Flavones from Combretum quadrangulare Growing in Vietnam and Their Alpha-Glucosidase Inhibitory Activity
Abstract
1. Introduction
2. Results and Discussion
2.1. Phytochemical Study and Derivatization of Compounds 2 and 3
2.2. Alpha-Glucosidase Inhibitory Activity of Isolated Compounds
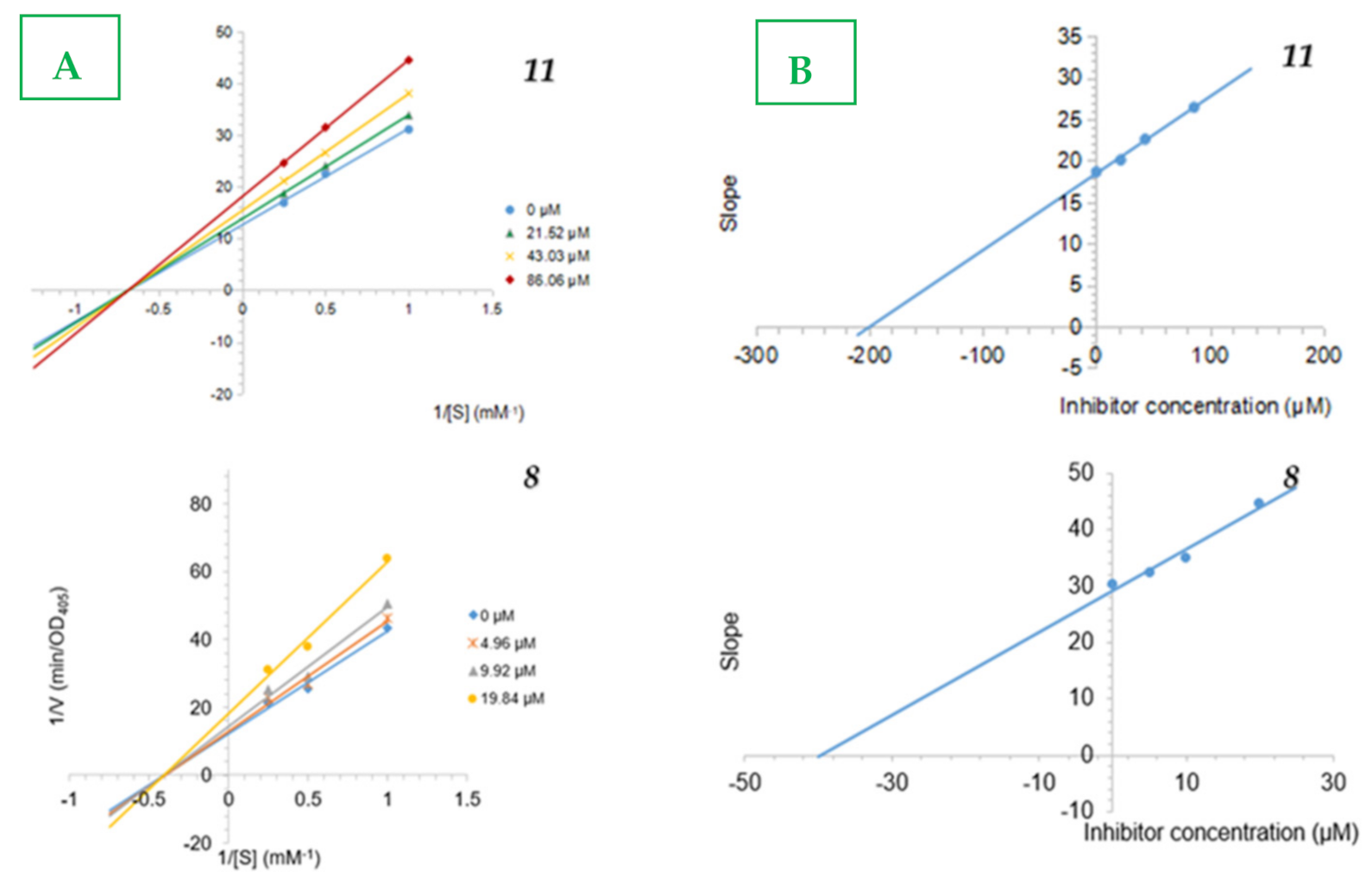
2.3. Inhibition Type and Inhibition Constants of the Compounds 8 and 11 on Alpha-Glucosidase
2.4. Antibacterial Activity of Isolated Compounds
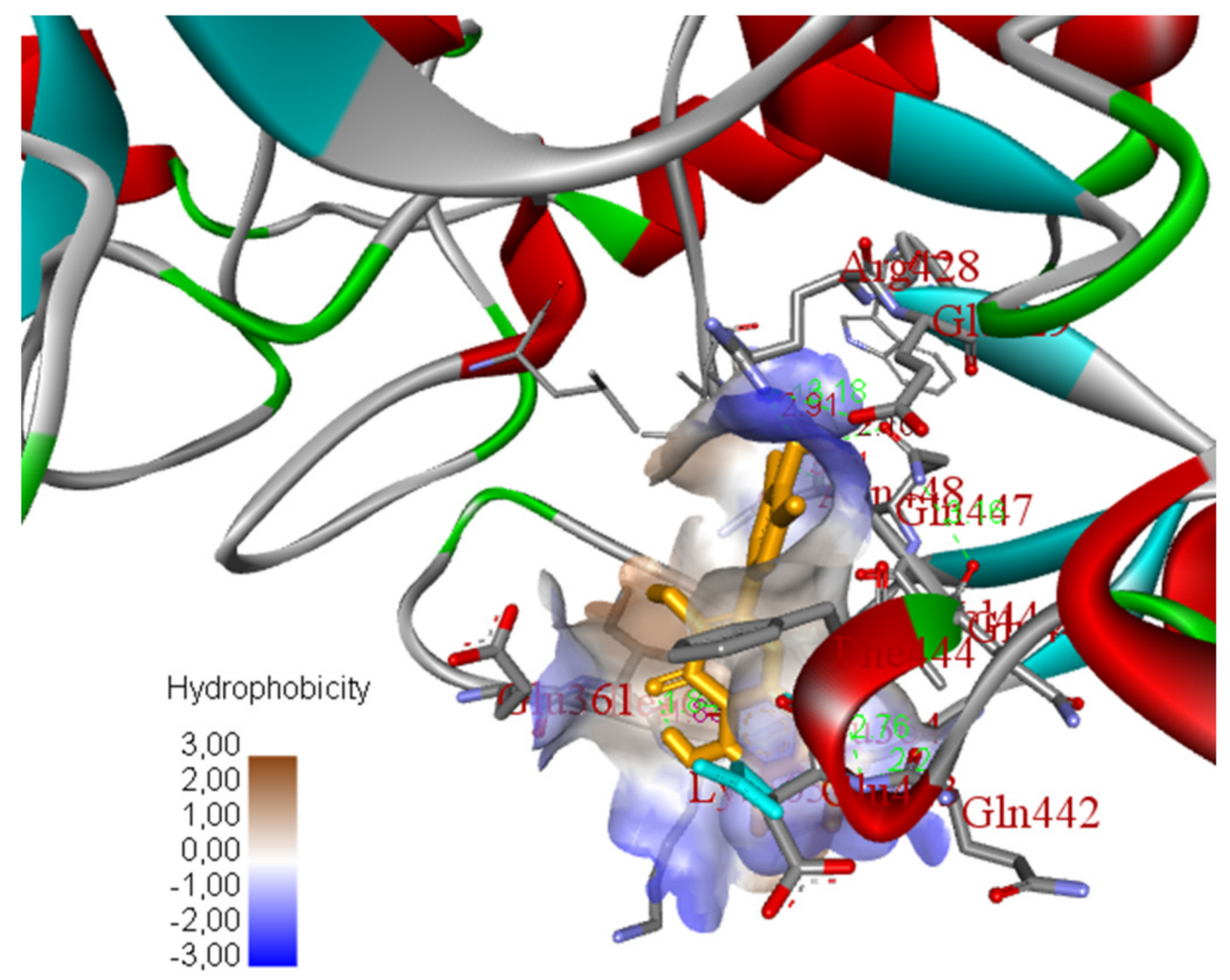
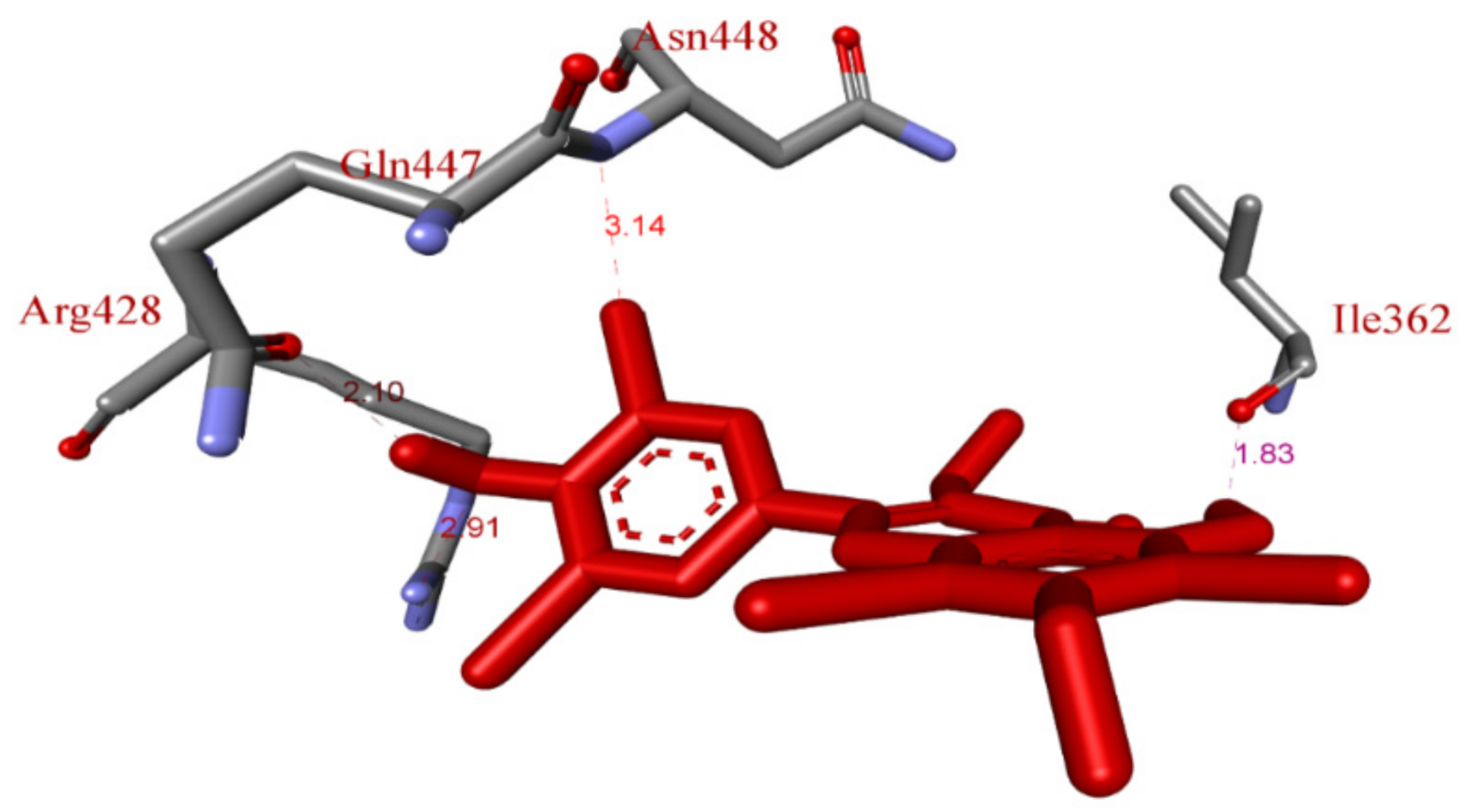
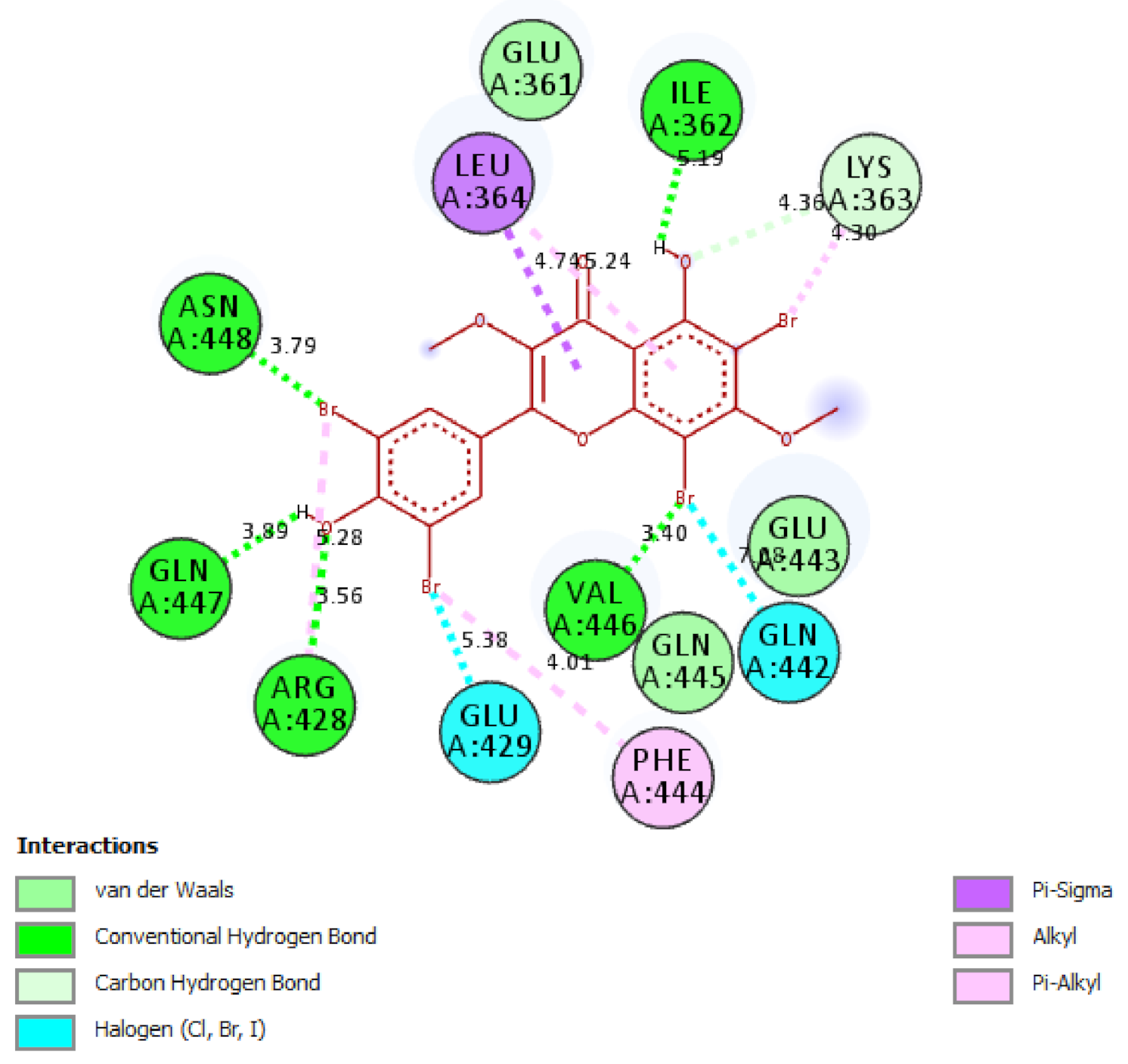
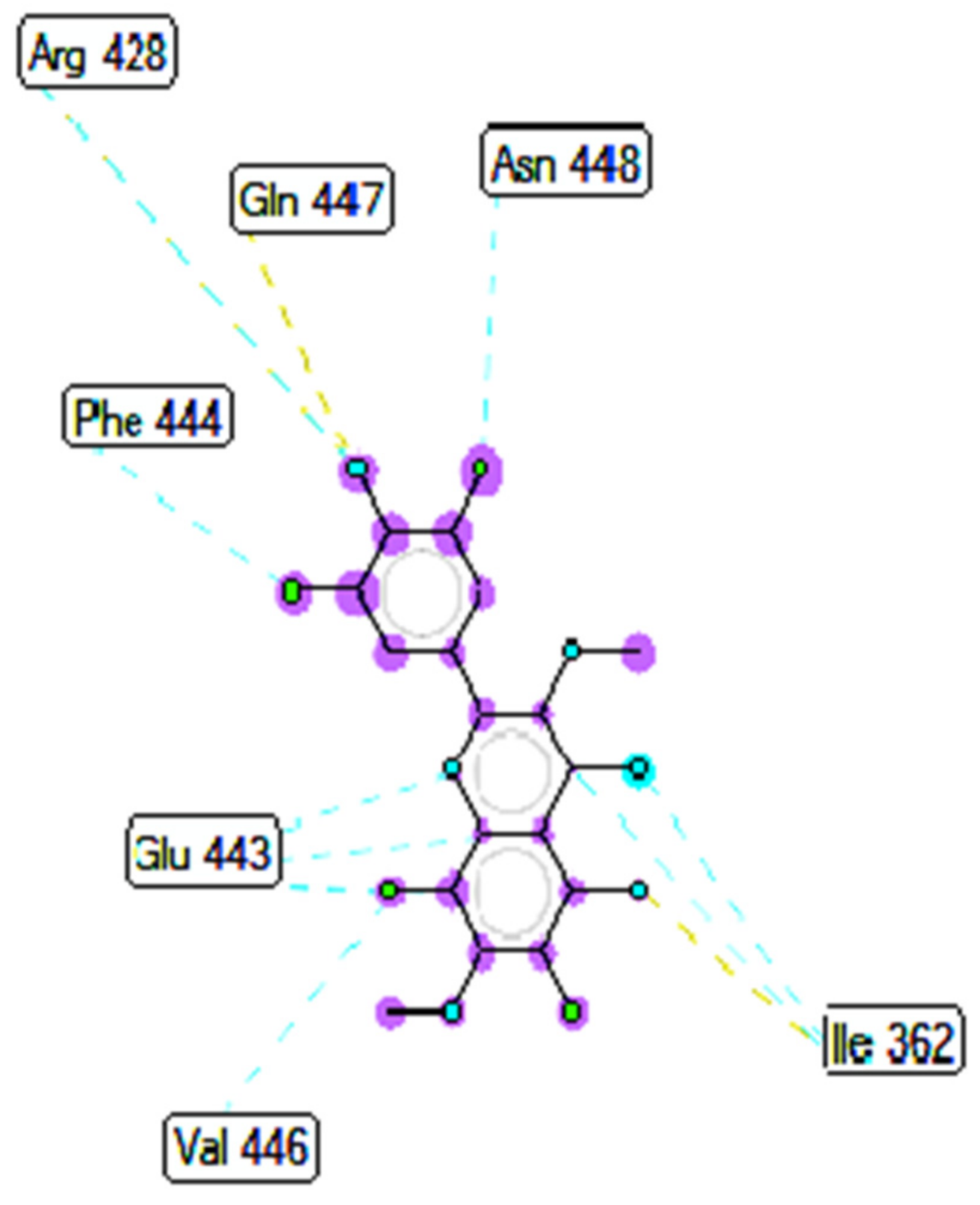
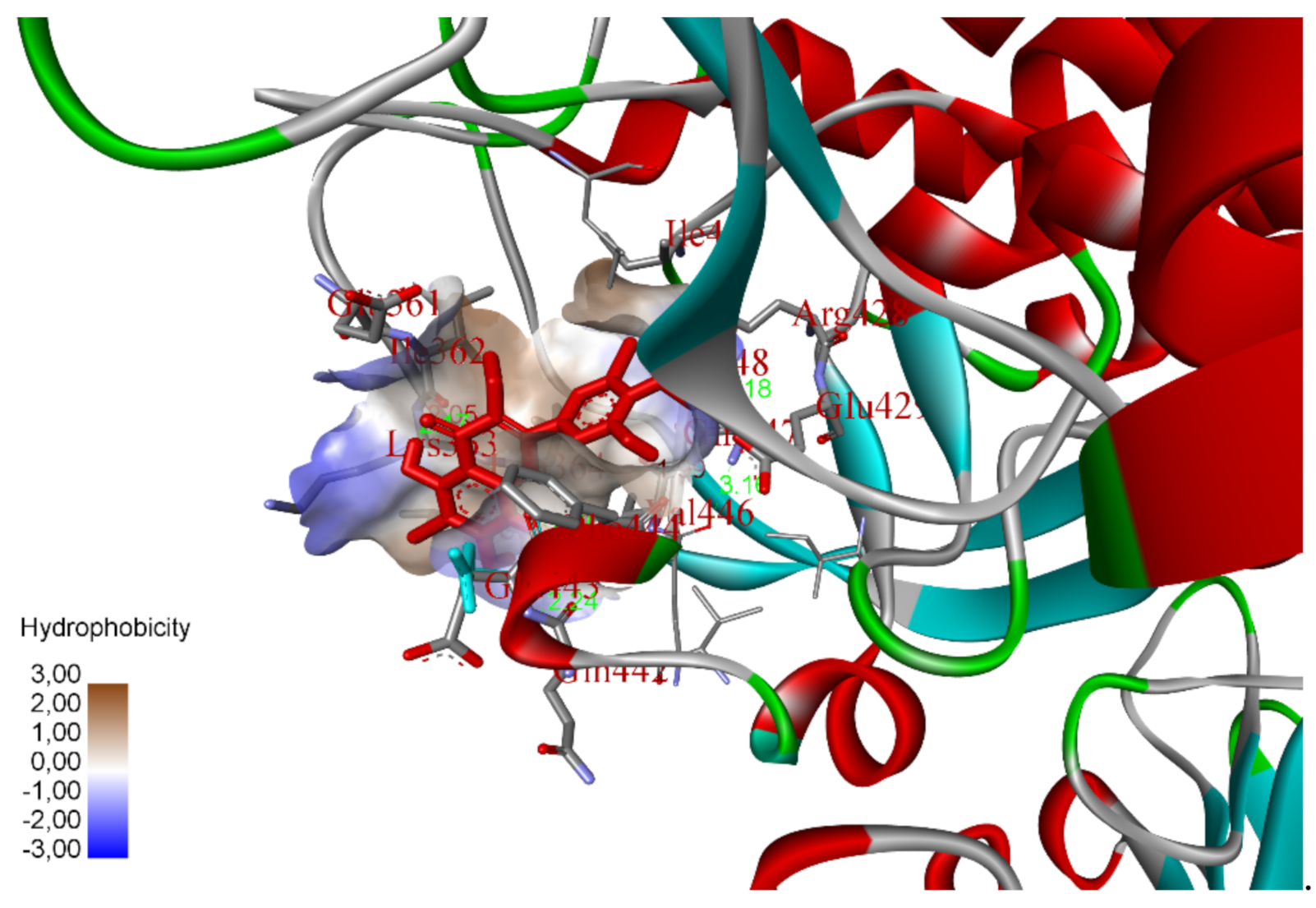
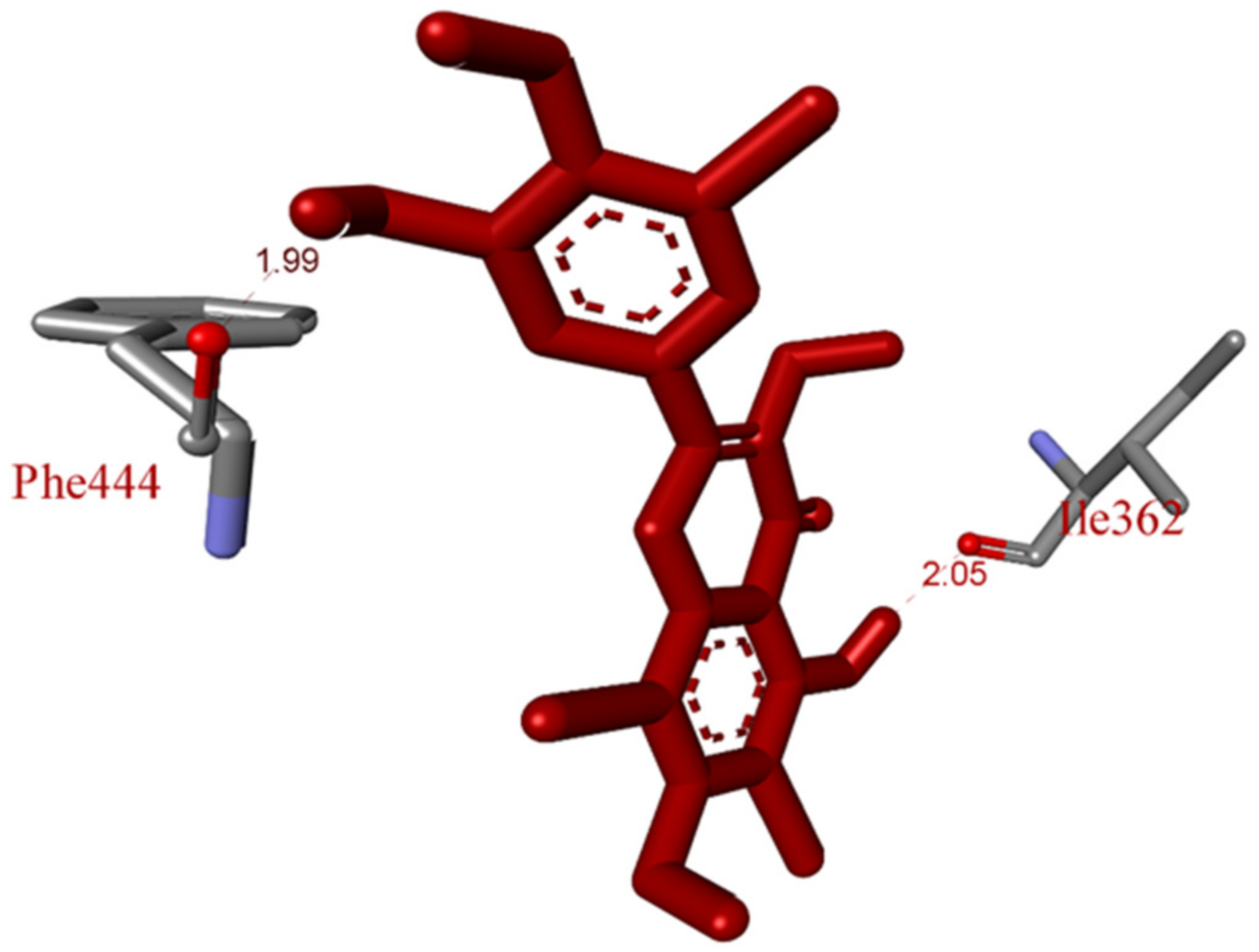
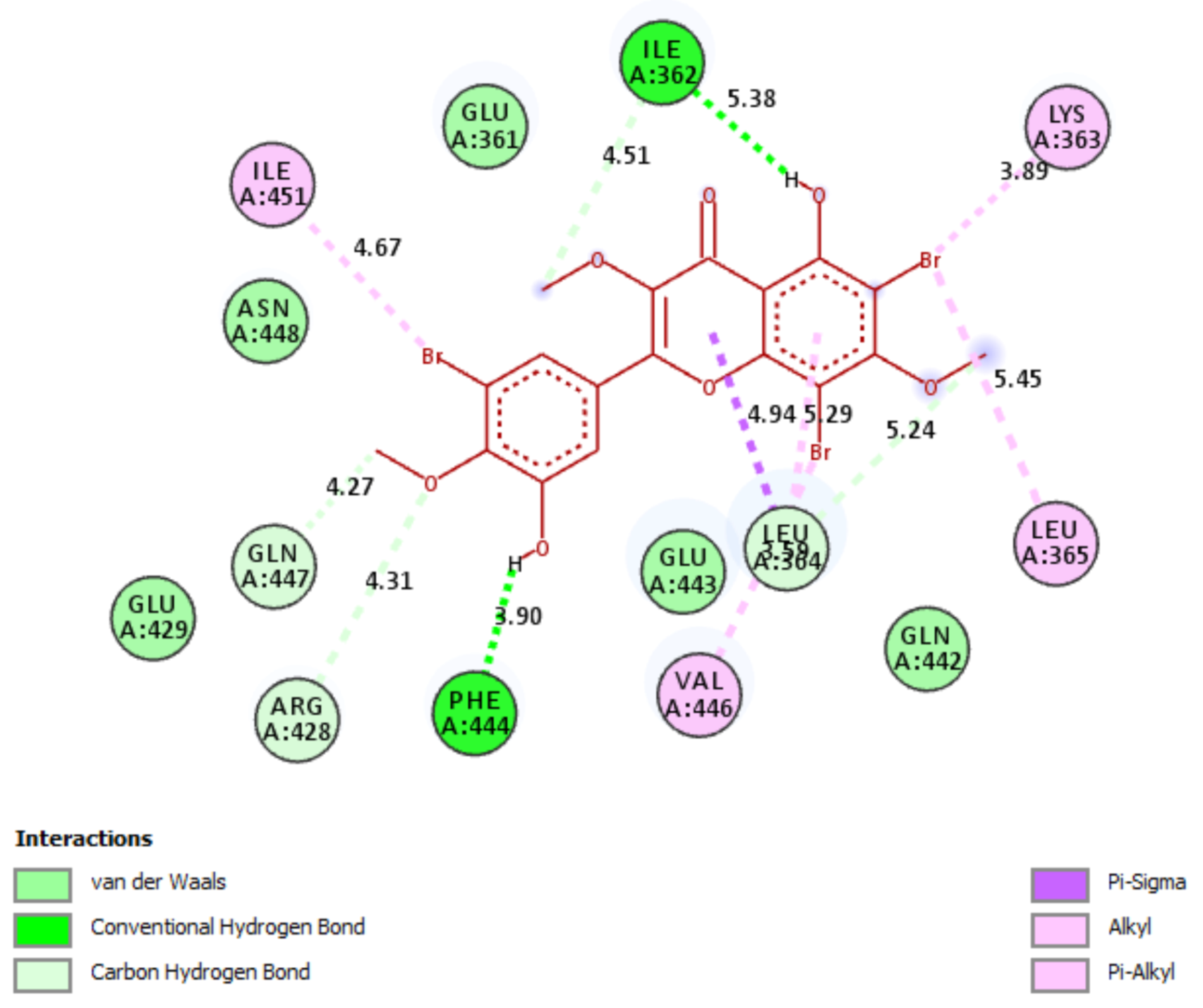
2.5. In Silico Molecular Docking Model
3. Materials and Methods
3.1. Source of the Plant Material
3.2. Isolation and Structure Elucidation of the Compounds
3.3. Isolation
3.4. General Procedure to Synthesize Compounds 8–11
3.4.1. 6,8,5′-Tribromoayanin (8)
3.4.2. 8-Bromokamatakenin (9)
3.4.3. 6-Bromokamatakenin (10)
3.4.4. 6,8,3′,5′-Tetrabromokamatakenin (11)
3.5. Alpha-Glucosidase Inhibition Assay
3.6. Inhibitory Type Assay of 8 and 11 on Alpha-Glucosidase
3.7. Antibacterial Activity Assay
3.8. Molecular Docking Study Method
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Adnyana, I.K.; Tezuka, Y.; Banskota, A.H.; Tran, K.Q.; Kadota, S. Hepatoprotective Constituents of the Seeds of Combretum quadrangulare. Biol. Pharm. Bull. 2000, 23, 1328–1332. [Google Scholar] [CrossRef] [PubMed]
- Banskota, A.; Tezuka, Y.; Tran, Q.; Kadota, S. Chemical Constituents and Biological Activities of Vietnamese Medicinal Plants. Curr. Top. Med. Chem. 2003, 3, 227–248. [Google Scholar] [CrossRef] [PubMed]
- Ganzera, M.; Ellmerer-Müller, E.P.; Stuppner, H. Cycloartane triterpenes from Combretum quadrangulare. Phytochemistry 1998, 49, 835–838. [Google Scholar] [CrossRef]
- Banskota, A.H.; Tezuka, Y.; Phung, L.K.; Tran, K.Q.; Saiki, I.; Miwa, Y.; Taga, T.; Kadota, S. Cytotoxic cycloartane-type triterpenes from Combretum quadrangulare. Bioorg. Med. Chem. Lett. 1998, 8, 3519–3524. [Google Scholar] [CrossRef]
- Banskota, A.H.; Tezuka, Y.; Tran, K.Q.; Tanaka, K.; Saiki, I.; Kadota, S. Thirteen novel cycloartane-type triterpenes from Combretum quadrangulare. J. Nat. Prod. 1999, 63, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Banskota, A.H.; Tezuka, Y.; Tran, K.Q.; Tanaka, K.; Saiki, I.; Kadota, S. Methyl Quadrangularates A-D and Related Triterpenes from Combretum quadrangulare. Chem. Pharm. Bull. 2000, 48, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Adnyana, I.K.; Tezuka, Y.; Awale, S.; Banskota, A.H.; Tran, K.Q.; Kadota, S. Quadranosides VI-XI, Six New Triterpene Glucosides from the Seeds of Combretum quadrangulare. Chem. Pharm. Bull. 2000, 48, 1114–1120. [Google Scholar] [CrossRef] [PubMed]
- Adnyana, I.K.; Tezuka, Y.; Banskota, A.H.; Tran, K.Q.; Kadota, S. Three new triterpenes from the seeds of Combretum quadrangulare and their hepatoprotective activity. J. Nat. Prod. 2001, 64, 360–363. [Google Scholar] [CrossRef]
- Banskota, A.H.; Tezuka, Y.; Adnyana, I.K.; Xiong, Q.; Hase, K.; Tran, K.Q.; Tanak, K.; Saiki, I.; Kadota, S. Hepatoprotective Effect of Combretum quadrangulare and Its Constituents. Biol. Pharm. Bull. 2000, 23, 456–460. [Google Scholar] [CrossRef]
- Toume, K.; Nakazawa, T.; Ohtsuki, T.; Arai, M.A.; Koyano, T.; Kowithayakorn, T.; Ishibashi, M. Cycloartane triterpenes isolated from Combretum quadrangulare in a screening program for death-receptor expression enhancing activity. J. Nat. Prod. 2011, 74, 249–255. [Google Scholar] [CrossRef]
- Duong, T.-H.; Bui, X.-H.; Le Pogam, P.; Nguyen, H.-H.; Tran, T.-T.; Nguyen, T.-A.-T.; Chavasiri, W.; Boustie, J.; Nguyen, K.-P.-P. Two novel diterpenes from the roots of Phyllanthus acidus (L.) Skeel. Tetrahedron 2017, 73, 5634–5638. [Google Scholar] [CrossRef]
- Duong, T.-H.; Beniddir, M.A.; Nguyen, V.-K.; Aree, T.; Gallard, J.-F.; Mac, D.-H.; Nguyen, H.-H.; Bui, X.-H.; Boustie, J.; Nguyen, K.-P.-P.; et al. Sulfonic Acid-Containing Flavonoids from the Roots of Phyllanthus acidus. J. Nat. Prod. 2018, 81, 2026–2031. [Google Scholar] [CrossRef]
- Nguyen, T.-A.-T.; Duong, T.-H.; Le Pogam, P.; Beniddir, M.A.; Nguyen, H.-H.; Do, T.-M.-L.; Nguyen, K.-P.-P. Two new triterpenoids from the roots of Phyllanthus emblica. Fitoterapia 2018, 130, 140–144. [Google Scholar] [CrossRef]
- Macedo, I.; Da Silva, J.H.; Da Silva, P.T.; Cruz, B.G.; Vale, J.P.D.; Dos Santos, H.S.; Bandeira, P.N.; De Souza, E.B.; Xavier, M.R.; Coutinho, H.D.; et al. Structural and Microbiological Characterization of 5-Hydroxy-3,7,4′-Trimethoxyflavone: A Flavonoid Isolated from Vitex gardneriana Schauer Leaves. Microb. Drug Resist. 2019, 25, 434–438. [Google Scholar] [CrossRef]
- Rahman, A.; Mahardika, I.; Saputri, R.D.; Tjahjandarie, T.S.; Tanjung, M. Isolasi, karakterisasi, dan uji aktivitas antioksidan senyawa turunan kuersetin dari kulit batang melicope quercifolia. J. Sains Dan Kesehat. 2020, 2, 413–417. [Google Scholar]
- A Castillo, Q.; Triana, J.; Eiroa, J.L.; Padrón, J.M.; Plata, G.B.; Abel-Santos, E.V.; Báez, L.A.; Rodríguez, D.C.; A Jiménez, M.; Pérez-Pujols, M.F. Flavonoids from Eupatorium illitum and Their Antiproliferative Activities. Pharmacogn. J. 2015, 7, 178–181. [Google Scholar] [CrossRef]
- Tu, Y.-C.; Lian, T.-W.; Yen, J.-H.; Chen, Z.-T.; Wu, M.-J. Antiatherogenic Effects of Kaempferol and Rhamnocitrin. J. Agric. Food Chem. 2007, 55, 9969–9976. [Google Scholar] [CrossRef]
- Salem, M.M.; Hussein, S.R.; El-Sharawy, R.; El-Khateeb, A.; Ragab, E.A.; Dawood, K.M.; El Negoumy, S.I.M. Antioxidant and antiviral activities of the aqueous alcoholic leaf extract of Boscia angustifolia A. Rich. (Capparaceae) and its major component ‘ombuin’. Egypt. Pharm. J. 2016, 15, 1. [Google Scholar]
- Kranjc, E.; Albreht, A.; Vovk, I.; Makuc, D.; Plavec, J. Non-targeted chromatographic analyses of cuticular wax flavonoids from Physalis alkekengi L. J. Chromatogr. A 2016, 1437, 95–106. [Google Scholar] [CrossRef]
- Zheng, J.; Song, M.; Dong, P.; Qiu, P.; Guo, S.; Zhong, Z.; Li, S.; Ho, C.-T.; Xiao, H. Identification of novel bioactive metabolites of 5-demethylnobiletin in mice. Mol. Nutr. Food Res. 2013, 57, 1999–2007. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, L.A.L.; Faqueti, L.G.; Reginatto, F.H.; Dos Santos, A.D.C.; Barison, A.; Biavatti, M.W. Phytochemical analysis of Vernonanthura tweedieana and a validated UPLC-PDA method for the quantification of eriodictyol. Rev. Bras. Farm. 2015, 25, 375–381. [Google Scholar] [CrossRef]
- Arriffin, N.M.; Jamil, S.; Basar, N.; Khamis, S.; Abdullah, S.A.; Lathiff, S.M.A. Phytochemical studies and antioxidant activities of Artocarpus scortechinii King. Rec. Nat. Prod 2017, 11, 299–303. [Google Scholar]
- Oladimeji, A.O.; Oladosu, I.A.; Ali, M.S.; Ahmed, Z. Flavonoids from the roots of Dioclea reflexa (Hook F.). Bull. Chem. Soc. Ethiop. 2015, 29, 441. [Google Scholar] [CrossRef]
- Peng, X.; Zheng, Z.; Cheng, K.-W.; Shan, F.; Ren, G.-X.; Chen, F.; Wang, M. Inhibitory effect of mung bean extract and its constituents vitexin and isovitexin on the formation of advanced glycation endproducts. Food Chem. 2008, 106, 475–481. [Google Scholar] [CrossRef]
- Peng, J.; Fan, G.; Wu, Y.; Chen, H. Isalation and separation isoorientin and isovitexin from Patrinia villosa juss by high speed counter-current chromatography. Fenxi Huaxue 2005, 33, 1389–1392. [Google Scholar]
- Narender, N.; Srinivasu, P.; Kulkarni, S.J.; Raghavan, K. V Liquid phase regioselective bromination of aromatic com-pounds over HZSM-5 catalyst. Synth. Commun. 2000, 30, 3669–3675. [Google Scholar] [CrossRef]
- Devi, A.P.; Duong, T.-H.; Ferron, S.; Beniddir, M.A.; Dinh, M.-H.; Nguyen, V.-K.; Pham, N.-K.-T.; Mac, D.-H.; Boustie, J.; Chavasiri, W.; et al. Salazinic Acid-Derived Depsidones and Diphenylethers with α-Glucosidase Inhibitory Activity from the Lichen Parmotrema dilatatum. Planta Medica 2020, 86, 1216–1224. [Google Scholar] [CrossRef]
- Tran, C.-L.; Dao, T.-B.-N.; Tran, T.-N.; Mai, D.-T.; Tran, T.-M.-D.; Tran, N.-M.-A.; Dang, V.-S.; Vo, T.-X.; Duong, T.-H.; Sichaem, J. Alpha-Glucosidase Inhibitory Diterpenes from Euphorbia antiquorum Growing in Vietnam. Molecules 2021, 26, 2257. [Google Scholar] [CrossRef]
- Prastiyanto, M.E.; Tama, P.D.; Ananda, N.; Wilson, W.; Mukaromah, A.H. Antibacterial Potential of Jatropha sp. Latex against Multidrug-Resistant Bacteria. Int. J. Microbiol. 2020, 2020, 1–6. [Google Scholar] [CrossRef]
- Duong, T.-H.; Devi, A.P.; Tran, N.-M.-A.; Phan, H.-V.-T.; Huynh, N.-V.; Sichaem, J.; Tran, H.-D.; Alam, M.; Nguyen, T.-P.; Nguyen, H.-H.; et al. Synthesis, α-glucosidase inhibition, and molecular docking studies of novel N-substituted hydrazide derivatives of atranorin as antidiabetic agents. Bioorg. Med. Chem. Lett. 2020, 30, 127359. [Google Scholar] [CrossRef]

| Compound | R1 | R2 | R3 | R4 | R5 | R6 | R7 |
|---|---|---|---|---|---|---|---|
| (1) 5-Hydroxy-3,7,4′-trimethoxyflavone | OMe | H | H | H | OMe | H | Me |
| (2) Ayanin | OMe | H | H | OH | OMe | H | Me |
| (3) Kuatakenin | OMe | H | H | H | OH | H | Me |
| (4) Rhamnocitrin | OH | H | H | H | OH | H | Me |
| (5) Ombuin | OH | H | H | OH | OMe | H | Me |
| (6) Myricetin-3,7,3′,5′-tetramethyl ether | OH | H | H | OMe | OH | OMe | Me |
| (7) Gardenin D | H | OMe | OMe | OH | OMe | H | Me |
| (8) 6,8,5′-Tribromoayanin | OMe | Br | Br | OH | OMe | Br | Me |
| (9) 8-Bromokumatakenin | OMe | H | Br | H | OH | H | Me |
| (10) 6-Bromokumatakenin | OMe | Br | H | H | OH | H | Me |
| (11) 6,8,3′,5′-Tetrabromokumatakenin | OMe | Br | Br | Br | OH | Br | Me |
| (12) Luteolin | H | H | H | OH | OH | H | H |
| (13) Apigenin | H | H | H | H | OH | H | H |
| (14) Mearnsetin | OH | H | H | OH | OMe | OH | H |
| (15) Vitexin | H | H | Glc | H | OH | H | H |
| (16) Isoorientin | H | Glc | H | OH | OH | H | H |

| Compounds | IC50 µM |
|---|---|
| 1 | >300 |
| 2 | 245.3 ± 3.7 |
| 3 | >300 |
| 4 | 206.7 ± 2.6 |
| 5 | >300 |
| 6 | 282.0 ± 2.9 |
| 7 | 188.0 ± 3.1 |
| 8 | 87.1 ± 2.3 |
| 9 | 275.0 ± 4.5 |
| 10 | >300 |
| 11 | 30.5 ± 1.9 |
| 12 | 97.7 ± 1.7 |
| 13 | 130.8 ± 3.9 |
| 14 | 205.3 ± 5.6 |
| 15 | >300 |
| 16 | >300 |
| Acarbose (positive control) | 332.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dao, T.-B.-N.; Nguyen, T.-M.-T.; Nguyen, V.-Q.; Tran, T.-M.-D.; Tran, N.-M.-A.; Nguyen, C.H.; Nguyen, T.-H.-T.; Nguyen, H.-H.; Sichaem, J.; Tran, C.-L.; et al. Flavones from Combretum quadrangulare Growing in Vietnam and Their Alpha-Glucosidase Inhibitory Activity. Molecules 2021, 26, 2531. https://doi.org/10.3390/molecules26092531
Dao T-B-N, Nguyen T-M-T, Nguyen V-Q, Tran T-M-D, Tran N-M-A, Nguyen CH, Nguyen T-H-T, Nguyen H-H, Sichaem J, Tran C-L, et al. Flavones from Combretum quadrangulare Growing in Vietnam and Their Alpha-Glucosidase Inhibitory Activity. Molecules. 2021; 26(9):2531. https://doi.org/10.3390/molecules26092531
Chicago/Turabian StyleDao, Thi-Bich-Ngoc, Truong-Minh-Tri Nguyen, Van-Quy Nguyen, Thi-Minh-Dinh Tran, Nguyen-Minh-An Tran, Chuong Hoang Nguyen, Thi-Hoai-Thu Nguyen, Huu-Hung Nguyen, Jirapast Sichaem, Cong-Luan Tran, and et al. 2021. "Flavones from Combretum quadrangulare Growing in Vietnam and Their Alpha-Glucosidase Inhibitory Activity" Molecules 26, no. 9: 2531. https://doi.org/10.3390/molecules26092531
APA StyleDao, T.-B.-N., Nguyen, T.-M.-T., Nguyen, V.-Q., Tran, T.-M.-D., Tran, N.-M.-A., Nguyen, C. H., Nguyen, T.-H.-T., Nguyen, H.-H., Sichaem, J., Tran, C.-L., & Duong, T.-H. (2021). Flavones from Combretum quadrangulare Growing in Vietnam and Their Alpha-Glucosidase Inhibitory Activity. Molecules, 26(9), 2531. https://doi.org/10.3390/molecules26092531






