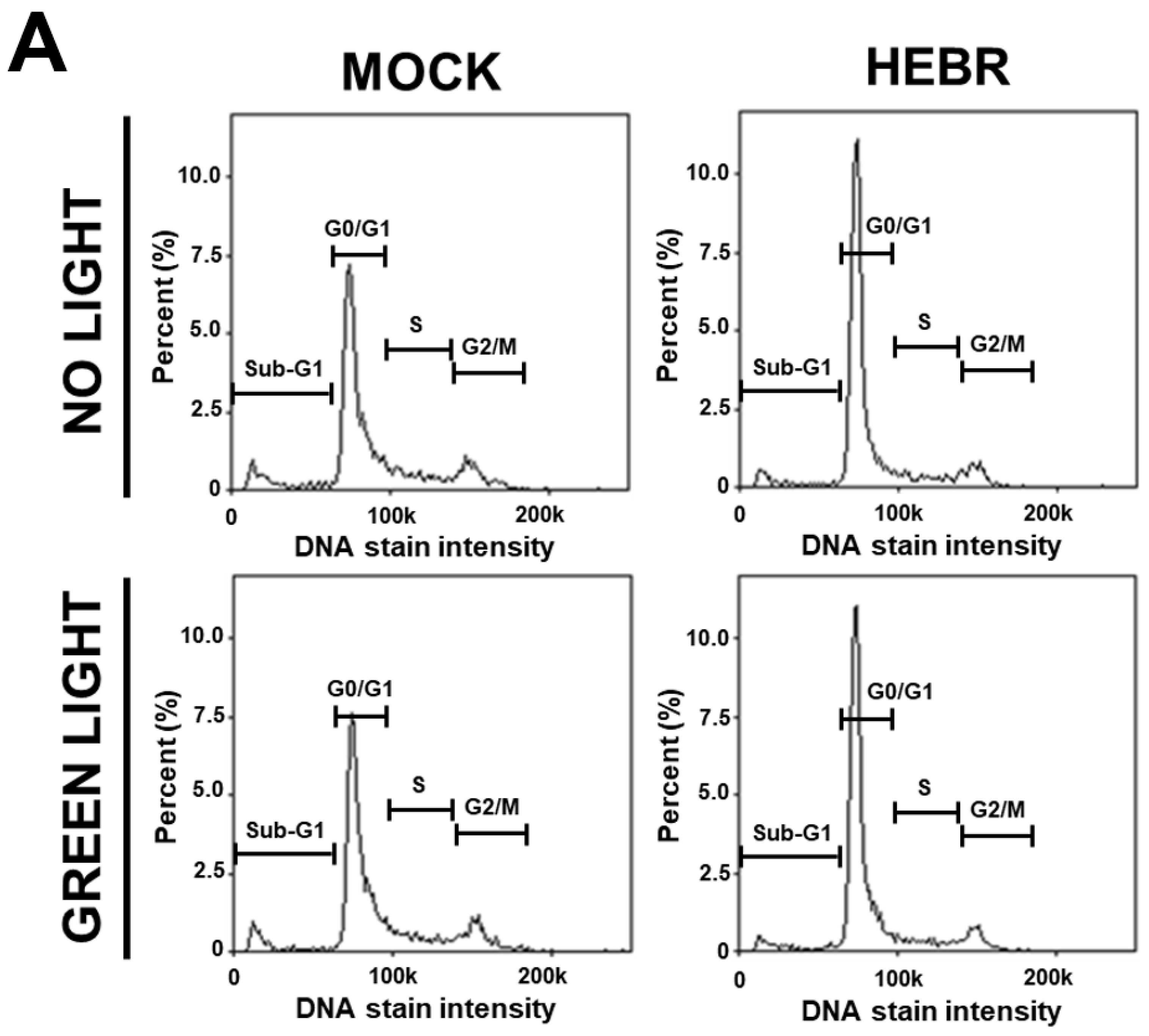
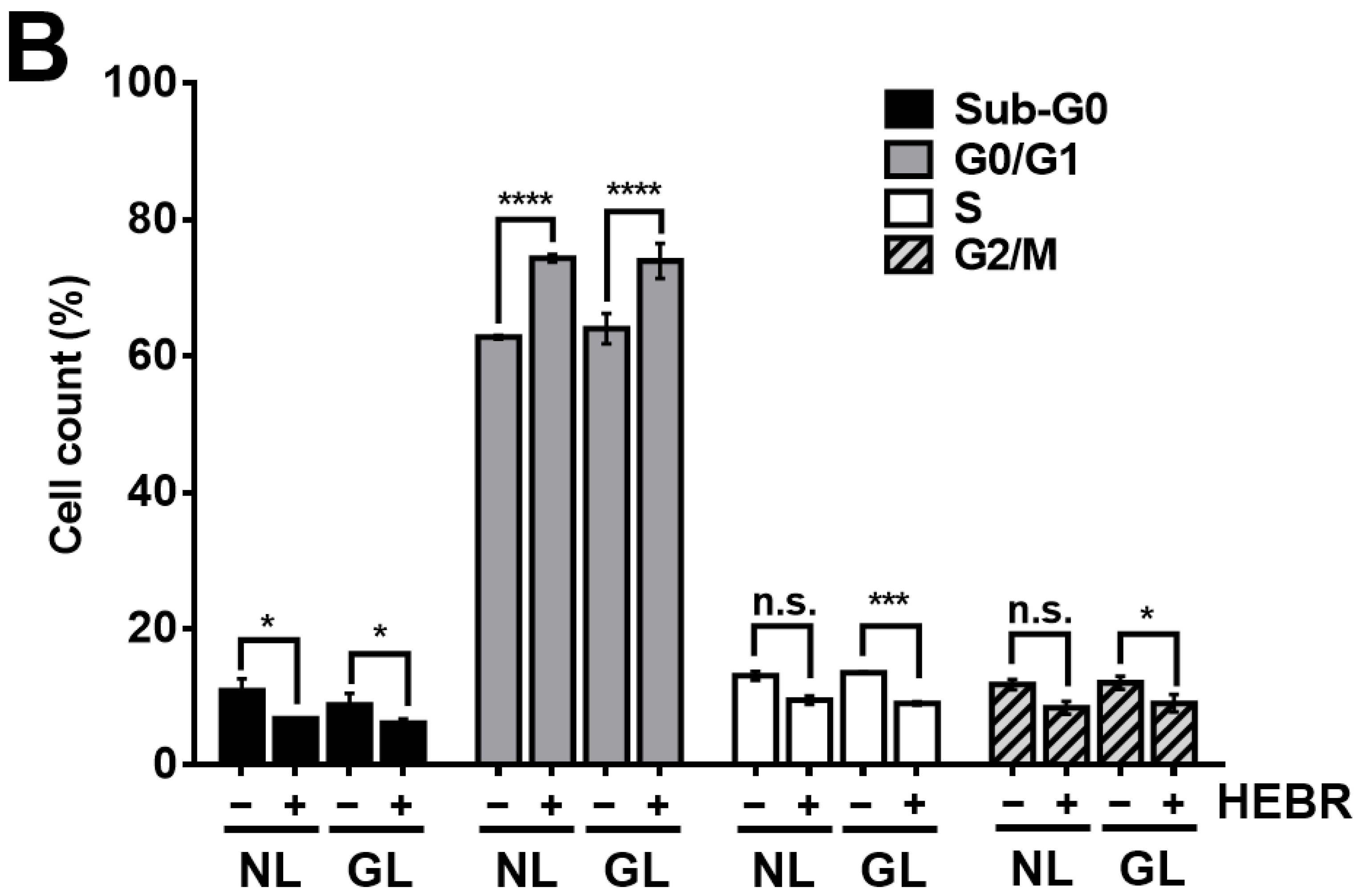
2.2. Cell Cycle Arrest in HEBR-Protein-Treated A549 Cells
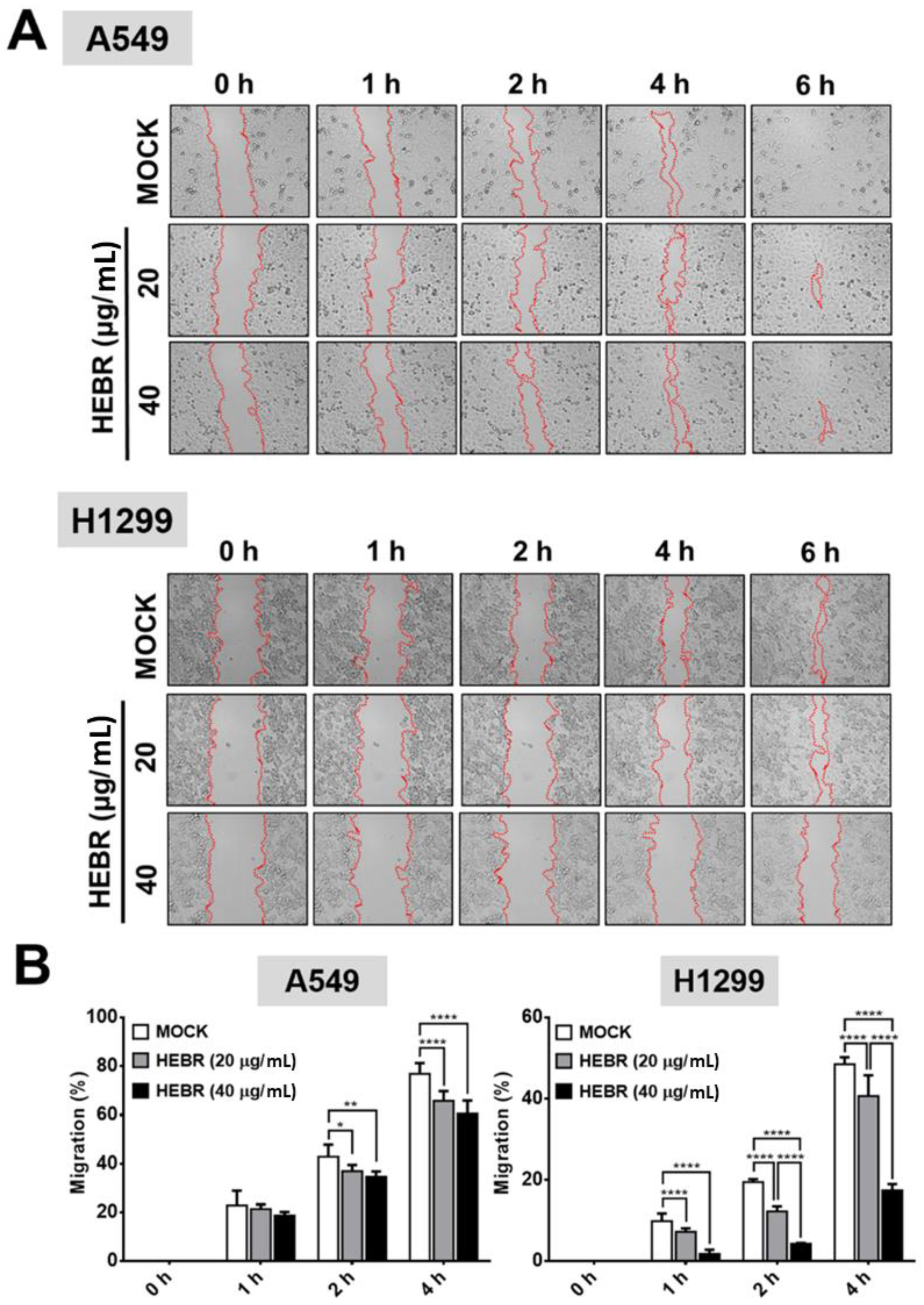
2.3. Scratch Assay on HEBR-Treated Cells
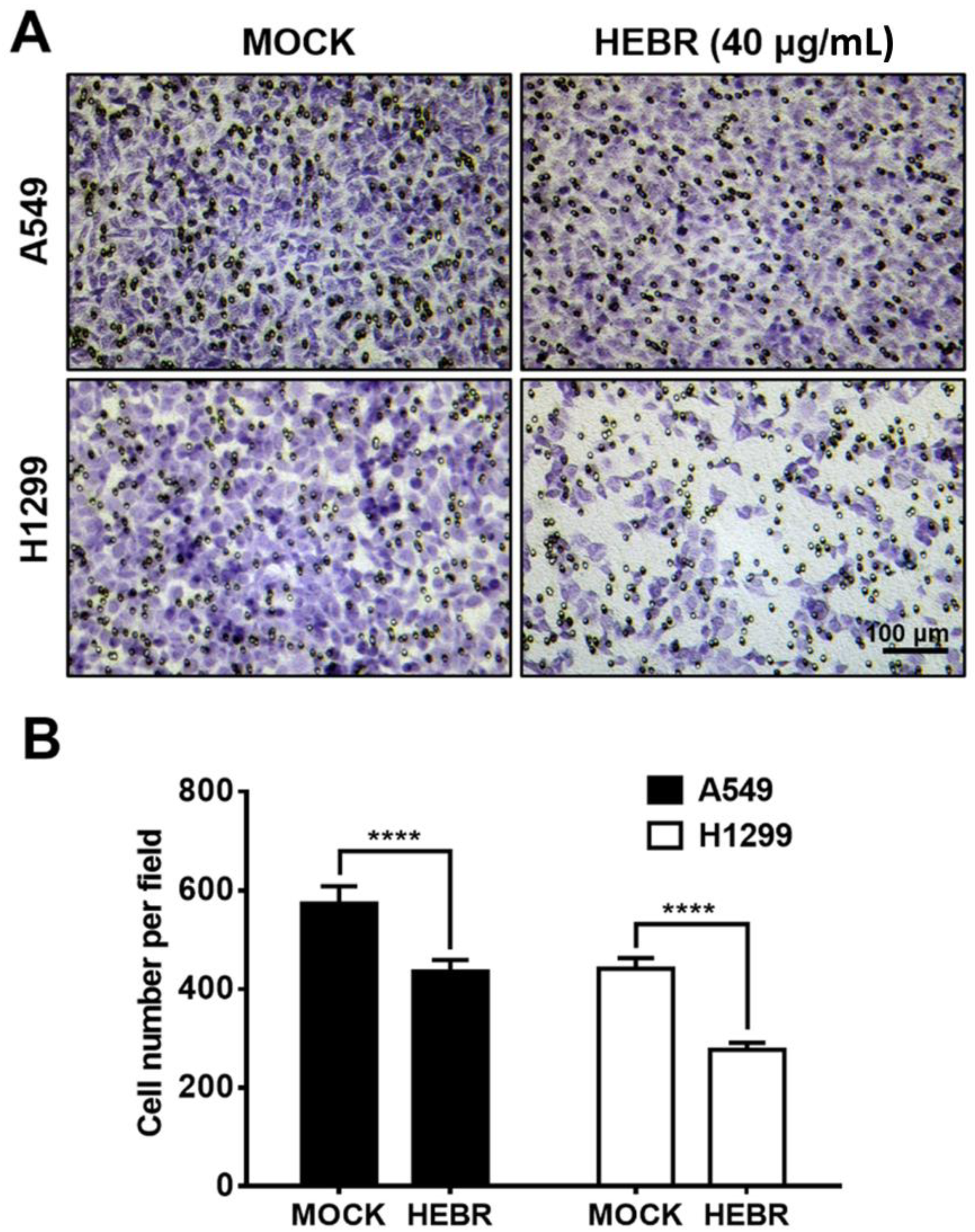
2.4. Transwell Assay of HEBR Protein on A549 and H1299 Cells
2.5. Gene Expression of A549 and H1299 Cells after Treatment with HEBR Protein
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Preparation of Purified HEBR Protein
4.3. Immunostaining for HEBR Protein in A549 and H1299 Cells
4.4. Cell Viability Analysis
4.5. Cell Cycle
4.6. Migration Assay
4.7. RNA Extraction and Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR) Analysis
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Rodríguez-Valera, F. Introduction to Saline Environments. In The Biology of Halophilic Bacteria; CRC Press: Boca Raton, FL, USA, 1993; pp. 1–23. [Google Scholar]
- Ventosa, A.; Nieto, J.J.; Oren, A. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 1998, 62, 504–544. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.K.; Walsh, D.A.; Bapteste, E.; Rodriguez-Valera, F.; Doolittle, W.F.; Papke, R.T. Evolution of rhodopsin ion pumps in haloarchaea. BMC Evol. Biol. 2007, 7, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oesterhelt, D.; Stoeckenius, W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. New Biol. 1971, 233, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Ernst, O.P.; Lodowski, D.T.; Elstner, M.; Hegemann, P.; Brown, L.S.; Kandori, H. Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chem. Rev. 2014, 114, 126–163. [Google Scholar] [CrossRef]
- Mukhina, T.; Gerelli, Y.; Hemmerle, A.; Koutsioubas, A.; Kovalev, K.; Teulon, J.-M.; Pellequer, J.-L.; Daillant, J.; Charitat, T.; Fragneto, G. Insertion and activation of functional Bacteriorhodopsin in a floating bilayer. J. Colloid Interface Sci. 2021, 597, 370–382. [Google Scholar] [CrossRef] [PubMed]
- Schobert, B.; Lanyi, J.K. Halorhodopsin is a light-driven chloride pump. J. Biol. Chem. 1982, 257, 10306–10313. [Google Scholar] [CrossRef]
- Feroz, H.; Ferlez, B.; Oh, H.; Mohammadiarani, H.; Ren, T.; Baker, C.S.; Gajewski, J.P.; Lugar, D.J.; Gaudana, S.B.; Butler, P.; et al. Liposome-based measurement of light-driven chloride transport kinetics of halorhodopsin. Biochim. Biophys. Acta BBA Biomembr. 2021, 1863, 183637. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, K.; Polovinkin, V.; Gushchin, I.; Alekseev, A.; Shevchenko, V.; Borshchevskiy, V.; Astashkin, R.; Balandin, T.; Bratanov, D.; Vaganova, S.; et al. Structure and mechanisms of sodium-pumping KR2 rhodopsin. Sci. Adv. 2019, 5, eaav2671. [Google Scholar] [CrossRef] [Green Version]
- Nagel, G.; Szellas, T.; Huhn, W.; Kateriya, S.; Adeishvili, N.; Berthold, P.; Ollig, D.; Hegemann, P.; Bamberg, E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA 2003, 100, 13940–13945. [Google Scholar] [CrossRef] [Green Version]
- Kato, H.E. Structure–function relationship of channelrhodopsins. In Optogenetics; Yawo, H., Kandori, H., Koizumi, A., Kageyama, R., Eds.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2021; Volume 1293, pp. 35–53. [Google Scholar] [CrossRef]
- Kühlbrandt, W. Bacteriorhodopsin—The movie. Nature 2000, 406, 569–570. [Google Scholar] [CrossRef]
- Miller, R.D.; Paré-Labrosse, O.; Sarracini, A.; Besaw, J.E. Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin in the multiphoton regime and biological relevance. Nat. Commun. 2020, 11, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, C.-H.; Yi, H.-P.; Hsieh, S.-Y.; Lin, H.-S.; Yang, C.-S. Overexpression of different types of microbial rhodopsins with a highly expressible bacteriorhodopsin from Haloarcula marismortui as a single protein in E. coli. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef]
- Hsu, M.-F.; Yu, T.-F.; Chou, C.-C.; Fu, H.-Y.; Yang, C.-S.; Wang, A.H.J. Using Haloarcula marismortui Bacteriorhodopsin as a fusion tag for enhancing and visible expression of integral membrane proteins in Escherichia coli. PLoS ONE 2013, 8, e56363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, H.-W.; Ko, L.-N.; Yang, C.-S.; Hsu, S.-h. Potential of engineered bacteriorhodopsins as photoactivated biomaterials in modulating neural stem cell behavior. ACS Biomater. Sci. Eng. 2019, 5, 3068–3078. [Google Scholar] [CrossRef] [PubMed]
- Mattingly, M.; Weineck, K.; Costa, J.; Cooper, R.L. Hyperpolarization by activation of halorhodopsin results in enhanced synaptic transmission: Neuromuscular junction and CNS circuit. PLoS ONE 2018, 13, e0200107. [Google Scholar] [CrossRef]
- Mittal, V. Epithelial mesenchymal transition in aggressive lung cancers. In Lung Cancer and Personalized Medicine: Novel Therapies and Clinical Management; Springer: Cham, Switzerland, 2016; pp. 37–56. [Google Scholar]
- Lim, J.; Thiery, J.P. Epithelial-mesenchymal transitions: Insights from development. Development 2012, 139, 3471–3486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-mesenchymal transition in cancer: A historical overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef]
- Yan, B.; Zhang, W.; Jiang, L.-Y.; Qin, W.-X.; Wang, X. Reduced E-Cadherin expression is a prognostic biomarker of non-small cell lung cancer: A meta-analysis based on 2395 subjects. Int. J. Clin. Exp. Med. 2014, 7, 4352. [Google Scholar]
- Loh, C.-Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-cadherin and N-cadherin switch in epithelial-to-mesenchymal transition: Signaling, therapeutic implications, and challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef] [Green Version]
- Georgakopoulos-Soares, I.; Chartoumpekis, D.V.; Kyriazopoulou, V.; Zaravinos, A. EMT factors and metabolic pathways in cancer. Front. Oncol. 2020, 10, 499. [Google Scholar] [CrossRef] [PubMed]
- Cano, A.; Pérez-Moreno, M.A.; Rodrigo, I.; Locascio, A.; Blanco, M.J.; del Barrio, M.G.; Portillo, F.; Nieto, M.A. The transcription factor snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000, 2, 76–83. [Google Scholar] [CrossRef]
- Lambert, A.W.; Weinberg, R.A. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat. Rev. Cancer 2021, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Saigusa, S.; Tanaka, K.; Toiyama, Y.; Yokoe, T.; Okugawa, Y.; Ioue, Y.; Miki, C.; Kusunoki, M. Correlation of CD133, OCT4, and SOX2 in rectal cancer and their association with distant recurrence after chemoradiotherapy. Ann. Surg. Oncol. 2009, 16, 3488–3498. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J., Jr.; Wu, Y.-L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef]
- Jones, G.S.; Baldwin, D.R. Recent advances in the management of lung cancer. Clin. Med. 2018, 18 (Suppl. S2), s41. [Google Scholar] [CrossRef]
- Jiang, L.; Huang, J.; Jiang, S.; Rong, W.; Shen, Y.; Li, C.; Tian, Y.; Ning, J.; Chen, X.; Yang, Y.; et al. The surgical perspective in neoadjuvant immunotherapy for resectable non-small cell lung cancer. Cancer Immunol. Immunother. 2021, 70, 2313–2321. [Google Scholar] [CrossRef]
- Serrano-Novillo, C.; Capera, J.; Colomer-Molera, M.; Condom, E.; Ferreres, J.C.; Felipe, A. Implication of voltage-gated potassium channels in neoplastic cell proliferation. Cancers 2019, 11, 287. [Google Scholar] [CrossRef] [Green Version]
- Luo, P.-W.; Han, H.-W.; Yang, C.-S.; Shrestha, L.K.; Ariga, K.; Hsu, S.-h. Optogenetic modulation and reprogramming of bacteriorhodopsin-transfected human fibroblasts on self-assembled fullerene C60 nanosheets. Adv. Biosyst. 2019, 3, 1800254. [Google Scholar] [CrossRef]
- Hsieh, F.-Y.; Han, H.-W.; Chen, X.-R.; Yang, C.-S.; Wei, Y.; Hsu, S.-h. Non-viral delivery of an optogenetic tool into cells with self-healing hydrogel. Biomaterials 2018, 174, 31–40. [Google Scholar] [CrossRef]
- Kozin, E.D.; Brown, M.C.; Lee, D.J.; Stankovic, K.M. Light-based neuronal activation: The future of cranial nerve stimulation. Otolaryngol. Clin. N. Am. 2020, 53, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Bostanciklioglu, M. Optogenetic stimulation of serotonin nuclei retrieve the lost memory in Alzheimer’s disease. J. Cell. Physiol. 2020, 235, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.W.; Ye, X.L.; Huang, T.; Yang, X.F.; Zou, L.Y. Optogenetics-induced activation of glutamate receptors improves memory function in mice with Alzheimer’s disease. Neural Regen. Res. 2019, 14, 2147–2155. [Google Scholar]
- Kielbus, M.; Czapinski, J.; Odrzywolski, A.; Stasiak, G.; Szymanska, K.; Kalafut, J.; Kos, M.; Giannopoulos, K.; Stepulak, A.; Rivero-Muller, A. Optogenetics in cancer drug discovery. Expert Opin. Drug Discov. 2018, 13, 459–472. [Google Scholar] [CrossRef]
- Hu, W.; Li, Q.; Li, B.; Ma, K.; Zhang, C.; Fu, X. Optogenetics sheds new light on tissue engineering and regenerative medicine. Biomaterials 2020, 227, 119546. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Lin, X.; Zhang, J.; Su, L.; Ma, M.; Ea, V.L.; Liu, X.; Wang, L.; Chang, J.; Li, X.; et al. Blue light-triggered optogenetic system for treating uveal melanoma. Oncogene 2020, 39, 2118–2124. [Google Scholar] [CrossRef]
- McCormick, J.W.; Pincus, D.; Resnekov, O.; Reynolds, K.A. Strategies for engineering and rewiring kinase regulation. Trends Biochem. Sci. 2019, 45, 259–271. [Google Scholar] [CrossRef]
- D’Antonio, C.; Passaro, A.; Gori, B.; Del Signore, E.; Migliorino, M.R.; Ricciardi, S.; Fulvi, A.; De Marinis, F. Bone and brain metastasis in lung cancer: Recent advances in therapeutic strategies. Ther. Adv. Med. Oncol. 2014, 6, 101–114. [Google Scholar] [CrossRef] [Green Version]
- Talmadge, J.E.; Fidler, I.J. AACR centennial series: The biology of cancer metastasis: Historical perspective. Cancer Res. 2010, 70, 5649–5669. [Google Scholar] [CrossRef] [Green Version]
- Batlle, E.; Sancho, E.; Francí, C.; Domínguez, D.; Monfar, M.; Baulida, J.; De Herreros, A.G. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000, 2, 84–89. [Google Scholar] [CrossRef]
- Hüser, L.; Novak, D.; Umansky, V.; Altevogt, P.; Utikal, J. Targeting SOX2 in anticancer therapy. Expert Opin. Ther. Targets 2018, 22, 983–991. [Google Scholar] [CrossRef]
- Xiang, R.; Liao, D.; Cheng, T.; Zhou, H.; Shi, Q.; Chuang, T.; Markowitz, D.; Reisfeld, R.; Luo, Y. Downregulation of transcription factor SOX2 in cancer stem cells suppresses growth and metastasis of lung cancer. Br. J. Cancer 2011, 104, 1410–1417. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhu, Z.; Li, L.; Ye, W.; Zeng, J.; Gao, J.; Wang, S.; Zhang, L.; Huang, Z. Effect of overexpression of Oct4 and Sox2 genes on the biological and oncological characteristics of gastric cancer cells. OncoTargets Ther. 2019, 12, 4667. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-H.; Fu, K.-Y.; Hong, P.-D.; Ma, H.; Liou, N.-H.; Ma, K.-H.; Liu, J.-C.; Huang, K.-L.; Dai, L.-G.; Chang, S.-C.; et al. The Effects of Microenvironment on Wound Healing by Keratinocytes Derived From Mesenchymal Stem Cells. Ann. Plast. Surg. 2013, 71, S67–S74. [Google Scholar] [CrossRef] [PubMed]
- Cormier, N.; Yeo, A.; Fiorentino, E.; Paxson, J. Optimization of the wound scratch assay to detect changes in murine mesenchymal stromal cell migration after damage by soluble cigarette smoke extract. J. Vis. Exp. 2015, 106, e53414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, C.-H.; Lee, S.-W.; Li, C.-F.; Wang, J.; Yang, W.-L.; Wu, C.-Y.; Wu, J.; Nakayama, K.I.; Kang, H.-Y.; Huang, H.-Y.; et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat. Cell Biol. 2010, 12, 457–467. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wong, C.-W.; Ko, L.-N.; Huang, H.-J.; Yang, C.-S.; Hsu, S.-h. Engineered Bacteriorhodopsin May Induce Lung Cancer Cell Cycle Arrest and Suppress Their Proliferation and Migration. Molecules 2021, 26, 7344. https://doi.org/10.3390/molecules26237344
Wong C-W, Ko L-N, Huang H-J, Yang C-S, Hsu S-h. Engineered Bacteriorhodopsin May Induce Lung Cancer Cell Cycle Arrest and Suppress Their Proliferation and Migration. Molecules. 2021; 26(23):7344. https://doi.org/10.3390/molecules26237344
Chicago/Turabian StyleWong, Chui-Wei, Ling-Ning Ko, Hung-Jin Huang, Chii-Shen Yang, and Shan-hui Hsu. 2021. "Engineered Bacteriorhodopsin May Induce Lung Cancer Cell Cycle Arrest and Suppress Their Proliferation and Migration" Molecules 26, no. 23: 7344. https://doi.org/10.3390/molecules26237344
APA StyleWong, C.-W., Ko, L.-N., Huang, H.-J., Yang, C.-S., & Hsu, S.-h. (2021). Engineered Bacteriorhodopsin May Induce Lung Cancer Cell Cycle Arrest and Suppress Their Proliferation and Migration. Molecules, 26(23), 7344. https://doi.org/10.3390/molecules26237344






