Inorganic Elements in Mytilus galloprovincialis Shells: Geographic Traceability by Multivariate Analysis of ICP-MS Data
Abstract
1. Introduction
2. Results and Discussion
2.1. Analytical Method
2.2. Chemometrics
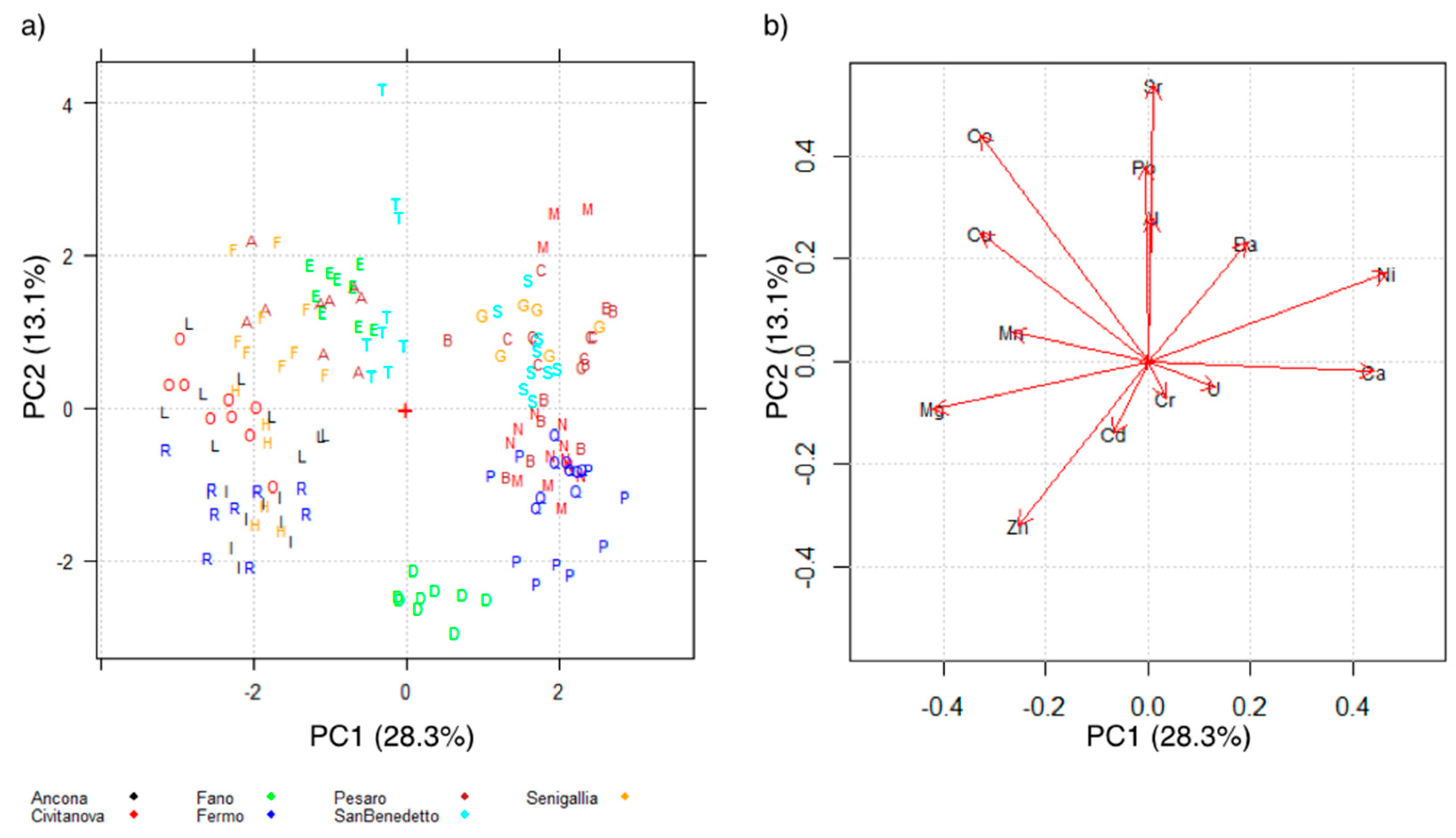
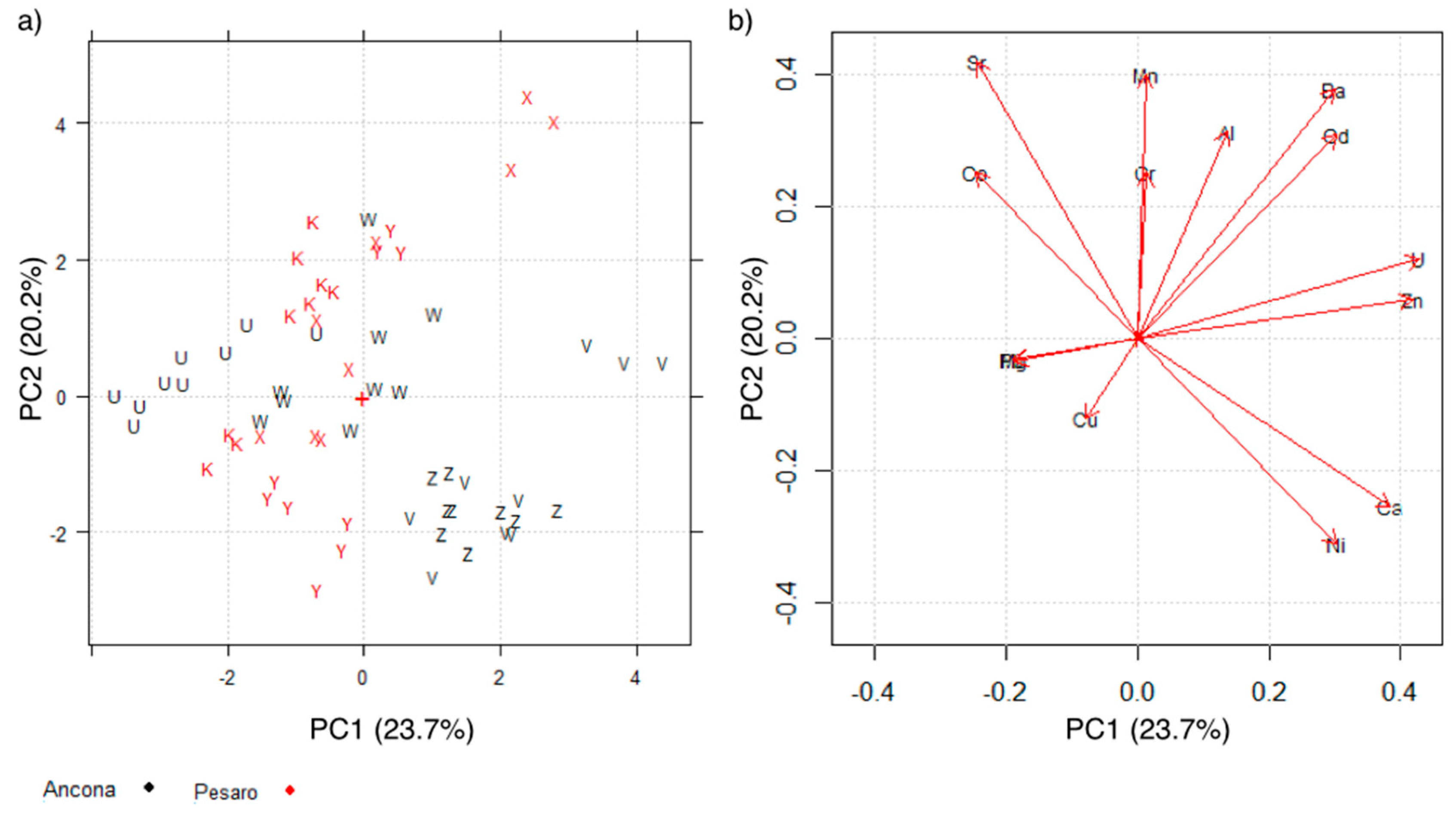
2.2.1. Principal Component Analysis
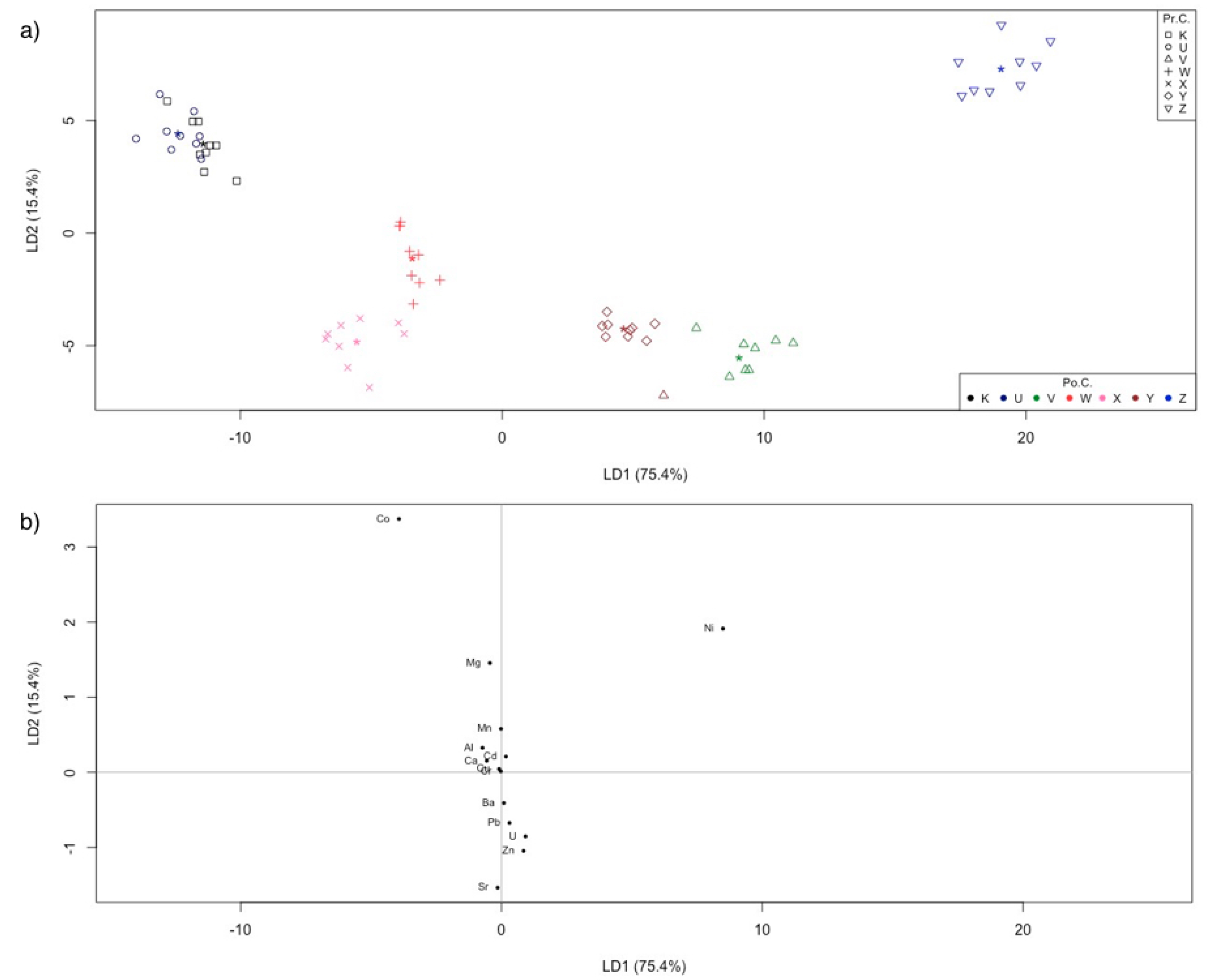
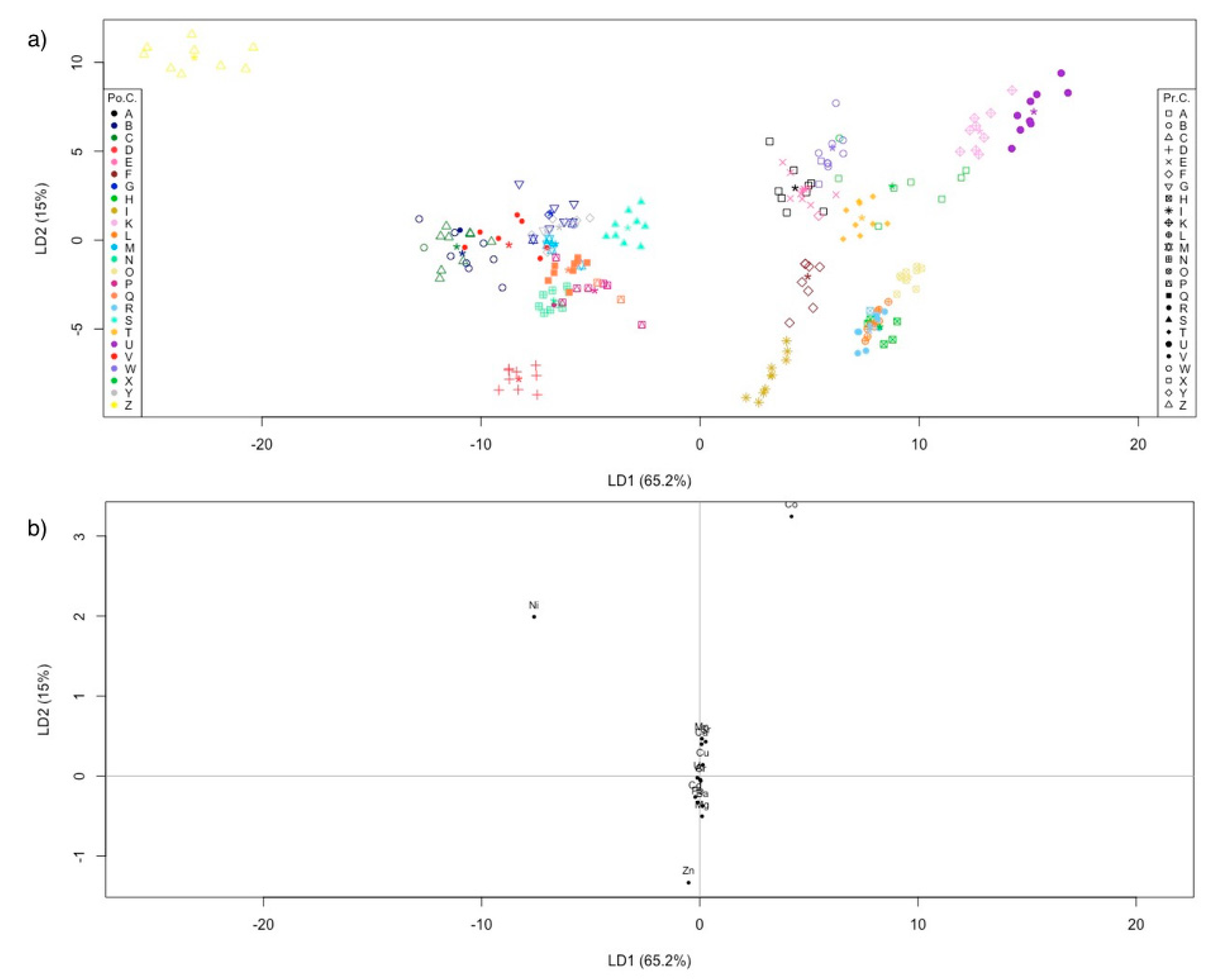
2.2.2. Linear Discriminant Analysis
Farms and Natural Banks
Farmed Samples
Samples from Natural Banks
All Samples
3. Materials and Methods
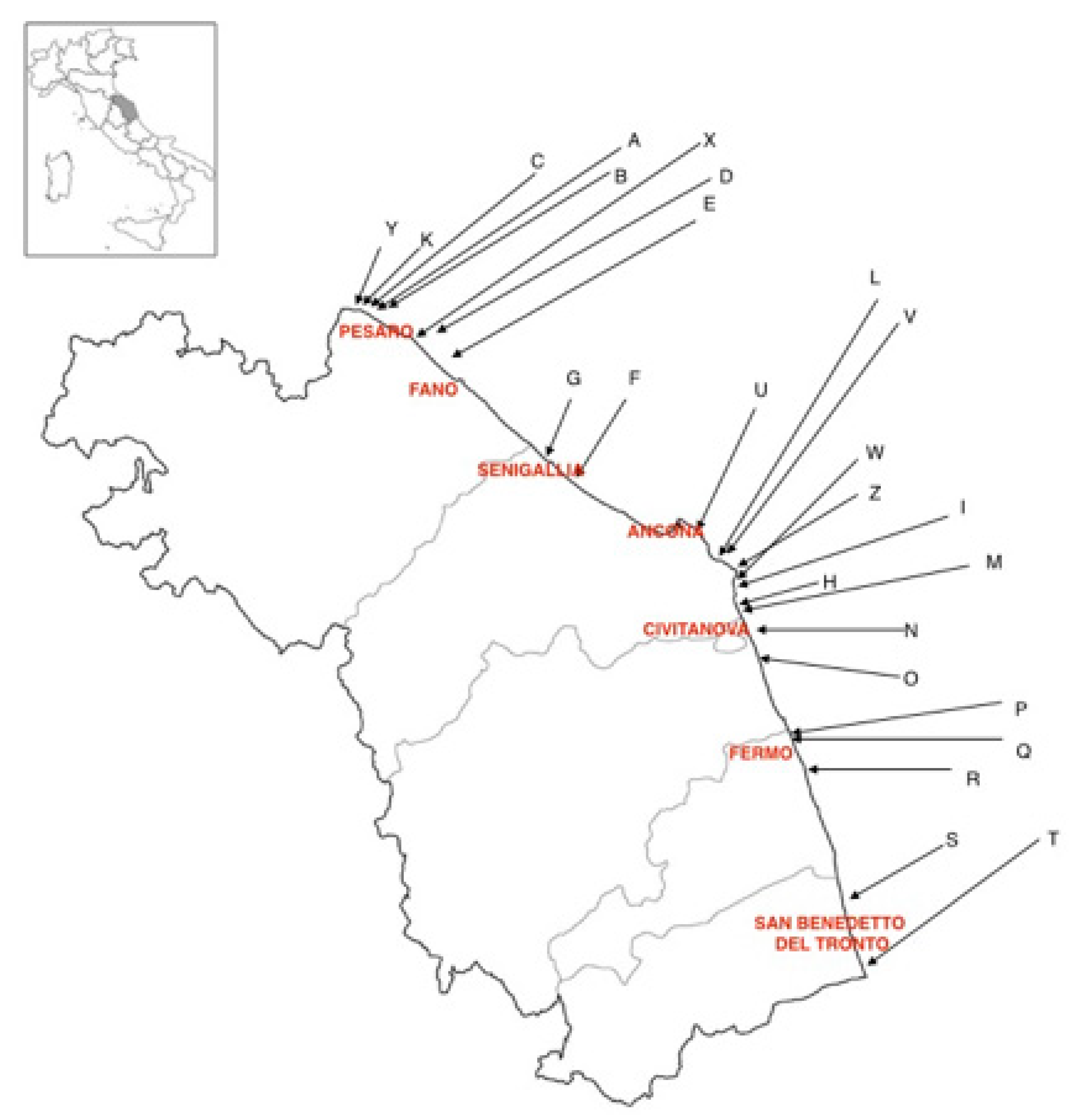
3.1. Samples and Datasets
3.2. Experimental
3.2.1. Reagents, Standards, and Certified Reference Materials
3.2.2. Sample Pretreatment and Preparation
3.2.3. Instrumental Analysis
3.2.4. Quality Assurance/Quality Control
3.3. Chemometrics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Astorga España, M.S.; Rodríguez Rodríguez, E.M.; Díaz Romero, C. Comparison of mineral and trace element concentrations in two molluscs from the Strait of Magellan (Chile). J. Food Compos. Anal. 2007. [Google Scholar] [CrossRef]
- Wilson, W.A.; Fritts, A.K.; Fritts, M.W.; Unrine, J.M.; Tweedy, B.N.; Casper, A.F. Freshwater mussel shells (Unionidae) describe anthropogenic changes to trace element cycling within a North American river. Sci. Total Environ. 2018. [Google Scholar] [CrossRef]
- Firth, D.C.; Salie, K.; O’Neill, B.; Hoffman, L.C. Monitoring of trace metal accumulation in two South African farmed mussel species, Mytilus galloprovincialis and Choromytilus meridionalis. Mar. Pollut. Bull. 2019. [Google Scholar] [CrossRef] [PubMed]
- Il Parlamento Europeo e il Consiglio dell’Unione Europea Regolamento (Ce) N. 852/2004. Gazz. Uff. dell’Unione Eur. 2004. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=LEGISSUM%3Af84002 (accessed on 12 March 2021).
- Regolamento CE REGOLAMENTO (CE) N. 853/2004. Gazz. Uff. dell’Unione Eur. 2004. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=LEGISSUM%3Af84002 (accessed on 12 March 2021).
- Reg. CE 854/2004 Reg. CE 854/2004Il Parlamento Europeo e il Consiglio dell’Unione Europea. Gazz. Uff. dell’Unione Eur. 2004. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=LEGISSUM%3Af84002 (accessed on 12 March 2021).
- Ricardo, F.; Pimentel, T.; Moreira, A.S.P.; Rey, F.; Coimbra, M.A.; Rosário Domingues, M.; Domingues, P.; Costa Leal, M.; Calado, R. Potential use of fatty acid profiles of the adductor muscle of cockles (Cerastoderma edule) for traceability of collection site. Sci. Rep. 2015. [Google Scholar] [CrossRef] [PubMed]
- Tynan, S.; Eggins, S.; Kinsley, L.; Welch, S.A.; Kirste, D. Mussel Shells as An environmental Tracers: An Example from the Loveday Basin. Regolith 2005 Ten Years CRC LEME Proc. CRC LEME Reg. Regolith Symp. 2005. Available online: https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.552.474&rep=rep1&type=pdf (accessed on 12 March 2021).
- Protasowicki, M.; Dural, M.; Jaremek, J. Trace metals in the shells of blue mussels (Mytilus edulis) from the Poland coast of Baltic sea. Environ. Monit. Assess. 2008. [Google Scholar] [CrossRef]
- Pourang, N.; Richardson, C.A.; Chenery, S.R.N.; Nasrollahzedeh, H. Assessment of trace elements in the shell layers and soft tissues of the pearl oyster Pinctada radiata using multivariate analyses: A potential proxy for temporal and spatial variations of trace elements. Environ. Monit. Assess. 2014. [Google Scholar] [CrossRef]
- Locatelli, C.; Melucci, D.; de Laurentiis, F.; Zappi, A. Determination of emerging metal pollutants and toxic metals in mussels and bivalve mollusks, very important food and environmental bio-monitoring species. In Mussels: Characteristics, Biology and Conservation; Nova Science Publisher: Hauppauge, NY, USA, 2018; pp. 33–84. ISBN 9781536134612. [Google Scholar]
- Bourgoin, B. Mytilus edulis shell as a bioindicator of lead pollution considerations on bioavailability and variability. Mar. Ecol. Prog. Ser. 1990. [Google Scholar] [CrossRef]
- Besada, V.; Manuel Andrade, J.; Schultze, F.; José González, J. Monitoring of heavy metals in wild mussels (Mytilus galloprovincialis) from the Spanish North-Atlantic coast. Cont. Shelf Res. 2011. [Google Scholar] [CrossRef]
- Besada, V.; Andrade, J.M.; Schultze, F.; Fumega, J.; Cambeiro, B.; González, J.J. Statistical comparison of trace metal concentrations in wild mussels (Mytilus galloprovincialis) in selected sites of Galicia and Gulf of Biscay (Spain). J. Mar. Syst. 2008. [Google Scholar] [CrossRef]
- Perošević, A.; Pezo, L.; Joksimović, D.; Đurović, D.; Milašević, I.; Radomirović, M.; Stanković, S. The impacts of seawater physicochemical parameters and sediment metal contents on trace metal concentrations in mussels—A chemometric approach. Environ. Sci. Pollut. Res. 2018. [Google Scholar] [CrossRef]
- Pérez-Mayol, S.; Blasco, J.; Tornero, V.; Morales-Nin, B.; Massanet, A.; Tovar-Sánchez, A. Are the shells of Scrobicularia plana useful for monitoring trace metal Pollution events? J. Environ. Biol. 2014, 35, 9–17. [Google Scholar] [PubMed]
- Ricardo, F.; Pimentel, T.; Génio, L.; Calado, R. Spatio-Temporal variability of trace elements fingerprints in cockle (Cerastoderma edule) shells and its relevance for tracing geographic origin. Sci. Rep. 2017. [Google Scholar] [CrossRef]
- Bennion, M.; Morrison, L.; Brophy, D.; Carlsson, J.; Abrahantes, J.C.; Graham, C.T. Trace element fingerprinting of blue mussel (Mytilus edulis)shells and soft tissues successfully reveals harvesting locations. Sci. Total Environ. 2019. [Google Scholar] [CrossRef] [PubMed]
- Szefer, P.; Frelek, K.; Szefer, K.; Lee, C.B.; Kim, B.S.; Warzocha, J.; Zdrojewska, I.; Ciesielski, T. Distribution and relationships of trace metals in soft tissue, byssus and shells of Mytilus edulis trossulus from the southern Baltic. Environ. Pollut. 2002. [Google Scholar] [CrossRef]
- Szefer, P.; Fowler, S.W.; Ikuta, K.; Osuna, F.P.; Ali, A.A.; Kim, B.S.; Fernandes, H.M.; Belzunce, M.J.; Guterstam, B.; Kunzendorf, H.; et al. A comparative assessment of heavy metal accumulation in soft parts and byssus of mussels from subarctic, temperate, subtropical and tropical marine environments. Environ. Pollut. 2006. [Google Scholar] [CrossRef]
- Cubadda, F.; Raggi, A.; Coni, E. Element fingerprinting of marine organisms by dynamic reaction cell inductively coupled plasma mass spectrometry. Anal. Bioanal. Chem. 2006. [Google Scholar] [CrossRef] [PubMed]
- Ricardo, F.; Génio, L.; Costa Leal, M.; Albuquerque, R.; Queiroga, H.; Rosa, R.; Calado, R. Trace element fingerprinting of cockle (Cerastoderma edule) shells can reveal harvesting location in adjacent areas. Sci. Rep. 2015. [Google Scholar] [CrossRef] [PubMed]
- Sorte, C.J.B.; Etter, R.J.; Spackman, R.; Boyle, E.E.; Hannigan, R.E. Elemental fingerprinting of mussel shells to predict population sources and redistribution potential in the Gulf of Maine. PLoS ONE 2013. [Google Scholar] [CrossRef] [PubMed]
- US EPA. Method 3052, Microwave Assisted acid Digestion of Siliceous and Organically Based Matrices; US EPA: Washington, CD, USA, 1996.
- US EPA. Method 3051A: Microwave Assisted Digestion of Sediments, Sludges, Soils and Oils. Test Methods; US EPA: Washington, DC, USA, 2007.
- Wenzl, T.; Haedrich, J.; Schaechtele, A.; Robouch, P.; Stroka, J. Guidance Document on the Estimation of LOD and LOQ for Measurements in the Field of Contaminants in Feed and Food. EUR 28099 EN; Publications Office of the European Union: Luxembourg, 2016. [Google Scholar]
- Todeschini, R. Introduzione alla Chemiometria; Edises: Napoli, Italy, 1998. [Google Scholar]
- Salinitro, M.; Tassoni, A.; Casolari, S.; de Laurentiis, F.; Zappi, A.; Melucci, D. Heavy Metals Bioindication Potential of the Common Weeds Senecio vulgaris L., Polygonum aviculare L. and Poa annua L. Molecules 2019, 24, 2813. [Google Scholar] [CrossRef]
- Bona, E.; Março, P.H.; Valderrama, P. Chemometrics Applied to Food Control. In Food Control and Biosecurity; Academic Press: Cambridge, UK, 2018; pp. 105–133. ISBN 9780128114971. [Google Scholar]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014. [Google Scholar] [CrossRef]
- Kumar, N.; Bansal, A.; Sarma, G.S.; Rawal, R.K. Chemometrics tools used in analytical chemistry: An overview. Talanta 2014. [Google Scholar] [CrossRef] [PubMed]
- Brereton, R.G. Chemometrics for Pattern Recognition; Wiley: Hoboken, NJ, USA, 2009; ISBN 9780470987254. [Google Scholar]
- Ballabio, D.; Todeschini, R. Multivariate Classification for Qualitative Analysis. In Infrared Spectroscopy for Food Quality Analysis and Control; Academic Press: Cambridge, UK, 2009; pp. 83–104. ISBN 9780123741363. [Google Scholar]
- Ballabio, D.; Grisoni, F.; Todeschini, R. Multivariate comparison of classification performance measures. Chemom. Intell. Lab. Syst. 2018, 174, 33–44. [Google Scholar] [CrossRef]
- Leardi, C.R.; Melzi, G.P. CAT (Chemometric Agile Tool). Available online: http://gruppochemiometria.it/index.php/software (accessed on 12 March 2021).

| Al | Cu | Cr | Zn | Mn | Cd | Co | U | Ba | Ni | Pb | Mg | Sr | Ca | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Al | 1.00 | 0.19 | −0.06 | −0.01 | −0.04 | −0.08 | 0.11 | −0.13 | −0.01 | 0.15 | 0.05 | −0.08 | 0.13 | 0.04 |
| Cu | 1.00 | 0.08 | 0.18 | 0.31 | −0.01 | 0.56 | 0.01 | −0.12 | −0.31 | 0.09 | 0.36 | 0.05 | −0.32 | |
| Cr | 1.00 | −0.05 | −0.03 | 0.07 | 0.01 | −0.07 | 0.08 | 0.04 | −0.18 | −0.1 | −0.07 | 0.05 | ||
| Zn | 1.00 | 0.41 | −0.17 | −0.12 | 0.06 | −0.21 | −0.36 | −0.11 | 0.35 | −0.24 | −0.28 | |||
| Mn | 1.00 | −0.00 | 0.14 | −0.00 | −0.08 | −0.18 | 0.41 | 0.45 | −0.03 | −0.17 | ||||
| Cd | 1.00 | 0.04 | 0.01 | 0.14 | −0.25 | 0.00 | 0.09 | −0.26 | −0.14 | |||||
| Co | 1.00 | −0.17 | −0.08 | −0.40 | 0.18 | 0.30 | 0.34 | −0.52 | ||||||
| U | 1.00 | 0.37 | 0.23 | −0.14 | −0.03 | 0.02 | 0.21 | |||||||
| Ba | 1.00 | 0.31 | 0.05 | −0.18 | 0.30 | 0.10 | ||||||||
| Ni | 1.00 | 0.17 | −0.62 | 0.1 | 0.79 | |||||||||
| Pb | 1.00 | −0.14 | 0.14 | 0.08 | ||||||||||
| Mg | 1.00 | 0.01 | −0.50 | |||||||||||
| Sr | 1.00 | −0.08 | ||||||||||||
| Ca | 1.00 |
| Al | Cu | Cr | Zn | Mn | Cd | Co | U | Ba | Ni | Pb | Mg | Sr | Ca | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Al | 1.00 | 0.05 | −0.01 | 0.15 | 0.57 | 0.29 | −0.07 | 0.29 | 0.44 | −0.10 | 0.18 | 0.10 | 0.15 | 0.03 |
| Cu | 1.00 | −0.06 | −0.00 | −0.18 | −0.09 | 0.06 | −0.03 | −0.27 | −0.01 | 0.08 | 0.14 | −0.14 | −0.12 | |
| Cr | 1.00 | −0.00 | 0.17 | 0.16 | 0.21 | 0.20 | 0.18 | −0.18 | −0.22 | −0.09 | 0.20 | −0.16 | ||
| Zn | 1.00 | −0.12 | 0.42 | −0.17 | 0.60 | 0.48 | 0.16 | −0.29 | −0.28 | −0.22 | 0.44 | |||
| Mn | 1.00 | 0.21 | 0.16 | 0.11 | 0.50 | −0.07 | 0.28 | 0.07 | 0.31 | −0.20 | ||||
| Cd | 1.00 | −0.05 | 0.51 | 0.50 | −0.01 | −0.26 | −0.33 | 0.19 | 0.09 | |||||
| Co | 1.00 | −0.06 | 0.11 | −0.30 | −0.05 | 0.52 | 0.31 | −0.60 | ||||||
| U | 1.00 | 0.45 | 0.33 | −0.33 | −0.09 | −0.33 | 0.35 | |||||||
| Ba | 1.00 | 0.10 | −0.13 | −0.09 | 0.17 | 0.15 | ||||||||
| Ni | 1.00 | −0.08 | 0.11 | −0.80 | 0.56 | |||||||||
| Pb | 1.00 | 0.19 | −0.02 | −0.10 | ||||||||||
| Mg | 1.00 | −0.10 | −0.11 | |||||||||||
| Sr | 1.00 | −0.49 | ||||||||||||
| Ca | 1.00 |
| Predicted (CV) | ||||
|---|---|---|---|---|
| Actual | F | NB | Sn | Sp |
| F | 150 | 3 | 0.980 | 0.932 |
| NB | 11 | 52 | 0.825 | 0.945 |
| NER | 0.935 | |||
| Sampling Site | A | B | C | D | E | F | G | H | I |
| Sn | 1.00 | 0.778 | 1.00 | 1.00 | 1.00 | 0.889 | 1.00 | 0.833 | 1.00 |
| Sp | 1.00 | 1.00 | 0.818 | 1.00 | 0.900 | 1.00 | 0.750 | 0.833 | 1.00 |
| Sampling Site | L | M | N | O | P | Q | R | S | T |
| Sn | 1.00 | 0.667 | 1.00 | 1.00 | 0.778 | 1.00 | 0.889 | 1.00 | 1.00 |
| Sp | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.818 | 0.889 | 1.00 | 1.00 |
| Sampling Site | K | U | V | W | X | Y | Z |
|---|---|---|---|---|---|---|---|
| Sn | 1.00 | 1.00 | 0.889 | 1.00 | 1.00 | 1.00 | 1.00 |
| Sp | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.900 | 1.00 |
| Sampling Site | A | B | C | D | E | F | G | H | I |
| Sn | 1.00 | 0.889 | 1.00 | 1.00 | 1.00 | 0.889 | 0.833 | 0.833 | 1.00 |
| Sp | 1.00 | 0.889 | 0.900 | 1.00 | 0.900 | 1.00 | 0.625 | 1.00 | 1.00 |
| Sampling Site | K | L | M | N | O | P | Q | R | |
| Sn | 1.00 | 1.00 | 0.667 | 1.00 | 1.00 | 0.778 | 1.00 | 1.00 | |
| Sp | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.875 | 0.818 | 0.900 | |
| Sampling Site | S | T | U | V | W | X | Y | Z | |
| Sn | 1.00 | 1.00 | 1.00 | 0.778 | 0.889 | 0.778 | 0.889 | 1.00 | |
| Sp | 1.00 | 1.00 | 1.00 | 1.00 | 0.800 | 0.875 | 0.889 | 1.00 |
| Matrix | Species | Areas (Site) (Analysed Elements) | Statistical Model * | Correct Assignment Rate (%) | Reference |
|---|---|---|---|---|---|
| shells, foot, periostracum | Mytilus edulis | Ireland coast (4) (As, Cd, Cu, Fe, Mn, Ni, Pb, Sr, Zn ) | Random forest analysis | 67.5 – periostracum 100 − shells + foot + periostracum | M. Bennion et al., 2019 [18] |
| shells | Amblema plicata, Quadrula quadrula | North American river (5) (Mn, Fe, Co, Ni, Cu, Zn, As, Cd, Se) | ANOVA PCA | W. A. Wilson et al., 2018 [2] | |
| shells | Cerastoderma edule | Portuguese Atlantic Coastline (8) (Mg/Ca, Mn/Ca, Sr/Ca, Ba/Ca) | MANOVA LDA | 90 | F. Ricardo et al., 2017 [17] |
| shells | Cerastoderma edule | Estuarine system Ria de Aveiro, Portugal (5) (Ba/Ca, Mg/Ca, Mn/Ca, Pb/Ca, Sr/Ca) | ANOSIM ANOVA SIMPER CAP | 92 | F. Ricardo et al., 2015 [22] |
| shells (juveniles and adults) | Mytilus edulis | Gulf of Maine, USA (7) (Ba/Ca, Cu/Ca, Pb/Ca, La/Ca, Sr/Ca, Mg/Ca, Zn/Ca) | LDA ANOVA | 68.4−juvenile mussels 57.3−adult mussels | C. J. B. Sorte et al., 2013 [23] |
| shells | Mytilus gallop. | Central Adriatic Sea Coast (Marche Region) (25: 18 farmed, 7 natural banks) (Al, Cu, Cr, Zn, Mn, Cd, Co, U, Ba, Ni, Pb, Mg, Sr, Ca) | PCA LDA | 98.4−natural banks mussels 94.1−farmed mussels | This paper |
| Sampling Area | Sampling Site | Sampling Period | Total Number of Samples | |||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | ||||
| Farmed | Pesaro | A | July | July | July | 3 |
| B | June | July | October | 3 | ||
| C | July | October | October | 3 | ||
| Fano | D | July | July | August | 3 | |
| E | July | August | September | 3 | ||
| Senigallia | F | July | August | October | 3 | |
| G | July | August | 2 | |||
| H | August | October | 2 | |||
| Ancona | I | July | July | August | 3 | |
| L | July | July | September | 3 | ||
| Civitanova | M | July | July | 2 | ||
| O | July | July | August | 3 | ||
| N | July | July | July | 3 | ||
| Fermo | P | July | July | August | 3 | |
| R | July | July | August | 3 | ||
| Q | July | July | August | 3 | ||
| San Benedetto del Tronto | S | July | August | August | 3 | |
| T | July | August | August | 3 | ||
| Natural Banks | Ancona | U | July | July | August | 3 |
| V | July | July | August | 3 | ||
| Z | July | July | August | 3 | ||
| W | July | July | August | 3 | ||
| Pesaro | K | July | July | August | 3 | |
| Y | July | July | August | 3 | ||
| X | July | July | August | 3 | ||
| Total | 72 | |||||
| ICP-MS PARAMETERS | |||||
|---|---|---|---|---|---|
| Plasma gas (L/min) | 15 | Sweeps/reading | 20 | ||
| AUX gas (L/min) | 0.8 ÷ 1.5 | Readings/replicate | 1 | ||
| RF power (W) | 1300 | Replicates | 3 | ||
| Nebulizer gas (L/min) | 0.75 ÷ 1.04 | Scan mode | Peak hopping | ||
| Rpa | 0 | Dwell time (ms) | 100 | ||
| Sample uptake (mL/min) | 0.96 | Dwell time Pb, Cd, U (ms) | 200 | ||
| m/z | Internal Standard | Acquisition Mode | RPq | Cell Gas: NH3 (mL/min) | |
| Ba | 138 | 103Rh | Std mode | 0.25 | 0 |
| Cd | 111 | ||||
| Co | 59 | ||||
| Ni | 60 | ||||
| Pb | 206 + 207 + 208 | 175Lu | |||
| Sr | 88 | 103Rh | |||
| U | 238 | ||||
| Al | 27 | 103Rh | DRC | 0.75 | 0.5 |
| Mg | 24 | 0.45 | 0.7 | ||
| Ca | 44 | 0.5 | 0.5 | ||
| Cr | 52 | 0.35 | 0.5 | ||
| Mn | 55 | 0.35 | 0.5 | ||
| Cu | 63 | 0.75 | 0.45 | ||
| Zn | 68 | 0.75 | 0.45 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forleo, T.; Zappi, A.; Melucci, D.; Ciriaci, M.; Griffoni, F.; Bacchiocchi, S.; Siracusa, M.; Tavoloni, T.; Piersanti, A. Inorganic Elements in Mytilus galloprovincialis Shells: Geographic Traceability by Multivariate Analysis of ICP-MS Data. Molecules 2021, 26, 2634. https://doi.org/10.3390/molecules26092634
Forleo T, Zappi A, Melucci D, Ciriaci M, Griffoni F, Bacchiocchi S, Siracusa M, Tavoloni T, Piersanti A. Inorganic Elements in Mytilus galloprovincialis Shells: Geographic Traceability by Multivariate Analysis of ICP-MS Data. Molecules. 2021; 26(9):2634. https://doi.org/10.3390/molecules26092634
Chicago/Turabian StyleForleo, Tiziana, Alessandro Zappi, Dora Melucci, Martina Ciriaci, Francesco Griffoni, Simone Bacchiocchi, Melania Siracusa, Tamara Tavoloni, and Arianna Piersanti. 2021. "Inorganic Elements in Mytilus galloprovincialis Shells: Geographic Traceability by Multivariate Analysis of ICP-MS Data" Molecules 26, no. 9: 2634. https://doi.org/10.3390/molecules26092634
APA StyleForleo, T., Zappi, A., Melucci, D., Ciriaci, M., Griffoni, F., Bacchiocchi, S., Siracusa, M., Tavoloni, T., & Piersanti, A. (2021). Inorganic Elements in Mytilus galloprovincialis Shells: Geographic Traceability by Multivariate Analysis of ICP-MS Data. Molecules, 26(9), 2634. https://doi.org/10.3390/molecules26092634