Quantitative Mass Spectrometry-Based Proteomics for Biomarker Development in Ovarian Cancer
Abstract
1. Overview of Ovarian Cancer
2. Targeted Therapies for Ovarian Cancer
3. Overview of Ovarian Cancer Biomarkers
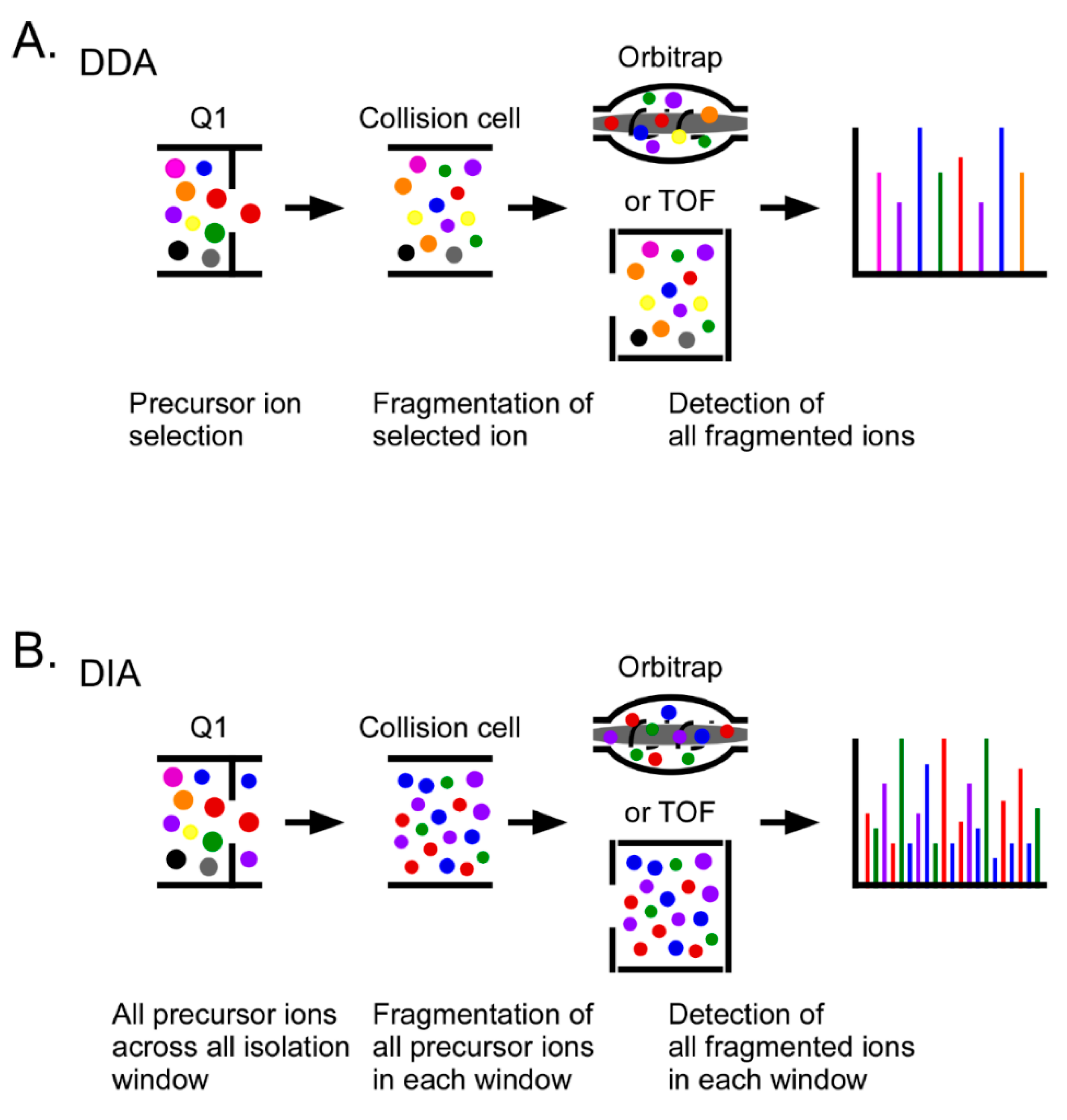
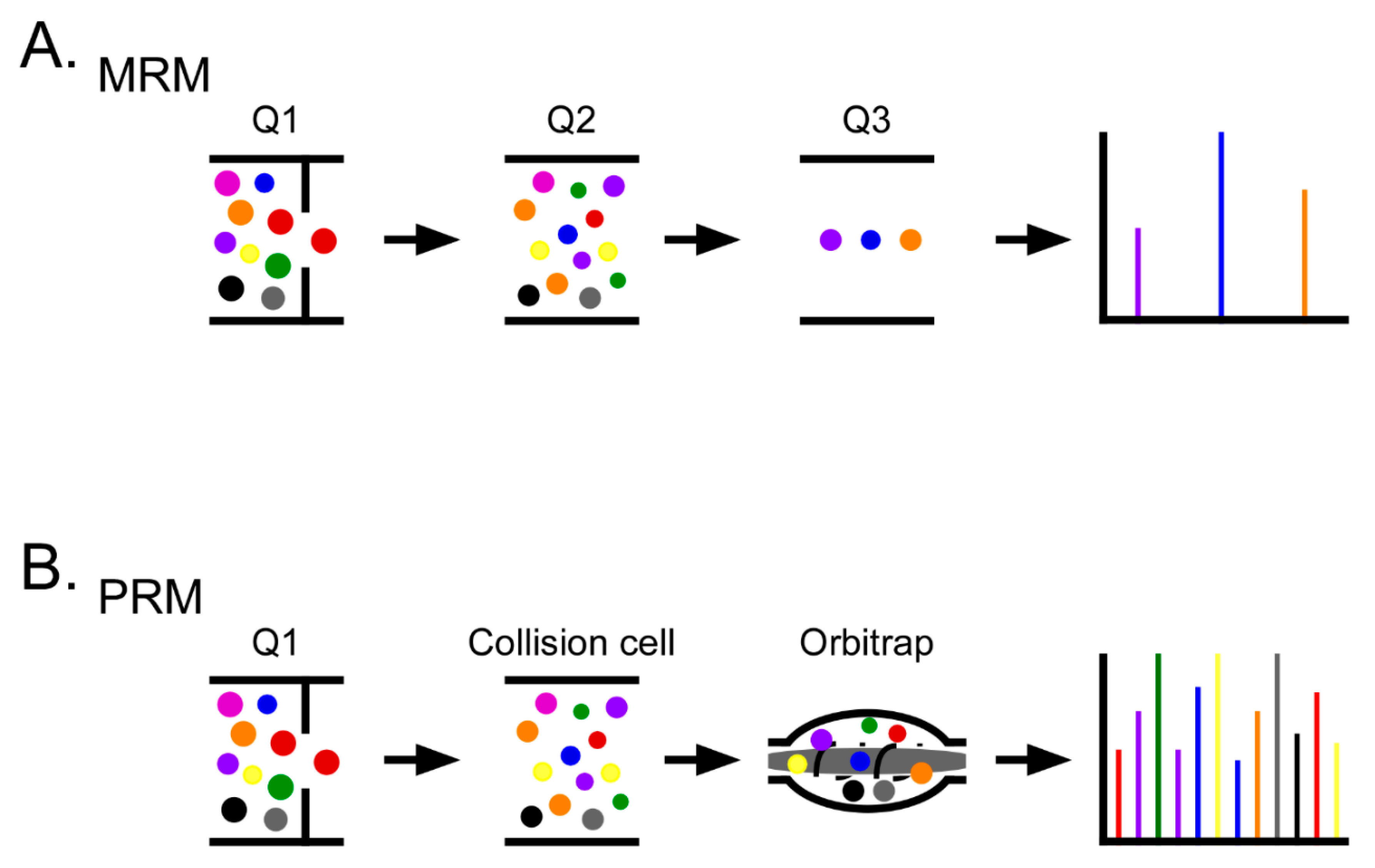
4. Proteomics in Ovarian Cancer
5. Mass Spectrometry for Biomarker Development
6. Future Perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roett, M.A.; Evans, P. Ovarian Cancer: An Overview. Am. Fam. Physician 2009, 80, 609–616. [Google Scholar] [PubMed]
- Stewart, C.; Ralyea, C.; Lockwood, S. Ovarian Cancer: An Integrated Review. Semin. Oncol. Nurs. 2019, 35, 151–156. [Google Scholar] [CrossRef]
- Elzek, M.A.; Rodland, K.D. Proteomics of ovarian cancer: Functional insights and clinical applications. Cancer Metastasis Rev. 2015, 34, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Thomson, C.; Caan, B. Breast and Ovarian Cancer. In Optimizing Women’s Health through Nutrition; Apple Academic Press: Palm Bay, FL, USA, 2007; Volume 60, pp. 229–263. [Google Scholar]
- Momenimovahed, Z.; Tiznobaik, A.; Taheri, S.; Salehiniya, H. Ovarian cancer in the world: Epidemiology and risk factors. Int. J. Womens Health 2019, 11, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Schumer, S.T.; Cannistra, S.A. Granulosa Cell Tumor of the Ovary. J. Clin. Oncol. 2003, 21, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Pokhriyal, R.; Hariprasad, R.; Kumar, L.; Hariprasad, G. Chemotherapy Resistance in Advanced Ovarian Cancer Patients. Biomark. Cancer 2019, 11, 1179299–1986081. [Google Scholar] [CrossRef]
- Prat, J. FIGO’s staging classification for cancer of the ovary, fallopian tube, and peritoneum: Abridged republication. J. Gynecol. Oncol. 2015, 26, 87–89. [Google Scholar] [CrossRef]
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Mph, K.D.M.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef]
- Jayson, G.C.; Kohn, E.C.; Kitchener, H.C.; Ledermann, J.A. Ovarian cancer. Lancet 2014, 384, 1376–1388. [Google Scholar] [CrossRef]
- Chien, J.; Poole, E.M. Ovarian Cancer Prevention, Screening, and Early Detection. Int. J. Gynecol. Cancer 2017, 27, S20–S22. [Google Scholar] [CrossRef] [PubMed]
- Wentzensen, N.; Poole, E.M.; Trabert, B.; White, E.; Arslan, A.A.; Patel, A.V.; Setiawan, V.W.; Visvanathan, K.; Weiderpass, E.; Adami, H.-O.; et al. Ovarian Cancer Risk Factors by Histologic Subtype: An Analysis From the Ovarian Cancer Cohort Consortium. J. Clin. Oncol. 2016, 34, 2888–2898. [Google Scholar] [CrossRef] [PubMed]
- Kurman, R.J.; Shih, I.M. The dualistic model of ovarian carcinogenesis revisited, revised, and expanded. Am. J. Pathol. 2016, 186, 733–747. [Google Scholar] [CrossRef]
- Smolle, E.; Taucher, V.; Pichler, M.; Petru, E.; Lax, S.; Haybaeck, J. Targeting Signaling Pathways in Epithelial Ovarian Cancer. Int. J. Mol. Sci. 2013, 14, 9536–9555. [Google Scholar] [CrossRef]
- Köbel, M.; Kalloger, S.E.; Boyd, N.; McKinney, S.; Mehl, E.; Palmer, C.; Leung, S.; Bowen, N.J.; Ionescu, D.N.; Rajput, A.; et al. Ovarian Carcinoma Subtypes Are Different Diseases: Implications for Biomarker Studies. PLoS Med. 2008, 5, e232. [Google Scholar] [CrossRef]
- Lheureux, S.; Msc, M.B.; Oza, A.M. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J. Clin. 2019, 69, 280–304. [Google Scholar] [CrossRef]
- Kurman, R.J.; Shih, I.-M. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer—Shifting the paradigm. Hum. Pathol. 2011, 42, 918–931. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, V.P.; Grassi, M.L.; Palma, C.D.S.; Carrara, H.H.A.; Faça, V.M.; Dos Reis, F.J.C.; Poersch, A. The contribution and perspectives of proteomics to uncover ovarian cancer tumor markers. Transl. Res. 2019, 206, 71–90. [Google Scholar] [CrossRef]
- Kurman, R.J.; Shih, I.-M. The Origin and Pathogenesis of Epithelial Ovarian Cancer: A Proposed Unifying Theory. Am. J. Surg. Pathol. 2010, 34, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Romero, I.; Bast, R.C. Minireview: Human Ovarian Cancer: Biology, Current Management, and Paths to Personalizing Therapy. Endocrinology 2012, 153, 1593–1602. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, K.C.; Shah, S.P.; Al-Agha, O.M.; Zhao, Y.; Al, E. ARID1A Mutations in Endometriosis-Associated Ovarian Carcinomas. N. Engl. J. Med. 2011, 363, 1532–1543. [Google Scholar] [CrossRef]
- Jones, S.; Wang, T.-L.; Shih, L.-M.; Mao, T.-L.; Nakayama, K.; Roden, R.; Glas, R.; Slamon, D.; Diaz, L.A., Jr.; Vogelstein, B.; et al. Frequent Mutations of Chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 2010, 330, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Shih, I.-M.; Kurman, R.J. Ovarian Tumorigenesis. Am. J. Pathol. 2004, 164, 1511–1518. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Etemadmoghadam, D.; Temple, J.; Lynch, A.G.; Riad, M.; Sharma, R.; Stewart, C.; Fereday, S.; Caldas, C.; DeFazio, A.; et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J. Pathol. 2010, 221, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Landen, C.N., Jr.; Birrer, M.J.; Sood, A.K. Early Events in the Pathogenesis of Epithelial Ovarian Cancer. J. Clin. Oncol. 2008, 26, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.K.; Pujade-Lauraine, E.; Aoki, D.; Mirza, M.R.; Lorusso, D.; Oza, A.M.; du Bois, A.; Vergote, I.; Reuss, A.; Bacon, M.; et al. Fifth Ovarian Cancer Consensus Conference of the Gynecologic Cancer InterGroup: Recurrent disease. Ann. Oncol. 2017, 28, 727–732. [Google Scholar] [CrossRef]
- Bookman, M.; Okamoto, A.; Stuart, G.; Yanaihara, N.; Aoki, D.; Bacon, M.; Fujiwara, K.; González-Martín, A.; Harter, P.; Kim, J.; et al. Harmonising clinical trials within the Gynecologic Cancer InterGroup: Consensus and unmet needs from the Fifth Ovarian Cancer Consensus Conference. Ann. Oncol. 2017, 28, viii30–viii35. [Google Scholar] [CrossRef]
- Palaia, I.; Tomao, F.; Sassu, C.M.; Musacchio, L.; Panici, P.B. Immunotherapy For Ovarian Cancer: Recent Advances And Combination Therapeutic Approaches. Onco Targets Ther. 2020, 13, 6109–6129. [Google Scholar] [CrossRef]
- Pujade-Lauraine, E. New treatments in ovarian cancer. Ann. Oncol. 2017, 28, viii57–viii60. [Google Scholar] [CrossRef]
- Wang, Q.; Peng, H.; Qi, X.; Wu, M.; Zhao, X. Targeted therapies in gynecological cancers: A comprehensive review of clinical evidence. Signal Transduct. Target. Ther. 2020, 5, 1–34. [Google Scholar] [CrossRef]
- Callens, C.; Vaur, D.; Soubeyran, I.; Rouleau, E.; Just, P.-A.; Guillerm, E.; Golmard, L.; Goardon, N.; Sevenet, N.; Cabaret, O.; et al. Concordance Between Tumor and Germline BRCA Status in High-Grade Ovarian Carcinoma Patients in the Phase III PAOLA-1/ENGOT-ov25 Trial. J. Natl. Cancer Inst. 2020. [Google Scholar] [CrossRef]
- Pignata, S.; Pisano, C.; Di Napoli, M.; Cecere, S.C.; Tambaro, R.; Attademo, L. Treatment of recurrent epithelial ovarian cancer. Cancer 2019, 125, 4609–4615. [Google Scholar] [CrossRef] [PubMed]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.J.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.D.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Ledermann, J.A.; Drew, Y.; Kristeleit, R.S. Homologous recombination deficiency and ovarian cancer. Eur. J. Cancer 2016, 60, 49–58. [Google Scholar] [CrossRef]
- D’Andrea, A.D. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair 2018, 71, 172–176. [Google Scholar] [CrossRef]
- Sakai, W.; Swisher, E.M.; Jacquemont, C.; Chandramohan, K.V.; Couch, F.J.; Langdon, S.P.; Wurz, K.; Higgins, J.; Villegas, E.; Taniguchi, T. Functional Restoration of BRCA2 Protein by Secondary BRCA2 Mutations in BRCA2-Mutated Ovarian Carcinoma. Cancer Res. 2009, 69, 6381–6386. [Google Scholar] [CrossRef]
- Barber, L.J.; Sandhu, S.; Chen, L.; Campbell, J.; Kozarewa, I.; Fenwick, K.; Assiotis, I.; Rodrigues, D.N.; Reis-Filho, J.S.; Moreno, V.; et al. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J. Pathol. 2013, 229, 422–429. [Google Scholar] [CrossRef]
- Quigley, D.; Alumkal, J.J.; Wyatt, A.W.; Kothari, V.; Foye, A.; Lloyd, P.; Aggarwal, R.; Kim, W.; Lu, E.; Schwartzman, J.; et al. Analysis of Circulating Cell-Free DNA Identifies Multiclonal Heterogeneity of BRCA2 Reversion Mutations Associated with Resistance to PARP Inhibitors. Cancer Discov. 2017, 7, 999–1005. [Google Scholar] [CrossRef]
- Drost, R.; Bouwman, P.; Rottenberg, S.; Boon, U.; Schut, E.; Klarenbeek, S.; Klijn, C.; van der Heijden, I.; van der Gulden, H.; Wientjens, E.; et al. BRCA1 RING Function Is Essential for Tumor Suppression but Dispensable for Therapy Resistance. Cancer Cell 2011, 20, 797–809. [Google Scholar] [CrossRef]
- Johnson, N.; Johnson, S.F.; Yao, W.; Li, Y.-C.; Choi, Y.-E.; Bernhardy, A.J.; Wang, Y.; Capelletti, M.; Sarosiek, K.A.; Moreau, L.A.; et al. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. Proc. Natl. Acad. Sci. USA 2013, 110, 17041–17046. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, P.; Aly, A.; Escandell, J.M.; Pieterse, M.; Bartkova, J.; Van Der Gulden, H.; Hiddingh, S.; Thanasoula, M.; Kulkarni, A.; Yang, Q.; et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 2010, 17, 688–695. [Google Scholar] [CrossRef]
- Jaspers, J.E.; Kersbergen, A.; Boon, U.; Sol, W.; Van Deemter, L.; Zander, S.A.; Drost, R.; Wientjens, E.; Ji, J.; Aly, A.; et al. Loss of 53BP1 Causes PARP Inhibitor Resistance in Brca1-Mutated Mouse Mammary Tumors. Cancer Discov. 2013, 3, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Boersma, V.; Moatti, N.; Segura-Bayona, S.; Peuscher, M.H.; van der Torre, J.; Wevers, B.A.; Orthwein, A.; Durocher, D.; Jacobs, J.J.L. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5′ end resection. Nature 2015, 521, 537–540. [Google Scholar] [CrossRef]
- Xu, G.; Chapman, J.R.; Brandsma, I.; Yuan, J.; Mistrik, M.; Bouwman, P.; Bartkova, J.; Gogola, E.; Warmerdam, D.; Barazas, M.; et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 2015, 521, 541–544. [Google Scholar] [CrossRef]
- Vaidyanathan, A.; Sawers, L.; Gannon, A.-L.; Chakravarty, P.; Scott, A.L.; Bray, S.E.; Ferguson, M.J.; Smith, G. ABCB1 (MDR1) induction defines a common resistance mechanism in paclitaxel- and olaparib-resistant ovarian cancer cells. Br. J. Cancer 2016, 115, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.R.; Callen, E.; Ding, X.; Gogola, E.; Duarte, A.A.; Lee, J.-E.; Wong, N.; Lafarga, V.; Calvo, J.A.; Panzarino, N.J.; et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 2016, 535, 382–387. [Google Scholar] [CrossRef]
- Gogola, E.; Duarte, A.A.; de Ruiter, J.R.; Wiegant, W.W.; Schmid, J.A.; de Bruijn, R.; James, D.I.; Llobet, S.G.; Vis, D.J.; Annunziato, S.; et al. Selective Loss of PARG Restores PARylation and Counteracts PARP Inhibitor-Mediated Synthetic Lethality. Cancer Cell 2018, 33, 1078–1093.e12. [Google Scholar] [CrossRef]
- Sun, C.; Fang, Y.; Yin, J.; Chen, J.; Ju, Z.; Zhang, D.; Chen, X.; Vellano, C.P.; Jeong, K.J.; Ng, P.K.-S.; et al. Rational combination therapy with PARP and MEK inhibitors capitalizes on therapeutic liabilities in RAS mutant cancers. Sci. Transl. Med. 2017, 9, eaal5148. [Google Scholar] [CrossRef]
- Tapodi, A.; Debreceni, B.; Hanto, K.; Bognar, Z.; Wittmann, I.; Gallyas, F.; Varbiro, G.; Sumegi, B. Pivotal Role of Akt Activation in Mitochondrial Protection and Cell Survival by Poly(ADP-ribose)polymerase-1 Inhibition in Oxidative Stress. J. Biol. Chem. 2005, 280, 35767–35775. [Google Scholar] [CrossRef]
- Champiat, S.; Ileana, E.; Giaccone, G.; Besse, B.; Mountzios, G.; Eggermont, A.; Soria, J.-C. Incorporating Immune-Checkpoint Inhibitors into Systemic Therapy of NSCLC. J. Thorac. Oncol. 2014, 9, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Alme, A.K.B.; Karir, B.S.; Faltas, B.M.; Drake, C.G.; Kimmel, S.; Kimmel, S.; Cancer, C.; Medicine, W.C. Blocking immune checkpoints in prostate, kidney and urothelial cancer. Urol. Oncol. 2016, 34, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Corbo, C.; Cevenini, A.; Salvatore, F. Biomarker discovery by proteomics-based approaches for early detection and personalized medicine in colorectal cancer. Proteom. Clin. Appl. 2017, 11, 15–17. [Google Scholar] [CrossRef]
- Kulasingam, V.; Diamandis, E.P. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat. Clin. Pract. Oncol. 2008, 5, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Califf, R.M. Biomarker definitions and their applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef]
- Paul, A.; Paul, S. The breast cancer susceptibility genes (BRCA) in breast and ovarian cancers. Front. Biosci. 2014, 19, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Crutchfield, C.A.; Thomas, S.N.; Sokoll, L.J.; Chan, D.W. Advances in mass spectrometry-based clinical biomarker discovery. Clin. Proteom. 2016, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Meyerson, M.; Gabriel, S.B.; Getz, G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 2010, 11, 685–696. [Google Scholar] [CrossRef]
- Hanash, S.M.; Baik, C.S.; Kallioniemi, O. Emerging molecular biomarkers—blood-based strategies to detect and monitor cancer. Nat. Rev. Clin. Oncol. 2011, 8, 142–150. [Google Scholar] [CrossRef]
- Maitland, M.L.; Schilsky, R.L. Clinical trials in the era of personalized oncology. CA Cancer J. Clin. 2011, 61, 365–381. [Google Scholar] [CrossRef] [PubMed]
- Simon, R. Critical Review of Umbrella, Basket, and Platform Designs for Oncology Clinical Trials. Clin. Pharmacol. Ther. 2017, 102, 934–941. [Google Scholar] [CrossRef] [PubMed]
- Letai, A. Functional precision cancer medicine—moving beyond pure genomics. Nat. Med. 2017, 23, 1028–1035. [Google Scholar] [CrossRef] [PubMed]
- West, H. No solid evidence, only hollow argument for universal tumor sequencing: Show me the data. JAMA Oncol. 2016, 2, 717–718. [Google Scholar] [CrossRef]
- Prasad, V. Perspective: The precision-oncology illusion. Nature 2016, 537, S63. [Google Scholar] [CrossRef] [PubMed]
- Hyman, D.M.; Taylor, B.S.; Baselga, J. Implementing Genome-Driven Oncology. Cell 2017, 168, 584–599. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Whiteaker, J.R.; Hoofnagle, A.N.; Baird, G.S.; Rodland, K.D.; Paulovich, A.G. Clinical potential of mass spectrometry-based proteogenomics. Nat. Rev. Clin. Oncol. 2019, 16, 256–268. [Google Scholar] [CrossRef]
- Goossens, N.; Nakagawa, S.; Sun, X.; Hoshida, Y. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015, 4, 256–269. [Google Scholar]
- Parkinson, D.R.; McCormack, R.T.; Keating, S.M.; Gutman, S.I.; Hamilton, S.R.; Mansfield, E.A.; Piper, M.A.; Deverka, P.; Frueh, F.W.; Jessup, J.M.; et al. Evidence of Clinical Utility: An Unmet Need in Molecular Diagnostics for Patients with Cancer. Clin. Cancer Res. 2014, 20, 1428–1444. [Google Scholar] [CrossRef]
- Dobbin, K.K.; Cesano, A.; Alvarez, J.; Hawtin, R.; Janetzki, S.; Kirsch, I.; Masucci, G.V.; Robbins, P.B.; Selvan, S.R.; Streicher, H.Z.; et al. Validation of biomarkers to predict response to immunotherapy in cancer: Volume II—clinical validation and regulatory considerations. J. Immunother. Cancer 2016, 4, 77. [Google Scholar] [CrossRef]
- Kraus, V.B. Biomarkers as drug development tools: Discovery, validation, qualification and use. Nat. Rev. Rheumatol. 2018, 14, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Bonifácio, V.D.B. Ovarian Cancer Biomarkers: Moving Forward in Early Detection. In Advances in Experimental Medicine and Biology; Metzler, J.B., Ed.; Springer Nature: Cham, Switzerland, 2020; Volume 1219, pp. 355–363. [Google Scholar]
- Bast, R.C.; Feeney, M.; Lazarus, H.; Nadler, L.M.; Colvin, R.B.; Knapp, R.C. Reactivity of a monoclonal antibody with human ovarian carcinoma. J. Clin. Investig. 1981, 68, 1331–1337. [Google Scholar] [CrossRef] [PubMed]
- Charkhchi, P.; Cybulski, C.; Gronwald, J.; Wong, F.O.; Narod, S.A.; Akbari, M.R. CA125 and Ovarian Cancer: A Comprehensive Review. Cancers 2020, 12, 3730. [Google Scholar] [CrossRef]
- Ueland, F.R. A Perspective on Ovarian Cancer Biomarkers: Past, Present and Yet-To-Come. Diagnostics 2017, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Arjmand, M.; Avval, F.Z. Clinical biomarkers for detection of ovarian cancer. J. Mol. Cancer 2019, 2, 3–7. [Google Scholar]
- Moss, E.L.; Hollingworth, J.; Reynolds, T.M. The role of CA125 in clinical practice. J. Clin. Pathol. 2005, 58, 308–312. [Google Scholar] [CrossRef]
- Boylan, K.L.M.; Geschwind, K.; Koopmeiners, J.S.; Geller, M.A.; Starr, T.K.; Skubitz, A.P.N. A multiplex platform for the identification of ovarian cancer biomarkers. Clin. Proteom. 2017, 14, 34. [Google Scholar] [CrossRef] [PubMed]
- Klein, T.; Wang, W.; Yu, L.; Wu, K.; Boylan, K.L.; Vogel, R.I.; Skubitz, A.P.; Wang, J.-P. Development of a multiplexed giant magnetoresistive biosensor array prototype to quantify ovarian cancer biomarkers. Biosens. Bioelectron. 2019, 126, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Boylan, K.L.M.; Afiuni-Zadeh, S.; Geller, M.A.; Argenta, P.A.; Griffin, T.J.; Skubitz, A.P.N. Evaluation of the potential of Pap test fluid and cervical swabs to serve as clinical diagnostic biospecimens for the detection of ovarian cancer by mass spectrometry-based proteomics. Clin. Proteom. 2021, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ueland, F.R.; DeSimone, C.P.; Seamon, L.G.; Miller, R.A.; Goodrich, S.; Podzielinski, I.; Sokoll, L.; Smith, A.; Van Nagell, J.R.; Zhang, Z. Effectiveness of a Multivariate Index Assay in the Preoperative Assessment of Ovarian Tumors. Obstet. Gynecol. 2011, 117, 1289–1297. [Google Scholar] [CrossRef]
- Zhang, Z.; Chan, D.W. The Road from Discovery to Clinical Diagnostics: Lessons Learned from the First FDA-Cleared In Vitro Diagnostic Multivariate Index Assay of Proteomic Biomarkers. Cancer Epidemiol. Biomarkers Prev. 2010, 19, 2995–2999. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.W.; Smith, A.; DeSimone, C.P.; Seamon, L.; Goodrich, S.; Podzielinski, I.; Sokoll, L.; Van Nagell, J.R.; Zhang, Z.; Ueland, F.R. Performance of the American College of Obstetricians and Gynecologists’ Ovarian Tumor Referral Guidelines With a Multivariate Index Assay. Obstet. Gynecol. 2011, 117, 1298–1306. [Google Scholar] [CrossRef]
- Heliström, I.; Raycraft, J.; Hayden-Ledbetter, M.; Ledbetter, J.A.; Schummer, M.; McIntosh, M.; Drescher, C.; Urban, N.; Hellström, K.E. The HE4 (WFDC2) protein is a biomarker for ovarian carcinoma. Cancer Res. 2003, 63, 3695–3700. [Google Scholar]
- Moore, R.G.; Brown, A.K.; Miller, M.C.; Skates, S.; Allard, W.J.; Verch, T.; Steinhoff, M.; Messerlian, G.; DiSilvestro, P.; Granai, C.; et al. The use of multiple novel tumor biomarkers for the detection of ovarian carcinoma in patients with a pelvic mass. Gynecol. Oncol. 2008, 108, 402–408. [Google Scholar] [CrossRef]
- Moore, R.G.; Miller, M.C.; DiSilvestro, P.; Landrum, L.M.; Gajewski, W.; Ball, J.J.; Skates, S.J. Evaluation of the Diagnostic Accuracy of the Risk of Ovarian Malignancy Algorithm in Women With a Pelvic Mass. Obstet. Gynecol. 2011, 118, 280–288. [Google Scholar] [CrossRef]
- Moore, R.G.; Jabre-Raughley, M.; Brown, A.K.; Robison, K.M.; Miller, M.C.; Allard, W.J.; Kurman, R.J.; Bast, R.C.; Skates, S.J. Comparison of a novel multiple marker assay vs the Risk of Malignancy Index for the prediction of epithelial ovarian cancer in patients with a pelvic mass. Am. J. Obstet. Gynecol. 2010, 203, 228.e1–228.e6. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, M.A.; Sandhu, N.; Høgdall, C.; Christensen, I.J.; Nedergaard, L.; Lundvall, L.; Engelholm, S.A.; Pedersen, A.T.; Hartwell, D.; Lydolph, M.; et al. Evaluation of HE4, CA125, risk of ovarian malignancy algorithm (ROMA) and risk of malignancy index (RMI) as diagnostic tools of epithelial ovarian cancer in patients with a pelvic mass. Gynecol. Oncol. 2012, 127, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Bast, R.C.; Lu, Z.; Han, C.Y.; Lu, K.H.; Anderson, K.S.; Drescher, C.W.; Skates, S.J. Biomarkers and Strategies for Early Detection of Ovarian Cancer. Cancer Epidemiol. Biomarkers Prev. 2020, 29, 2504–2512. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.L.; Herzog, T.J.; Chan, D.W.; Munroe, D.G.; Pappas, T.C.; Smith, A.; Zhang, Z.; Wolf, J. Validation of a second-generation multivariate index assay for malignancy risk of adnexal masses. Am. J. Obstet. Gynecol. 2016, 215, 82.e1–82.e11. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-L.; Lu, Z.; Bast, R.C., Jr. The role of biomarkers in the management of epithelial ovarian cancer. Expert Rev. Mol. Diagn. 2017, 17, 577–591. [Google Scholar] [CrossRef]
- Cramer, D.W.; Bast, R.C.; Berg, C.D.; Diamandis, E.P.; Godwin, A.K.; Hartge, P.; Lokshin, A.E.; Lu, K.H.; McIntosh, M.W.; Mor, G.; et al. Ovarian Cancer Biomarker Performance in Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial Specimens. Cancer Prev. Res. 2011, 4, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Savitski, M.M.; Reinhard, F.B.M.; Franken, H.; Werner, T.; Savitski, M.F.; Eberhard, D.; Molina, D.M.; Jafari, R.; Dovega, R.B.; Klaeger, S.; et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 2014, 346, 1255784. [Google Scholar] [CrossRef] [PubMed]
- Mertins, P.; Cptac, N.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef]
- Manes, N.P.; Angermann, B.R.; Koppenol-Raab, M.; An, E.; Sjoelund, V.H.; Sun, J.; Ishii, M.; Germain, R.N.; Meier-Schellersheim, M.; Nita-Lazar, A. Targeted Proteomics-Driven Computational Modeling of Macrophage S1P Chemosensing. Mol. Cell. Proteom. 2015, 14, 2661–2681. [Google Scholar] [CrossRef]
- Swiatly, A.; Horala, A.; Matysiak, J.; Hajduk, J.; Nowak-Markwitz, E.; Kokot, Z.J. Understanding Ovarian Cancer: iTRAQ-Based Proteomics for Biomarker Discovery. Int. J. Mol. Sci. 2018, 19, 2240. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-L.; Ye, F.; Cai, W.-J.; Hu, H.-D.; Hu, P.; Ren, H.; Zhu, F.-F.; Zhang, D.-Z. Quantitative proteome analysis of multidrug resistance in human ovarian cancer cell line. J. Cell. Biochem. 2010, 109, 625–633. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, T.; Zhang, Z.; Payne, S.H.; Zhang, B.; McDermott, J.E.; Zhou, J.-Y.; Petyuk, V.A.; Chen, L.; Ray, D.; et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 2016, 166, 755–765. [Google Scholar] [CrossRef]
- Aggarwal, K.; Choe, L.H.; Lee, K.H. Shotgun proteomics using the iTRAQ isobaric tags. Brief. Funct. Genom. Proteom. 2006, 5, 112–120. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhang, C.; Gan, L.; Jia, Y.; Xiong, Y.; Chen, Y.; Wang, Z.; Wang, L.; Luo, H.; Li, J.; et al. iTRAQ-Based Quantitative Proteomics Approach Identifies Novel Diagnostic Biomarkers That Were Essential for Glutamine Metabolism and Redox Homeostasis for Gastric Cancer. Proteom. Clin. Appl. 2019, 13, e1800038. [Google Scholar] [CrossRef]
- Xiao, K.; Yu, L.; Zhu, L.; Wu, Z.; Weng, X.; Qiu, G. Urine Proteomics Profiling and Functional Characterization of Knee Osteoarthritis Using iTRAQ Technology. Horm. Metab. Res. 2019, 51, 735–740. [Google Scholar] [CrossRef]
- Zhang, L.; Elias, J.E. Relative Protein Quantification Using Tandem Mass Tag Mass Spectrometry. In Methods in Molecular Biology; Springer Nature: Cham, Switzerland, 2017; Volume 1550, pp. 185–198. [Google Scholar]
- Liang, H.-C.; Lahert, E.; Pike, I.; Ward, M. Quantitation of protein post-translational modifications using isobaric tandem mass tags. Bioanalysis 2015, 7, 383–400. [Google Scholar] [CrossRef]
- Huang, A.; Zhang, M.; Li, T.; Qin, X. Serum Proteomic Analysis by Tandem Mass Tags (TMT) Based Quantitative Proteomics in Gastric Cancer Patients. Clin. Lab. 2018, 64, 855–866. [Google Scholar] [CrossRef]
- Ma, S.; Wang, C.; Zhao, B.; Ren, X.; Tian, S.; Wang, J.; Zhang, C.; Shao, Y.; Qiu, M.; Wang, X. Tandem mass tags labeled quantitative proteomics to study the effect of tobacco smoke exposure on the rat lung. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.-E.; Blagoev, B.; Kratchmarova, I.; Kristensen, D.B.; Steen, H.; Pandey, A.; Mann, M. Stable Isotope Labeling by Amino Acids in Cell Culture, SILAC, as a Simple and Accurate Approach to Expression Proteomics. Mol. Cell. Proteom. 2002, 1, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Blagoev, B.; Kratchmarova, I.; Ong, S.-E.; Nielsen, M.; Foster, L.J.; Mann, M. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 2003, 21, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.-E.; Mann, M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 2006, 1, 2650–2660. [Google Scholar] [CrossRef] [PubMed]
- Hoedt, E.; Zhang, G.; Neubert, T.A. Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) for Quantitative Proteomics. Adv. Exp. Med. Biol. 2019, 1140, 531–539. [Google Scholar] [CrossRef]
- Li, K.W.; Gonzalez-Lozano, M.A.; Koopmans, F.; Smit, A.B. Recent Developments in Data Independent Acquisition (DIA) Mass Spectrometry: Application of Quantitative Analysis of the Brain Proteome. Front. Mol. Neurosci. 2020, 13, 564446. [Google Scholar] [CrossRef]
- Meier, F.; Geyer, P.E.; Winter, S.V.; Cox, J.; Mann, M. BoxCar acquisition method enables single-shot proteomics at a depth of 10,000 proteins in 100 minutes. Nat. Methods 2018, 15, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Haverland, N.A.; Fox, H.S.; Ciborowski, P. Quantitative Proteomics by SWATH-MS Reveals Altered Expression of Nucleic Acid Binding and Regulatory Proteins in HIV-1-Infected Macrophages. J. Proteome Res. 2014, 13, 2109–2119. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, J.; Sethi, A.; Li, Q.K.; Chen, L.; Collins, B.; Gillet, L.C.J.; Wollscheid, B.; Zhang, H.; Aebersold, R. Glycoproteomic Analysis of Prostate Cancer Tissues by SWATH Mass Spectrometry Discovers N-acylethanolamine Acid Amidase and Protein Tyrosine Kinase 7 as Signatures for Tumor Aggressiveness. Mol. Cell. Proteom. 2014, 13, 1753–1768. [Google Scholar] [CrossRef]
- Meier, F.; Brunner, A.-D.; Frank, M.; Ha, A.; Bludau, I.; Voytik, E.; Kaspar-Schoenefeld, S.; Lubeck, M.; Raether, O.; Bache, N.; et al. diaPASEF: Parallel accumulation–serial fragmentation combined with data-independent acquisition. Nat. Methods 2020, 17, 1229–1236. [Google Scholar] [CrossRef]
- Distler, U.; Kuharev, J.; Navarro, P.; Tenzer, S. Label-free quantification in ion mobility–enhanced data-independent acquisition proteomics. Nat. Protoc. 2016, 11, 795–812. [Google Scholar] [CrossRef]
- Huang, L.; Wickramasekara, S.I.; Akinyeke, T.; Stewart, B.S.; Jiang, Y.; Raber, J.; Maier, C.S. Ion mobility-enhanced MSE-based label-free analysis reveals effects of low-dose radiation post contextual fear conditioning training on the mouse hippocampal proteome. J. Proteom. 2016, 140, 24–36. [Google Scholar] [CrossRef][Green Version]
- Moseley, M.A.; Hughes, C.J.; Juvvadi, P.R.; Soderblom, E.J.; Lennon, S.; Perkins, S.R.; Thompson, J.W.; Steinbach, W.J.; Geromanos, S.J.; Wildgoose, J.; et al. Scanning Quadrupole Data-Independent Acquisition, Part A: Qualitative and Quantitative Characterization. J. Proteome Res. 2018, 17, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.; Leitner, A.; Aebersold, R. Mass Spectrometry Applied to Bottom-Up Proteomics: Entering the High-Throughput Era for Hypothesis Testing. Annu. Rev. Anal. Chem. 2016, 9, 449–472. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.N.; Zhang, H. Targeted proteomic assays for the verification of global proteomics insights. Expert Rev. Proteom. 2016, 13, 897–899. [Google Scholar] [CrossRef] [PubMed]
- Uzozie, A.C.; Aebersold, R. Advancing translational research and precision medicine with targeted proteomics. J. Proteom. 2018, 189, 1–10. [Google Scholar] [CrossRef]
- Shi, T.; Song, E.; Nie, S.; Rodland, K.D.; Liu, T.; Qian, W.-J.; Smith, R.D. Advances in targeted proteomics and applications to biomedical research. Proteomics 2016, 16, 2160–2182. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.; Navarro, P.; Tate, S.; Röst, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted Data Extraction of the MS/MS Spectra Generated by Data-independent Acquisition: A New Concept for Consistent and Accurate Proteome Analysis. Mol. Cell. Proteom. 2012, 11. [Google Scholar] [CrossRef]
- Addona, T.A.; Abbatiello, S.E.; Schilling, B.; Skates, S.J.; Mani, D.R.; Bunk, D.M.; Spiegelman, C.H.; Zimmerman, L.J.; Ham, A.-J.L.; Keshishian, H.; et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring–based measurements of proteins in plasma. Nat. Biotechnol. 2009, 27, 633–641. [Google Scholar] [CrossRef]
- Thomas, S.N.; Harlan, R.; Chen, J.; Aiyetan, P.; Liu, Y.; Sokoll, L.J.; Aebersold, R.; Chan, D.W.; Zhang, H. Multiplexed Targeted Mass Spectrometry-Based Assays for the Quantification of N-Linked Glycosite-Containing Peptides in Serum. Anal. Chem. 2015, 87, 10830–10838. [Google Scholar] [CrossRef] [PubMed]
- Lange, V.; Picotti, P.; Domon, B.; Aebersold, R. Selected reaction monitoring for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2008, 4, 222. [Google Scholar] [CrossRef]
- Shi, T.; Su, D.; Liu, T.; Tang, K.; Camp, D.G.; Qian, W.-J.; Smith, R.D. Advancing the sensitivity of selected reaction monitoring-based targeted quantitative proteomics. Proteomics 2012, 12, 1074–1092. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.C.; Russell, J.D.; Bailey, D.J.; Westphall, M.S.; Coon, J.J. Parallel Reaction Monitoring for High Resolution and High Mass Accuracy Quantitative, Targeted Proteomics. Mol. Cell. Proteom. 2012, 11, 1475–1488. [Google Scholar] [CrossRef]
- Rauniyar, N. Parallel Reaction Monitoring: A Targeted Experiment Performed Using High Resolution and High Mass Accuracy Mass Spectrometry. Int. J. Mol. Sci. 2015, 16, 28566–28581. [Google Scholar] [CrossRef]
- Liebler, D.C.; Zimmerman, L.J. Targeted Quantitation of Proteins by Mass Spectrometry. Biochemistry 2013, 52, 3797–3806. [Google Scholar] [CrossRef] [PubMed]
- Gillette, M.A.; Carr, S.A. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat. Methods 2013, 10, 28–34. [Google Scholar] [CrossRef]
- Schubert, O.T.; Röst, H.L.; Collins, B.C.; Rosenberger, G.; Aebersold, R. Quantitative proteomics: Challenges and opportunities in basic and applied research. Nat. Protoc. 2017, 12, 1289–1294. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, C.; Croucher, D.R.; Soliman, M.A.; St-Denis, N.; Pasculescu, A.; Taylor, L.; Tate, S.A.; Hardy, W.R.; Colwill, K.; et al. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature 2013, 499, 166–171. [Google Scholar] [CrossRef]
- Altvater, M.; Chang, Y.; Melnik, A.; Occhipinti, L.; Schütz, S.; Rothenbusch, U.; Picotti, P.; Panse, V.G. Targeted proteomics reveals compositional dynamics of 60S pre-ribosomes after nuclear export. Mol. Syst. Biol. 2012, 8, 628. [Google Scholar] [CrossRef]
- De Graaf, E.L.; Kaplon, J.; Mohammed, S.; Vereijken, L.A.M.; Duarte, D.P.; Gallego, L.R.; Heck, A.J.R.; Peeper, D.S.; Altelaar, A.F.M. Signal Transduction Reaction Monitoring Deciphers Site-Specific PI3K-mTOR/MAPK Pathway Dynamics in Oncogene-Induced Senescence. J. Proteome Res. 2015, 14, 2906–2914. [Google Scholar] [CrossRef]
- Gallien, S.; Domon, B. Advances in high-resolution quantitative proteomics: Implications for clinical applications. Expert Rev. Proteom. 2015, 12, 489–498. [Google Scholar] [CrossRef]
- Liu, Y.; Hüttenhain, R.; Collins, B.; Aebersold, R. Mass spectrometric protein maps for biomarker discovery and clinical research. Expert Rev. Mol. Diagn. 2013, 13, 811–825. [Google Scholar] [CrossRef] [PubMed]
- Picotti, P.; Aebersold, R. Selected reaction monitoring–based proteomics: Workflows, potential, pitfalls and future directions. Nat. Methods 2012, 9, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Nie, B.; Sahm, H.; Brown, D.P.G.; Tegeler, T.; You, J.-S.; Wang, M. Targeted quantitative analysis of superoxide dismutase 1 in cisplatin-sensitive and cisplatin-resistant human ovarian cancer cells. J. Chromatogr. B 2010, 878, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, M.; Tomita, T.; Vogelstein, J.T.; Zhou, S.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Selected reaction monitoring approach for validating peptide biomarkers. Proc. Natl. Acad. Sci. USA 2017, 114, 13519–13524. [Google Scholar] [CrossRef]
- Hüttenhain, R.; Choi, M.; de la Fuente, L.M.; Oehl, K.; Chang, C.-Y.; Zimmermann, A.-K.; Malander, S.; Olsson, H.; Surinova, S.; Clough, T.; et al. A Targeted Mass Spectrometry Strategy for Developing Proteomic Biomarkers: A Case Study of Epithelial Ovarian Cancer. Mol. Cell. Proteom. 2019, 18, 1836–1850. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.-Y.; Beer, L.A.; Tanyi, J.L.; Zhang, R.; Liu, Q.; Speicher, D.W. Protein isoform-specific validation defines multiple chloride intracellular channel and tropomyosin isoforms as serological biomarkers of ovarian cancer. J. Proteom. 2013, 89, 165–178. [Google Scholar] [CrossRef]
- Shi, T.; Gao, Y.; Quek, S.I.; Fillmore, T.L.; Nicora, C.D.; Su, D.; Zhao, R.; Kagan, J.; Srivastava, S.; Rodland, K.D.; et al. A Highly Sensitive Targeted Mass Spectrometric Assay for Quantification of AGR2 Protein in Human Urine and Serum. J. Proteome Res. 2014, 13, 875–882. [Google Scholar] [CrossRef]
- He, J.; Sun, X.; Shi, T.; Schepmoes, A.A.; Fillmore, T.L.; Petyuk, V.A.; Xie, F.; Zhao, R.; Gritsenko, M.A.; Yang, F.; et al. Antibody-independent targeted quantification of TMPRSS2-ERG fusion protein products in prostate cancer. Mol. Oncol. 2014, 8, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Sertamo, K.; Pierrard, M.-A.; Mesmin, C.; Kim, S.Y.; Schlesser, M.; Berchem, G.; Domon, B. Verification of the Biomarker Candidates for Non-small-cell Lung Cancer Using a Targeted Proteomics Approach. J. Proteome Res. 2015, 14, 1412–1419. [Google Scholar] [CrossRef]
- Kim, Y.J.; Gallien, S.; El-Khoury, V.; Goswami, P.; Sertamo, K.; Schlesser, M.; Berchem, G.; Domon, B. Quantification of SAA1 and SAA2 in lung cancer plasma using the isotype-specific PRM assays. Proteomics 2015, 15, 3116–3125. [Google Scholar] [CrossRef] [PubMed]
- Kume, H.; Muraoka, S.; Kuga, T.; Adachi, J.; Narumi, R.; Watanabe, S.; Kuwano, M.; Kodera, Y.; Matsushita, K.; Fukuoka, J.; et al. Discovery of Colorectal Cancer Biomarker Candidates by Membrane Proteomic Analysis and Subsequent Verification using Selected Reaction Monitoring (SRM) and Tissue Microarray (TMA) Analysis. Mol. Cell. Proteom. 2014, 13, 1471–1484. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.; Wang, E.; Chisholm, K.A.; Kostyleva, R.; O’Connor-McCourt, M.; Pinto, D.M. A mass spectrometry-based plasma protein panel targeting the tumor microenvironment in patients with breast cancer. J. Proteom. 2013, 81, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, K.; Yu, S.J.; Jang, E.S.; Yu, J.; Cho, G.; Yoon, J.-H.; Kim, Y. Development of Biomarkers for Screening Hepatocellular Carcinoma Using Global Data Mining and Multiple Reaction Monitoring. PLoS ONE 2013, 8, e63468. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kim, J.Y.; Park, G.W.; Cheon, M.H.; Kwon, K.-H.; Ahn, Y.H.; Moon, M.H.; Lee, H.; Paik, Y.K.; Yoo, J.S. Targeted Mass Spectrometric Approach for Biomarker Discovery and Validation with Nonglycosylated Tryptic Peptides from N-linked Glycoproteins in Human Plasma. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef]
- Zhao, Y.; Jia, W.; Sun, W.; Jin, W.; Guo, L.; Wei, J.; Ying, W.; Zhang, Y.; Xie, Y.; Jiang, Y.; et al. Combination of Improved 18O Incorporation and Multiple Reaction Monitoring: A Universal Strategy for Absolute Quantitative Verification of Serum Candidate Biomarkers of Liver Cancer. J. Proteome Res. 2010, 9, 3319–3327. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.H.; Shin, P.M.; Oh, N.R.; Park, G.W.; Kim, H.; Yoo, J.S. A lectin-coupled, targeted proteomic mass spectrometry (MRM MS) platform for identification of multiple liver cancer biomarkers in human plasma. J. Proteom. 2012, 75, 5507–5515. [Google Scholar] [CrossRef]
- Pan, S.; Chen, R.; Brand, R.E.; Hawley, S.; Tamura, Y.; Gafken, P.R.; Milless, B.P.; Goodlett, D.R.; Rush, J.; Brentnall, T.A. Multiplex Targeted Proteomic Assay for Biomarker Detection in Plasma: A Pancreatic Cancer Biomarker Case Study. J. Proteome Res. 2012, 11, 1937–1948. [Google Scholar] [CrossRef]
- Yoneyama, T.; Ohtsuki, S.; Ono, M.; Ohmine, K.; Uchida, Y.; Yamada, T.; Tachikawa, M.; Terasaki, T. Quantitative Targeted Absolute Proteomics-Based Large-Scale Quantification of Proline-Hydroxylated α-Fibrinogen in Plasma for Pancreatic Cancer Diagnosis. J. Proteome Res. 2013, 12, 753–762. [Google Scholar] [CrossRef] [PubMed]
| Biomarker * | Type | Phase(s) | Clinical Utility | Note § |
|---|---|---|---|---|
| FDA-approved biomarkers | ||||
| CancerSEEK | Gene | 4 | Detection of genetic mutations | 2019 FDA breakthrough device |
| CA125 | Protein | 4 | Monitoring | Curated for phase 3 in breast |
| HE4 | Protein | 3 | Early detection | |
| OVA1 | Protein panel | 3 | Prediction | |
| Overa | Protein | 5 | Prediction | |
| ROMA | Protein panel | 3 | Prediction | |
| Biomarker candidates | ||||
| APC | Gene | 3, 2 | Under review in breast, lung, and prostate ¥ | |
| CDKN2A (p16) | Gene | 2 | Under review for phase 3 in breast and esophagus; under review for phase 1 in lung and prostate | |
| EGFR | Gene | 3 | Curated for phase 3 in breast; under review for phase 3 in lung; under review for phase 1 in prostate | |
| NID2 | Gene | 1 | Curated for phase 1 in head and neck | |
| p14/ARF | Gene | 3, 2 | Under review in prostate and ovary ¥ | |
| SMA4 | Gene | 3 | ||
| Cramer 5 marker panel | Protein panel | 3 | HE4, CA15-3, CA125, VTCN1, and CA72-4; Early detection | |
| 9 microsatellites | Protein | 2 | ||
| ACKR3 | Protein | 2 | ||
| ACTR3 | Protein | 2 | ||
| ADAM12 | Protein | 2 | ||
| AFP | Protein | 2 | Certified by FDA in liver | |
| AGRN | Protein | 1 | ||
| AKT1 | Protein | 2 | ||
| AMBP | Protein | 1 | ||
| AMY2A | Protein | 3 | ||
| ANXA2 | Protein | 1 | ||
| APCS | Protein | 3 | ||
| APOA1 | Protein | 3 | Under review for phase 2 in breast and pancreas | |
| APOB | Protein | 3, 2 | Under review in breast and ovary ¥ | |
| APOC4 | Protein | 3 | ||
| ARID1A | Protein | 2 | ||
| ATP6AP2 | Protein | 1 | ||
| B2M | Protein | 3 | ||
| BCAM | Protein | 3 | ||
| BLVRB | Protein | 3 | ||
| BRAF | Protein | 2 | ||
| BRCA1 | Protein | 2 | Under review for phase 2 in breast | |
| BRCA2 | Protein | 2 | ||
| C3 | Protein | 1 | ||
| CA15-3 | Protein | 3 | In ovarian cancer, used with CA125 for monitoring; Curated for phase 3 in breast; under review for phase 2 in lung; under review for phase 1 in prostate | |
| CA19-9 | Protein | 3 | In ovarian cancer, used with CA125 for monitoring; Curated for phase 3 in breast; under review for phase 3 in pancreas; under review for phase 1 in prostate | |
| CA72-4 | Protein | 3 | In ovarian cancer, used with CA125 for monitoring; Under review for phase 2 in breast | |
| CADM1 | Protein | 3 | ||
| CBLC | Protein | 3, 2 | Under review in lung and ovary ¥ | |
| CCDC102B | Protein | 2 | ||
| CCL11 | Protein | 3 | Curated for phase 3 in breast | |
| CD248 | Protein | 1 | ||
| CD59 | Protein | 1 | ||
| CDCP1 | Protein | 2 | ||
| CEACAM5 | Protein | 3 | Curated for phase 3 in breast; under review for phase 2 in colon, lung, and pancreas; under review for phase 1 in prostate | |
| CHI3L1 | Protein | 2 | ||
| CKM | Protein | 1, 3 | Under review in lung and ovary ¥ | |
| CPA4 | Protein | 1 | ||
| CRIP1 | Protein | 3 | ||
| CRIP2 | Protein | 2 | ||
| CRTAC1 | Protein | 1, 3 | Under review in prostate and ovary ¥ | |
| CST6 | Protein | 1 | ||
| CTCFL | Protein | 3 | ||
| CTGF | Protein | 1 | ||
| CTNNB1 | Protein | 2 | Under review in breast, pancreas, and ovary ¥ | |
| CXCL8 | Protein | 3 | Curated for phase 2 in bladder; curated for phase 3 in breast; under review for phase 2 in lung; under review for phase 1 in prostate | |
| DAG1 | Protein | 1 | ||
| DAPL1 | Protein | 3 | ||
| DEFB1 | Protein | 2 | ||
| DKK3 | Protein | 1 | ||
| DSC2 | Protein | 1, 2 | Under review in prostate and ovary ¥ | |
| DSG2 | Protein | 1, 3 | Under review in prostate and ovary ¥ | |
| ECM1 | Protein | 1 | ||
| EFEMP1 | Protein | 1 | Under review for phase 3 in lung; under review for phase 1 for prostate | |
| EFR3A | Protein | 1 | ||
| EGFL6 | Protein | 2 | ||
| EMILIN2 | Protein | 1 | ||
| EPB41L3 | Protein | 2 | ||
| EPCAM | Protein | 1 | Target for cancer immunotherapy | |
| EPSTI1 | Protein | 2 | ||
| ERBB2 | Protein | 3 | Curated for phase 3 in breast; under review for phase 2 in colon and lung | |
| ESM1 | Protein | 3 | ||
| FAM83H | Protein | 2 | ||
| FAS | Protein | 3 | ||
| FBLN1 | Protein | 1 | ||
| FBXW7 | Protein | 2 | ||
| FGFR2 | Protein | 2 | ||
| FGFR4 | Protein | 3 | ||
| FJX1 | Protein | 2 | ||
| FNDC3A | Protein | 1 | ||
| FOLH1B | Protein | 1 | Under review for phase 1 in prostate | |
| FOLR1 | Protein | 1 | ||
| FSH | Protein | 3 | ||
| FSTL1 | Protein | 1 | ||
| FZD10 | Protein | 2 | ||
| GDF15 | Protein | 2 | ||
| GFPT1 | Protein | 3 | ||
| GH1 | Protein | 3 | Under review for phase 2 in breast | |
| GLOD4 | Protein | 1 | ||
| GM2A | Protein | 1 | ||
| GPM6B | Protein | 2 | Under review for phase 1 in prostate | |
| GPR158 | Protein | 3 | ||
| GPR39 | Protein | 1 | ||
| GPR65 | Protein | 2 | ||
| GRN | Protein | 1 | ||
| H2AFJ | Protein | 3 | ||
| H2AFV | Protein | 3 | ||
| HAMP | Protein | 3 | ||
| HAPLN1 | Protein | 1 | Under review for phase 1 in lung | |
| HIST1H2AA | Protein | 3 | ||
| HMGB1 | Protein | 3 | ||
| HOXA9 | Protein | 2 | Curated for phase 1 in head and neck; under review for phase 1 in prostate | |
| HSPG2 | Protein | 1 | ||
| HTRA1 | Protein | 1 | ||
| ICAM1 | Protein | 2 | Curated for phase 3 in breast; under review for phase 3 in prostate | |
| IDH1 | Protein | 3 | ||
| IFI27 | Protein | 1 | ||
| IGF2 | Protein | 3 | ||
| IGFBP1 | Protein | 3 | Under review for phase 2 in breast | |
| IGFBP2 | Protein | 3 | Under review for phase 2 in breast and colon | |
| IGFBP3 | Protein | 1, 2 | Under review in pancreas and ovary ¥ | |
| IGFBP4 | Protein | 1, 3 | Under review in pancreas and ovary ¥ | |
| IGF-II | Protein | 2 | ||
| IL10 | Protein | 3 | ||
| IL2RA | Protein | 3 | ||
| IL6 | Protein | 2 | Under review for phase 2 in breast | |
| IL6R | Protein | 3 | ||
| ITIH4 | Protein | 3 | ||
| KCP | Protein | 3 | ||
| KLHL14 | Protein | 3 | ||
| KLK6 | Protein | 3 | Used with CA125 for monitoring | |
| KLK8 | Protein | 3 | Under review for phase 2 in breast and lung | |
| KRAS | Protein | 1, 3, 2 | Under review in colon, lung, pancreas, and ovary ¥ | |
| KRT19 | Protein | 3 | Under review in prostate ¥ | |
| KRT8 | Protein | 1 | ||
| LAMA5 | Protein | 3 | ||
| LAMB2 | Protein | 3 | ||
| LAPTM4B | Protein | 1 | ||
| LEP | Protein | 3 | Under review for phase 2 in breast | |
| LGALS3BP | Protein | 1 | ||
| LHB | Protein | 3 | ||
| LPAR3 | Protein | 1 | ||
| LRG1 | Protein | 1, 2 | Under review in breast, pancreas, and ovary ¥ | |
| LRRC47 | Protein | 3 | ||
| LTBP1 | Protein | 1 | ||
| LTBP2 | Protein | 1 | ||
| LY6G6C | Protein | 3 | ||
| LZTS1 | Protein | 1 | ||
| MAPK1 | Protein | 2 | ||
| MIF | Protein | 3 | Under review for phase 1 in lung | |
| MLH1 | Protein | 2 | Curated for phase 3 in breast | |
| MMP2 | Protein | 3 | ||
| MMP3 | Protein | 3 | Curated for phase 3 in breast; under review for phase 1 in lung | |
| MMP7 | Protein | 3 | ||
| MMP9 | Protein | 3 | Curated for phase 2 in bladder; curated for phase 3 in breast; under review for phase in lung | |
| MPO | Protein | 3, 2 | Under review in breast and ovary ¥ | |
| MPPED2 | Protein | 2 | Under review for phase 1 in prostate | |
| MPZL2 | Protein | 2 | Under review for phase 1 in prostate | |
| MSH2 | Protein | 2 | ||
| MSLN | Protein | 3 | ||
| MXRA5 | Protein | 1 | ||
| NID1 | Protein | 3 | ||
| NMU | Protein | 3 | ||
| NPC2 | Protein | 1 | ||
| NRAS | Protein | 2 | ||
| NUCB1 | Protein | 1 | ||
| OLFML2B | Protein | 1 | ||
| Osteopontin | Protein | 3 | Under review for phase 3 in breast; under review for phase 2 in liver; under review for phase 1 in lung | |
| OVGP1 | Protein | 1 | ||
| P2RY14 | Protein | 2 | ||
| PCDH17 | Protein | 1 | ||
| PCOLCE | Protein | 1 | ||
| PCSK9 | Protein | 3 | ||
| PEBP1 | Protein | 1 | ||
| PFAS | Protein | 3 | ||
| PGGHG | Protein | 3 | ||
| PI3 | Protein | 1 | ||
| PIK3CA | Protein | 2 | ||
| PIK3R1 | Protein | 2 | ||
| PLEC | Protein | 1 | ||
| PLTP | Protein | 1 | ||
| PLXNB1 | Protein | 3 | ||
| PNP | Protein | 3 | ||
| POLE | Protein | 2 | ||
| POLQ | Protein | 3 | ||
| POSTN | Protein | 3 | ||
| PPBP | Protein | 3 | ||
| PPP2R1A | Protein | 2 | ||
| PRDX6 | Protein | 3 | ||
| PRL | Protein | 3 | Under review for phase 3 in breast | |
| PRMT1 | Protein | 3 | ||
| PROS1 | Protein | 1 | ||
| PSAP | Protein | 1 | ||
| PTEN | Protein | 2 | ||
| PTH2R | Protein | 3 | ||
| PTK7 | Protein | 3 | ||
| PTPRS | Protein | 3 | ||
| QSOX1 | Protein | 1 | ||
| RMND5A | Protein | 2 | ||
| RNF43 | Protein | 2 | ||
| SCGB2A1 | Protein | 2 | Under review for phase 1 in prostate | |
| SCNN1A | Protein | 1, 3 | Under review in prostate and ovary ¥ | |
| SDC1 | Protein | 2 | Curated for phase 2 in bladder | |
| SEC23B | Protein | 2 | ||
| SECTM1 | Protein | 1 | ||
| SELENBP1 | Protein | 3 | ||
| SERPINA6 | Protein | 1 | ||
| SERPINE1 | Protein | 3 | Curated for phase 2 in bladder; under review for phase 2 in breast | |
| SLAMF8 | Protein | 2 | ||
| SLC11A1 | Protein | 1 | ||
| SLC30A6 | Protein | 2 | ||
| SLPI | Protein | 3 | Under review for phase 1 in lung and prostate | |
| SMRP | Protein | 2 | Under review for phase 4 in lung | |
| SOD3 | Protein | 3 | ||
| SPINT2 | Protein | 1 | ||
| SPON1 | Protein | 3 | ||
| SPON2 | Protein | 3 | Under review for phase 2 in colon | |
| SPP2 | Protein | 3 | ||
| ST13 | Protein | 3 | ||
| ST14 | Protein | 2 | ||
| TAGLN2 | Protein | 1 | ||
| TF | Protein | 3 | Under review for phase 1 in prostate | |
| TNF | Protein | 3, 2 | Under review in breast, lung, and ovary ¥ | |
| TNFAIP1 | Protein | 2 | ||
| TNFAIP6 | Protein | 2 | ||
| TNFRSF1A | Protein | 3 | ||
| TNFRSF1B | Protein | 3 | ||
| TNFRSF21 | Protein | 3 | ||
| TNFRSF6B | Protein | 3, 2 | Under review in colon and ovary ¥ | |
| TP53 | Protein | 1, 2 | Under review in breast, colon, lung, pancreas, prostate, and ovary | |
| Transthyretin | Protein | 3 | ||
| TSHB | Protein | 3 | ||
| TSSK4 | Protein | 3 | ||
| VCAM1 | Protein | 3 | Curated for phase 3 in breast | |
| VCAN | Protein | 3 | ||
| VTA1 | Protein | 2 | ||
| VTCN1 | Protein | 3 | Used with CA125 for monitoring | |
| VWF | Protein | 1 | ||
| WNT10A | Protein | 3 | ||
| WWC1 | Protein | 1 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ryu, J.; Thomas, S.N. Quantitative Mass Spectrometry-Based Proteomics for Biomarker Development in Ovarian Cancer. Molecules 2021, 26, 2674. https://doi.org/10.3390/molecules26092674
Ryu J, Thomas SN. Quantitative Mass Spectrometry-Based Proteomics for Biomarker Development in Ovarian Cancer. Molecules. 2021; 26(9):2674. https://doi.org/10.3390/molecules26092674
Chicago/Turabian StyleRyu, Joohyun, and Stefani N. Thomas. 2021. "Quantitative Mass Spectrometry-Based Proteomics for Biomarker Development in Ovarian Cancer" Molecules 26, no. 9: 2674. https://doi.org/10.3390/molecules26092674
APA StyleRyu, J., & Thomas, S. N. (2021). Quantitative Mass Spectrometry-Based Proteomics for Biomarker Development in Ovarian Cancer. Molecules, 26(9), 2674. https://doi.org/10.3390/molecules26092674







