Transcriptomic and Proteomic Analysis of Clear Cell Foci (CCF) in the Human Non-Cirrhotic Liver Identifies Several Differentially Expressed Genes and Proteins with Functions in Cancer Cell Biology and Glycogen Metabolism
Abstract
1. Introduction
2. Results
2.1. More RNAs Have Reduced Expression in CCF Compared to Controls
2.2. More Proteins Have Reduced Expression in CCF Compared to Control Samples
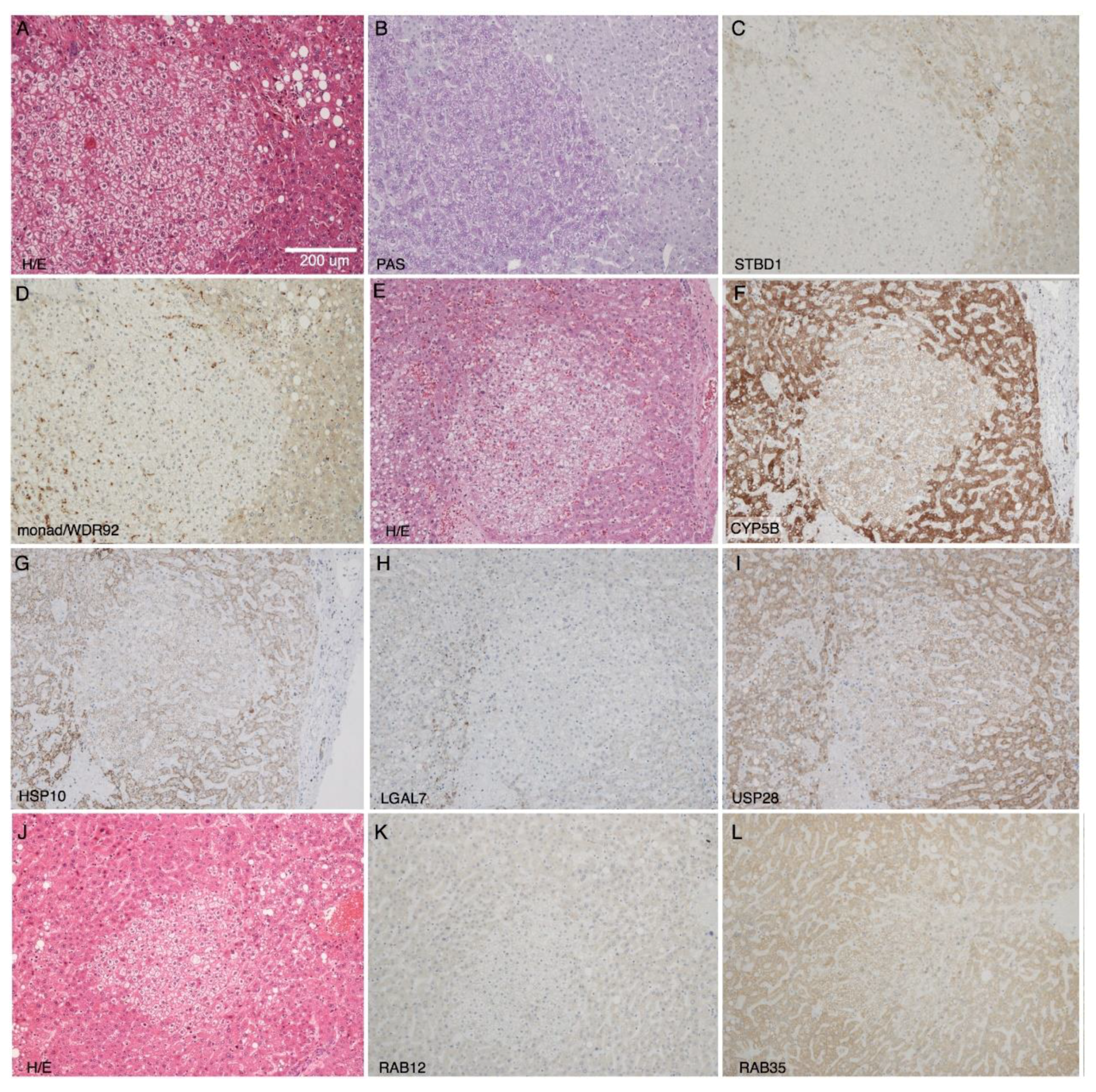
2.3. Immunohistochemical Validation of Differential Expression of Monad/WDR92, USP28, STBD1, CYB5B, and HSPE1 in CCF
2.4. Loss of STBD1 in Mice Does Not Cause Glycogen Accumulation in the Liver
2.5. Diethylnitrosamine (DEN)-Induced Hepatocellular Carcinomas in Usp28-KO Mice Do Not Accumulate More Glycogen than in Control Mice
3. Discussion
3.1. Non-Coding RNAs
3.2. STBD1
3.3. USP28
4. Conclusions
5. Materials and Methods
5.1. Human Liver Specimens
5.2. Cryosectioning
5.3. Laser Micro-Dissection
5.4. RNA and Protein Isolation
5.5. Microarray Analysis
5.6. Gel Electrophoresis and Silver Staining
5.7. Proteomics Sample Preparation and LC-MS/MS
5.8. Cluster Analysis and Plotting of Omics Data
5.9. Immunohistochemistry and Histochemistry
5.10. Mouse Models
5.11. Glycogen Quantification in Mouse Liver
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Franco, L.M.; Krishnamurthy, V.; Bali, D.; Weinstein, D.A.; Arn, P.; Clary, B.; Boney, A.; Sullivan, J.; Frush, D.P.; Chen, Y.-T.; et al. Hepatocellular carcinoma in glycogen storage disease type Ia: A case series. J. Inherit. Metab. Dis. 2005, 28, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Marengo, A.; Rosso, C.; Bugianesi, E. Liver Cancer: Connections with Obesity, Fatty Liver, and Cirrhosis. Annu. Rev. Med. 2016, 67, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Davila, J.A.; Morgan, R.O.; Shaib, Y.; McGlynn, K.A.; El-Serag, H.B. Diabetes increases the risk of hepatocellular carcinoma in the United States: A population based case control study. Gut 2005, 54, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Vigneri, R.; Goldfine, I.D.; Frittitta, L. Insulin, insulin receptors, and cancer. J. Endocrinol. Invest. 2016, 39, 1365–1376. [Google Scholar] [CrossRef] [PubMed]
- Libbrecht, L.; Desmet, V.; Roskams, T. Preneoplastic lesions in human hepatocarcinogenesis. Liver Int. 2005, 25, 16–27. [Google Scholar] [CrossRef]
- Evert, M.; Dombrowski, F. Hepatocellular carcinoma in the non-cirrhotic liver. Pathologe 2008, 29, 47–52. [Google Scholar] [CrossRef]
- Bannasch, P.; Ribback, S.; Su, Q.; Mayer, D. Clear cell hepatocellular carcinoma: Origin, metabolic traits and fate of glycogenotic clear and ground glass cells. HBPD INT 2017, 16, 570–594. [Google Scholar] [CrossRef]
- Bannasch, P. Pathogenesis of hepatocellular carcinoma: Sequential cellular, molecular, and metabolic changes. Prog Liver Dis 1996, 14, 161–197. [Google Scholar]
- Bannasch, P.; Mayer, D.; Hacker, H.J. Hepatocellular glycogenosis and hepatocarcinogenesis. Biochim. Biophys. Acta 1980, 605, 217–245. [Google Scholar] [CrossRef]
- Williams, G.M. The pathogenesis of rat liver cancer caused by chemical carcinogens. Biochim. Biophys. Acta 1980, 605, 167–189. [Google Scholar] [CrossRef]
- Pitot, H.C. Altered hepatic foci: Their role in murine hepatocarcinogenesis. Annu. Rev. Pharmacol. Toxicol. 1990, 30, 465–500. [Google Scholar] [CrossRef] [PubMed]
- Dombrowski, F.; Filsinger, E.; Bannasch, P.; Pfeifer, U. Altered liver acini induced in diabetic rats by portal vein islet isografts resemble preneoplastic hepatic foci in their enzymic pattern. Am. J. Pathol. 1996, 148, 1249–1256. [Google Scholar] [PubMed]
- Dombrowski, F.; Mathieu, C.; Evert, M. Hepatocellular neoplasms induced by low-number pancreatic islet transplants in autoimmune diabetic BB/Pfd rats. Cancer Res. 2006, 66, 1833–1843. [Google Scholar] [CrossRef] [PubMed]
- Dombrowski, F.; Bannasch, P.; Pfeifer, U. Hepatocellular neoplasms induced by low-number pancreatic islet transplants in streptozotocin diabetic rats. Am. J. Pathol. 1997, 150, 1071–1087. [Google Scholar]
- Dombrowski, F.; Jost, C.M.; Manekeller, S.; Evert, M. Cocarcinogenic effects of islet hormones and N-nitrosomorpholine in hepatocarcinogenesis after intrahepatic transplantation of pancreatic islets in streptozotocin-diabetic rats. Cancer Res. 2005, 65, 7013–7022. [Google Scholar] [CrossRef][Green Version]
- Ribback, S.; Cigliano, A.; Kroeger, N.; Pilo, M.G.; Terracciano, L.; Burchardt, M.; Bannasch, P.; Calvisi, D.F.; Dombrowski, F. PI3K/AKT/mTOR pathway plays a major pathogenetic role in glycogen accumulation and tumor development in renal distal tubules of rats and men. Oncotarget 2015, 6, 13036–13048. [Google Scholar] [CrossRef]
- Evert, M.; Calvisi, D.F.; Evert, K.; De Murtas, V.; Gasparetti, G.; Mattu, S.; Destefanis, G.; Ladu, S.; Zimmermann, A.; Delogu, S.; et al. V-AKT murine thymoma viral oncogene homolog/mammalian target of rapamycin activation induces a module of metabolic changes contributing to growth in insulin-induced hepatocarcinogenesis. Hepatology 2012, 55, 1473–1484. [Google Scholar] [CrossRef]
- Calvisi, D.F.; Wang, C.; Ho, C.; Ladu, S.; Lee, S.A.; Mattu, S.; Destefanis, G.; Delogu, S.; Zimmermann, A.; Ericsson, J.; et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology 2011, 140, 1071–1083. [Google Scholar] [CrossRef]
- Calvisi, D.F.; Ladu, S.; Gorden, A.; Farina, M.; Conner, E.A.; Lee, J.-S.; Factor, V.M.; Thorgeirsson, S.S. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology 2006, 130, 1117–1128. [Google Scholar] [CrossRef]
- Ribback, S.; Calvisi, D.F.; Cigliano, A.; Sailer, V.; Peters, M.; Rausch, J.; Heidecke, C.-D.; Birth, M.; Dombrowski, F. Molecular and metabolic changes in human liver clear cell foci resemble the alterations occurring in rat hepatocarcinogenesis. J. Hepatol. 2013, 58, 1147–1156. [Google Scholar] [CrossRef]
- Ribback, S.; Sonke, J.; Lohr, A.; Frohme, J.; Peters, K.; Holm, J.; Peters, M.; Cigliano, A.; Calvisi, D.F.; Dombrowski, F. Hepatocellular glycogenotic foci after combined intraportal pancreatic islet transplantation and knockout of the carbohydrate responsive element binding protein in diabetic mice. Oncotarget 2017, 8, 104315–104329. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Che, L.; Tharp, K.M.; Park, H.-M.; Pilo, M.G.; Cao, D.; Cigliano, A.; Latte, G.; Xu, Z.; Ribback, S.; et al. Differential requirement for de novo lipogenesis in cholangiocarcinoma and hepatocellular carcinoma of mice and humans. Hepatology 2016, 63, 1900–1913. [Google Scholar] [CrossRef] [PubMed]
- Che, L.; Pilo, M.G.; Cigliano, A.; Latte, G.; Simile, M.M.; Ribback, S.; Dombrowski, F.; Evert, M.; Chen, X.; Calvisi, D.F. Oncogene dependent requirement of fatty acid synthase in hepatocellular carcinoma. Cell Cycle 2017, 16, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Sun, D.; Li, W.; Shen, H.; Zhu, Y.; Li, C.; Chen, Y.; Lu, L.; Li, W.; Zhang, J.; et al. A c-Myc-MicroRNA functional feedback loop affects hepatocarcinogenesis. Hepatology 2013, 57, 2378–2389. [Google Scholar] [CrossRef]
- Richter, K.; Paakkola, T.; Mennerich, D.; Kubaichuk, K.; Konzack, A.; Kippari, H.A.; Kozlova, N.; Koivunen, P.; Haapasaari, K.-M.; Jukkola-Vuorinen, A.; et al. USP28 Deficiency Promotes Breast and Liver Carcinogenesis as well as Tumor Angiogenesis in a HIF-independent Manner. Mol. Cancer Res. 2018, 16, 1000–1012. [Google Scholar] [CrossRef]
- Uwamizu, A.; Inoue, A.; Suzuki, K.; Okudaira, M.; Shuto, A.; Shinjo, Y.; Ishiguro, J.; Makide, K.; Ikubo, M.; Nakamura, S.; et al. Lysophosphatidylserine analogues differentially activate three LysoPS receptors. J. Biochem. 2015, 157, 151–160. [Google Scholar] [CrossRef]
- Makide, K.; Uwamizu, A.; Shinjo, Y.; Ishiguro, J.; Okutani, M.; Inoue, A.; Aoki, J. Novel lysophosphoplipid receptors: Their structure and function. J. Lipid Res. 2014, 55, 1986–1995. [Google Scholar] [CrossRef]
- Chu, X.; Shen, M.; Xie, F.; Miao, X.-J.; Shou, W.-H.; Liu, L.; Yang, P.-P.; Bai, Y.-N.; Zhang, K.-Y.; Yang, L.; et al. An X chromosome-wide association analysis identifies variants in GPR174 as a risk factor for Graves’ disease. J. Med. Genet. 2013, 50, 479–485. [Google Scholar] [CrossRef]
- Napier, C.; Mitchell, A.L.; Gan, E.; Wilson, I.; Pearce, S.H.S. Role of the X-linked gene GPR174 in autoimmune Addison’s disease. J. Clin. Endocrinol. Metab. 2015, 100, E187–E190. [Google Scholar] [CrossRef]
- Andergassen, U.; Liesche, F.; Kölbl, A.C.; Ilmer, M.; Hutter, S.; Friese, K.; Jeschke, U. Glycosyltransferases as Markers for Early Tumorigenesis. Biomed Res Int 2015, 2015, 792672. [Google Scholar] [CrossRef]
- Lin, T.-C.; Chen, S.-T.; Huang, M.-C.; Huang, J.; Hsu, C.-L.; Juan, H.-F.; Lin, H.-H.; Chen, C.-H. GALNT6 expression enhances aggressive phenotypes of ovarian cancer cells by regulating EGFR activity. Oncotarget 2017, 8, 42588–42601. [Google Scholar] [CrossRef]
- Lin, J.; Chung, S.; Ueda, K.; Matsuda, K.; Nakamura, Y.; Park, J.-H. GALNT6 Stabilizes GRP78 Protein by O-glycosylation and Enhances its Activity to Suppress Apoptosis Under Stress Condition. Neoplasia 2017, 19, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Zhang, Y.; Fan, S.; Chen, L.; Tang, L.; Chen, X.; Lyu, J. GALNT6 Promotes Tumorigenicity and Metastasis of Breast Cancer Cell via β-catenin/MUC1-C Signaling Pathway. Int. J. Biol. Sci. 2019, 15, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Tarhan, Y.E.; Kato, T.; Jang, M.; Haga, Y.; Ueda, K.; Nakamura, Y.; Park, J.-H. Morphological Changes, Cadherin Switching, and Growth Suppression in Pancreatic Cancer by GALNT6 Knockdown. Neoplasia 2016, 18, 265–272. [Google Scholar] [CrossRef] [PubMed]
- von Morgen, P.; Hořejší, Z.; Macurek, L. Substrate recognition and function of the R2TP complex in response to cellular stress. Front. Genet. 2015, 6, 69. [Google Scholar]
- Saeki, M.; Egusa, H.; Kamano, Y.; Kakihara, Y.; Houry, W.A.; Yatani, H.; Noguchi, S.; Kamisaki, Y. Exosome-bound WD repeat protein Monad inhibits breast cancer cell invasion by degrading amphiregulin mRNA. PLoS ONE 2013, 8, e67326. [Google Scholar] [CrossRef]
- Saeki, M.; Irie, Y.; Ni, L.; Yoshida, M.; Itsuki, Y.; Kamisaki, Y. Monad, a WD40 repeat protein, promotes apoptosis induced by TNF-alpha. Biochem. Biophys. Res. Commun. 2006, 342, 568–572. [Google Scholar] [CrossRef]
- Chen, G.; Yu, C.; Tang, Z.; Liu, S.; An, F.; Zhu, J.; Wu, Q.; Cao, J.; Zhan, Q.; Zhang, S. Metformin suppresses gastric cancer progression through calmodulin-like protein 3 secreted from tumor-associated fibroblasts. Oncol. Rep. 2019, 41, 405–414. [Google Scholar] [CrossRef]
- Paul, N.R.; Allen, J.L.; Chapman, A.; Morlan-Mairal, M.; Zindy, E.; Jacquemet, G.; Fernandez del Ama, L.; Ferizovic, N.; Green, D.M.; Howe, J.D.; et al. α5β1 integrin recycling promotes Arp2/3-independent cancer cell invasion via the formin FHOD3. J. Cell Biol. 2015, 210, 1013–1031. [Google Scholar] [CrossRef]
- Zhao, C.; Inoue, J.; Imoto, I.; Otsuki, T.; Iida, S.; Ueda, R.; Inazawa, J. POU2AF1, an amplification target at 11q23, promotes growth of multiple myeloma cells by directly regulating expression of a B-cell maturation factor, TNFRSF17. Oncogene 2008, 27, 63–75. [Google Scholar] [CrossRef]
- Jiang, S.; Heller, B.; Tagliabracci, V.S.; Zhai, L.; Irimia, J.M.; Depaoli-Roach, A.A.; Wells, C.D.; Skurat, A.V.; Roach, P.J. Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J. Biol. Chem. 2010, 285, 34960–34971. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Wells, C.D.; Roach, P.J. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem. Biophys. Res. Commun. 2011, 413, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Yi, H.; Yang, C.; Kishnani, P.S.; Sun, B. Starch Binding Domain-containing Protein 1 Plays a Dominant Role in Glycogen Transport to Lysosomes in Liver. J. Biol. Chem. 2016, 291, 16479–16484. [Google Scholar] [CrossRef] [PubMed]
- Lambrus, B.G.; Daggubati, V.; Uetake, Y.; Scott, P.M.; Clutario, K.M.; Sluder, G.; Holland, A.J. A USP28-53BP1-p53-p21 signaling axis arrests growth after centrosome loss or prolonged mitosis. J. Cell Biol. 2016, 214, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Fong, C.S.; Mazo, G.; Das, T.; Goodman, J.; Kim, M.; O’Rourke, B.P.; Izquierdo, D.; Tsou, M.-F.B. 53BP1 and USP28 mediate p53-dependent cell cycle arrest in response to centrosome loss and prolonged mitosis. Elife 2016, 5, E1491. [Google Scholar] [CrossRef]
- Meitinger, F.; Anzola, J.V.; Kaulich, M.; Richardson, A.; Stender, J.D.; Benner, C.; Glass, C.K.; Dowdy, S.F.; Desai, A.; Shiau, A.K.; et al. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J. Cell Biol. 2016, 214, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Latgé, G.; Poulet, C.; Bours, V.; Josse, C.; Jerusalem, G. Natural Antisense Transcripts: Molecular Mechanisms and Implications in Breast Cancers. Int J Mol Sci 2018, 19, 123. [Google Scholar] [CrossRef]
- Youssef, I.; Ricort, J.-M. Deciphering the role of protein kinase D1 (PKD1) in cellular proliferation. Mol. Cancer Res. 2019, 17, 1961–1974. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Bouju, S.; Lignon, M.F.; Piétu, G.; Le Cunff, M.; Léger, J.J.; Auffray, C.; Dechesne, C.A. Molecular cloning and functional expression of a novel human gene encoding two 41-43 kDa skeletal muscle internal membrane proteins. Biochem. J. 1998, 335 Pt 3, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Janeček, Š. A motif of a microbial starch-binding domain found in human genethonin. Bioinformatics 2002, 18, 1534–1537. [Google Scholar] [CrossRef] [PubMed]
- Prats, C.; Graham, T.E.; Shearer, J. The dynamic life of the glycogen granule. J. Biol. Chem. 2018, 293, 7089–7098. [Google Scholar] [CrossRef]
- Klimek, F.; Mayer, D.; Bannasch, P. Biochemical microanalysis of glycogen content and glucose-6-phosphate dehydrogenase activity in focal lesions of the rat liver induced by N-nitrosomorpholine. Carcinogenesis 1984, 5, 265–268. [Google Scholar] [CrossRef]
- Favaro, E.; Bensaad, K.; Chong, M.G.; Tennant, D.A.; Ferguson, D.J.P.; Snell, C.; Steers, G.; Turley, H.; Li, J.-L.; Günther, U.L.; et al. Glucose Utilization via Glycogen Phosphorylase Sustains Proliferation and Prevents Premature Senescence in Cancer Cells. Cell Metabolism 2012, 16, 751–764. [Google Scholar] [CrossRef]
- Valero, R.; Bayés, M.; Francisca Sánchez-Font, M.; González-Angulo, O.; Gonzàlez-Duarte, R.; Marfany, G. Characterization of alternatively spliced products and tissue-specific isoforms of USP28 and USP25. Genome Biol. 2001, 2, RESEARCH0043. [Google Scholar] [CrossRef]
- Zhang, D.; Zaugg, K.; Mak, T.W.; Elledge, S.J. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 2006, 126, 529–542. [Google Scholar] [CrossRef]
- Popov, N.; Wanzel, M.; Madiredjo, M.; Zhang, D.; Beijersbergen, R.; Bernards, R.; Moll, R.; Elledge, S.J.; Eilers, M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol 2007, 9, 765–774. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Yang, X.H.; Kang, T.; Zhao, Y.; Wang, C.; Evers, B.M.; Zhou, B.P. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep 2013, 5, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Han, H.; Sun, Q.; Liu, K.; Lin, N.; Xu, C.; Zhao, Z.; Zhao, W. USP28 regulates deubiquitination of histone H2A and cell proliferation. Exp. Cell Res. 2019, 379, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Flügel, D.; Görlach, A.; Kietzmann, T. GSK-3β regulates cell growth, migration, and angiogenesis via Fbw7 and USP28-dependent degradation of HIF-1α. Blood 2012, 119, 1292–1301. [Google Scholar] [CrossRef] [PubMed]
- Mazzucco, A.E.; Smogorzewska, A.; Kang, C.; Luo, J.; Schlabach, M.R.; Xu, Q.; Patel, R.; Elledge, S.J. Genetic interrogation of replicative senescence uncovers a dual role for USP28 in coordinating the p53 and GATA4 branches of the senescence program. Genes Dev. 2017, 31, 1933–1938. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, Z.; Zhang, L.; Yang, Z.; Chen, X.; Luo, J.; Zhou, Z.; Mei, X.; Yu, X.; Shao, Z.; et al. Targeting deubiquitinase USP28 for cancer therapy. Cell Death Dis. 2018, 9, 186. [Google Scholar] [CrossRef]
- Bolstad, B.M.; Irizarry, R.A.; Astrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef]
- Yi, H.; Thurberg, B.L.; Curtis, S.; Austin, S.; Fyfe, J.; Koeberl, D.D.; Kishnani, P.S.; Sun, B. Characterization of a canine model of glycogen storage disease type IIIa. Dis Model Mech 2012, 5, 804–811. [Google Scholar] [CrossRef]
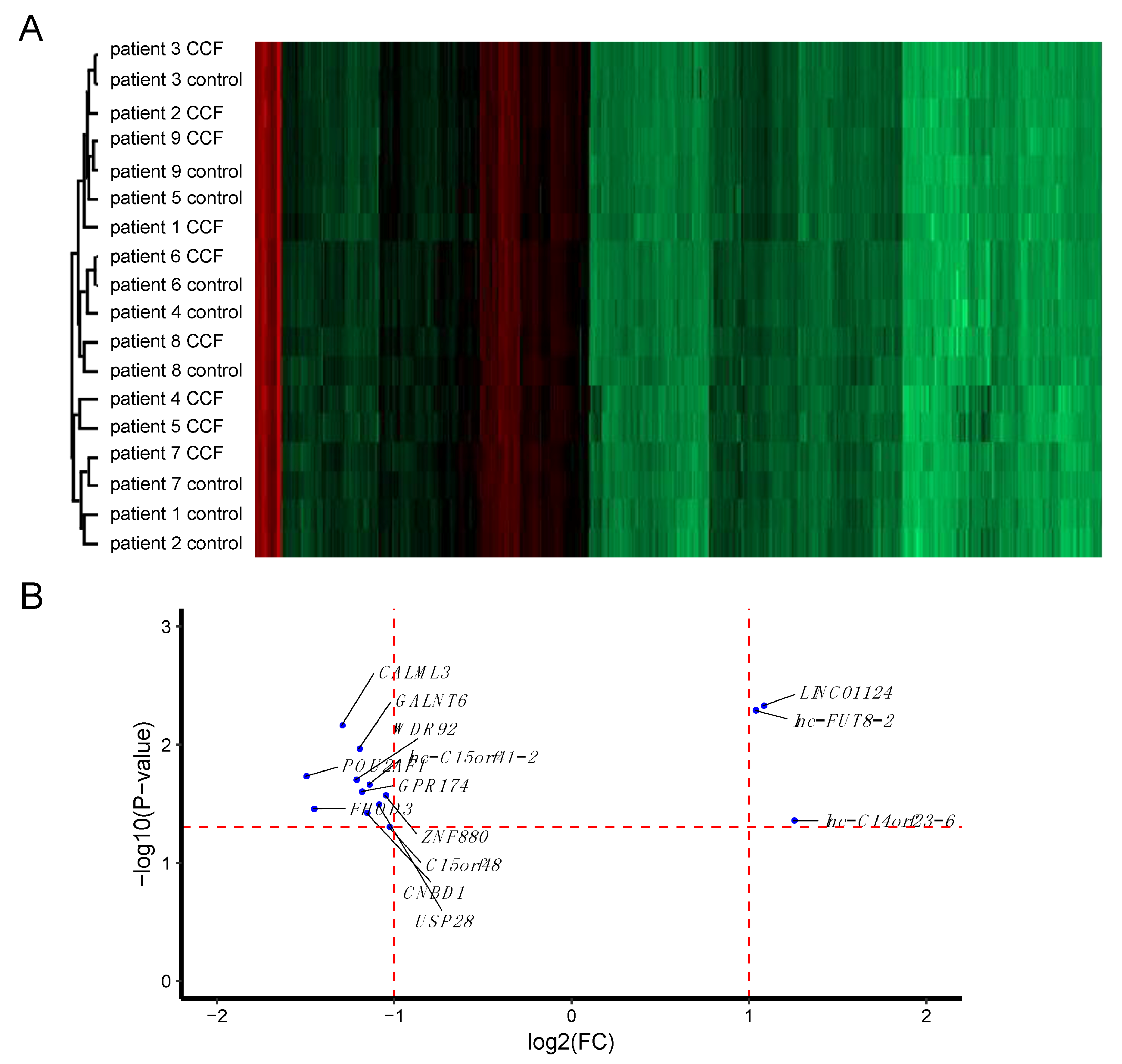

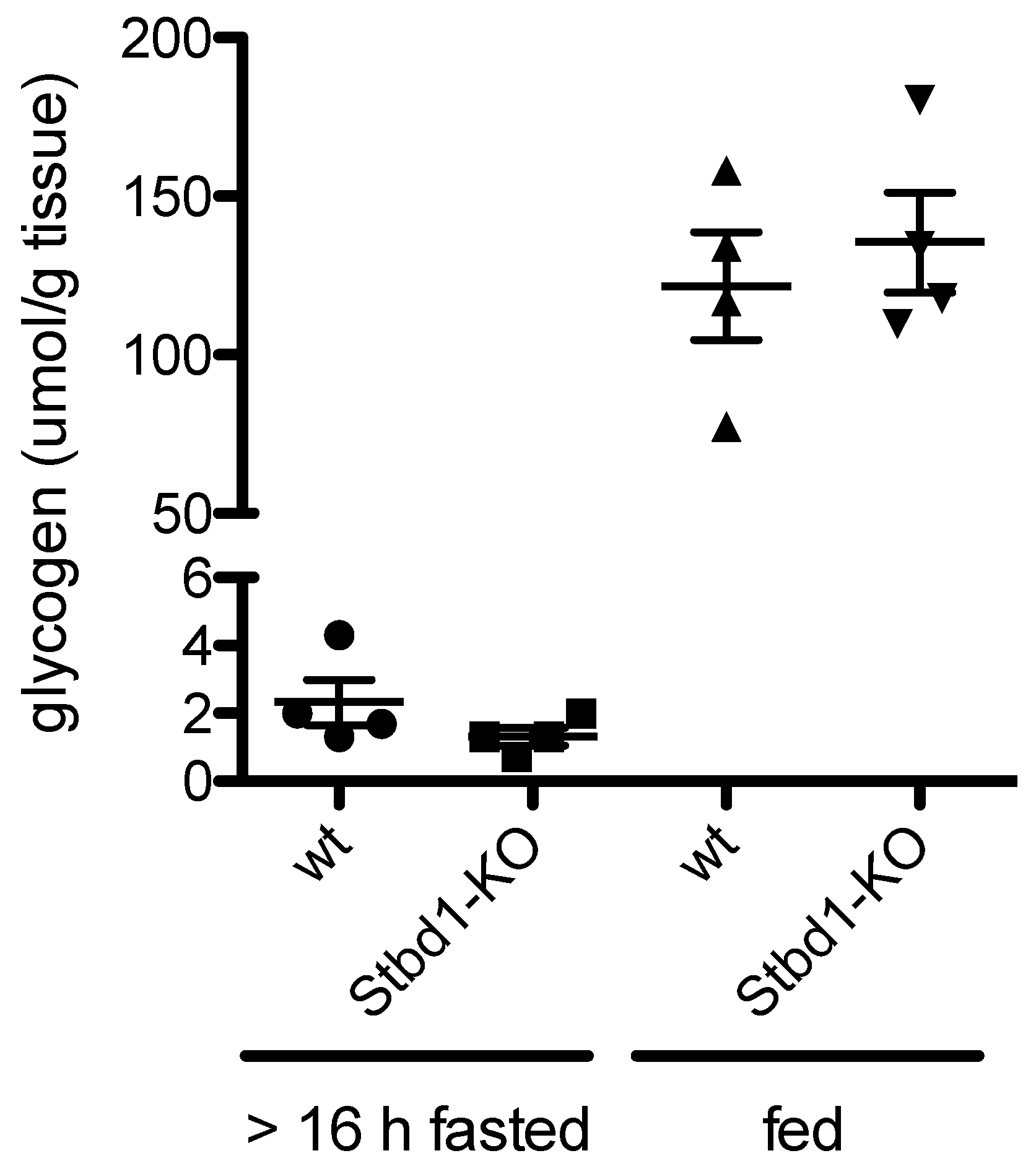

| Target Name | Gene Symbol | Gene Product | log2(FC) | p-Value | Function |
|---|---|---|---|---|---|
| lnc-C14orf23-6:1 lnc-FOXG1-6:17(LNCIPEDIA v5.2) | lnc-C14orf23-6 | lnc-FOXG1-6, (ENSG00000257120.1; CTD-2503I6.1; OTTHUMG00000170489.1; AL356756.1) | 1.25 | 0.0439 | lncRNA; antisense to PRKD1; +strand; 2 exons; 551 bp; in stringent set |
| NR_027433.1 LINC01124:6 (LNCIPEDIA v5.2) | LINC01124 | long intergenic non-protein coding RNA 1124 | 1.08 | 0.0047 | lncRNA; bidirectional; -strand; 1 exon; 2129 bp; in stringent set |
| lnc-FUT8-2:12 LINC02290:27 (LNCIPEDIA v5.2) | lnc-FUT8-2 LINC02290 (LNCIPEDIA v5.2) | XLOC_010856; linc-GPHN-2 | 1.04 | 0.0051 | lncRNA; intergenic; +strand; 4 exons; 463 bp; not in stringent set |
| NM_032413.3 | C15orf48 | chromosome 15 open reading frame 48 | −1.03 | 0.0496 | no reported function |
| NM_001145434.1 | ZNF880 | zinc finger protein 880 | −1.05 | 0.0269 | no reported function |
| NM_001301029.1 | USP28 | Ubiquitin carboxyl-terminal hydrolase 28 | −1.09 | 0.0320 | protein deubiquitination; c-Myc stabilization and hepatocarcinogenesis [24,25] |
| lnc-C15orf41-2:1 (LNCIPEDIA V5.2) | lnc-C15orf41-2 | lincRNA | −1.14 | 0.0217 | lncRNA; intergenic; +strand; 2 exons; 393 bp; in stringent set |
| NM_173538.2 | CNBD1 | cyclic nucleotide binding domain containing 1 | −1.16 | 0.0378 | |
| NM_032553.1 | GPR174 | G protein-coupled receptor 174 | −1.19 | 0.0250 | lysophosphatidylserine receptor [26,27]; SNPs are risk factors for Graves’ disease [28]; autoimmune Addison´s disease [29]; |
| NM_007210.3 | GALNT6 | polypeptide N-acetylgalactosaminyltransferase 6 | −1.20 | 0.0108 | marker for early tumorigenesis in breast cancer [30]; suppressor of colorectal cancer progression 30662801; enhancer of aggression of ovarian cancer cells [31]; protection from apoptosis under stress conditions [32]; promotion of tumorigenicity and metastasis in breast cancer [33]; growth suppression in pancreatic cancer [34] |
| NM_001256476.1 | WDR92 | monad/WD repeat domain 92 | −1.22 | 0.0198 | part of the R2TP/prefoldin-like complex [35]; inhibits breast cancer cell invasion [36]; TNFalpha/cycloheximide-mediated apoptosis [37] |
| NM_005185.3 | CALML3 | calmodulin-like 3 | −1.30 | 0.0069 | calcium homeostasis (by similarity); suppression of gastric cancer progression by secreted CALML3 after metformin treatment [38] |
| NM_001281739.1 | FHOD3 | Formin Homology 2 Domain Containing 3 | −1.46 | 0.0350 | actin binding 24914801; cancer cell invasiveness [39] |
| NM_006235.2 | POU2AF1 | POU class 2 associating factor 1 | −1.50 | 0.0185 | B cell maturation 17621271 |
| Accession | Description (Gene Symbol) | log2 Ratio (CCF/Control) | t-Test p-Value |
|---|---|---|---|
| higher expression in CCF | |||
| Q15286 | Ras-related protein Rab-35 (RAB35) | 0.9087 | 0.0447 |
| Q6IQ22 | Ras-related protein Rab-12 (RAB12) | 0.7768 | 0.0439 |
| Q03013 | Glutathione S-transferase Mu 4 (GSTM4) | 0.6028 | 0.0066 |
| lower expression in CCF | |||
| Q12797 | Aspartyl/asparaginyl beta-hydroxylase (ASPH) | −0.6097 | 0.0254 |
| Q15008 | 26S proteasome non-ATPase regulatory subunit 6 (PSMD6) | −0.6155 | 0.0121 |
| P84098 | 60S ribosomal protein L19 (RPL19) | −0.6163 | 0.0086 |
| P61247 | 40S ribosomal protein S3a (RPS3A) | −0.6171 | 0.0474 |
| Q13451 | Peptidyl-prolyl cis-trans isomerase FKBP5 (FKBP) | −0.6322 | 0.0489 |
| Q9UNF0 | Protein kinase C and casein kinase substrate in neurons protein 2 (PACSIN2) | −0.6591 | 0.0327 |
| O95810 | Serum deprivation-response protein (SDPR) | −0.6682 | 0.0357 |
| Q16836 | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial (HADH) | −0.6846 | 0.0146 |
| O75821 | Eukaryotic translation initiation factor 3 subunit G (EIF3G) | −0.7156 | 0.0333 |
| Q9BVK6 | Transmembrane emp24 domain-containing protein 9 (TMED9) | −0.7615 | 0.021 |
| P63208 | S-phase kinase-associated protein 1 (SKP1) | −0.7715 | 0.0282 |
| Q15436 | Protein transport protein Sec23A (SEC23A) | −0.7936 | 0.0441 |
| Q16629 | Serine/arginine-rich splicing factor 7 (SRSF7) | −0.8146 | 0.026 |
| Q95604 | HLA class I histocompatibility antigen, Cw-17 alpha chain (HLA-C) | −0.8195 | 0.0429 |
| O75436 | Vacuolar protein sorting-associated protein 26A (VPS26A) | −0.878 | 0.0217 |
| Q9NRV9 | Heme-binding protein 1 (HEBP1) | −0.9163 | 0.034 |
| O95210 | Starch-binding domain-containing protein 1 (STBD1) | −1.1576 | 0.0035 |
| P61604 | 10 kDa heat shock protein, mitochondrial (HSPE1) | −1.1642 | 0.0326 |
| O43169 | Cytochrome b5 type B (CYB5B) | −1.211 | 0.0157 |
| CCF vs. Control | ||
|---|---|---|
| Protein | High-Throughput Analysis | Immunohistochemistry |
| CYB5B | ↓ (protein) | ↑ 0 ←→ 2 ↓ 9 |
| HSPE1 | ↓ (protein) | ↑ 0 ←→ 5 ↓ 1 ↓ 4 |
| monad/WDR92 | ↓ (mRNA) | ↑ 0 ←→ 2 ↓ 9 |
| RAB12 | ↑ (protein) | ↑ 0 ←→ 7 ↓ 3 |
| RAB35 | ↑ (protein) | ↑ 0 ←→ 7 ↓ 3 |
| STBD1 | ↓ (protein) | ↑ 0 ←→ 0 ↓ 11 |
| USP28 | ↓ (mRNA) | ↑ 0 ←→ 1 ↓ 6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Metzendorf, C.; Wineberger, K.; Rausch, J.; Cigliano, A.; Peters, K.; Sun, B.; Mennerich, D.; Kietzmann, T.; Calvisi, D.F.; Dombrowski, F.; et al. Transcriptomic and Proteomic Analysis of Clear Cell Foci (CCF) in the Human Non-Cirrhotic Liver Identifies Several Differentially Expressed Genes and Proteins with Functions in Cancer Cell Biology and Glycogen Metabolism. Molecules 2020, 25, 4141. https://doi.org/10.3390/molecules25184141
Metzendorf C, Wineberger K, Rausch J, Cigliano A, Peters K, Sun B, Mennerich D, Kietzmann T, Calvisi DF, Dombrowski F, et al. Transcriptomic and Proteomic Analysis of Clear Cell Foci (CCF) in the Human Non-Cirrhotic Liver Identifies Several Differentially Expressed Genes and Proteins with Functions in Cancer Cell Biology and Glycogen Metabolism. Molecules. 2020; 25(18):4141. https://doi.org/10.3390/molecules25184141
Chicago/Turabian StyleMetzendorf, Christoph, Katharina Wineberger, Jenny Rausch, Antonio Cigliano, Kristin Peters, Baodong Sun, Daniela Mennerich, Thomas Kietzmann, Diego F. Calvisi, Frank Dombrowski, and et al. 2020. "Transcriptomic and Proteomic Analysis of Clear Cell Foci (CCF) in the Human Non-Cirrhotic Liver Identifies Several Differentially Expressed Genes and Proteins with Functions in Cancer Cell Biology and Glycogen Metabolism" Molecules 25, no. 18: 4141. https://doi.org/10.3390/molecules25184141
APA StyleMetzendorf, C., Wineberger, K., Rausch, J., Cigliano, A., Peters, K., Sun, B., Mennerich, D., Kietzmann, T., Calvisi, D. F., Dombrowski, F., & Ribback, S. (2020). Transcriptomic and Proteomic Analysis of Clear Cell Foci (CCF) in the Human Non-Cirrhotic Liver Identifies Several Differentially Expressed Genes and Proteins with Functions in Cancer Cell Biology and Glycogen Metabolism. Molecules, 25(18), 4141. https://doi.org/10.3390/molecules25184141







