Production of Biodegradable Polymer from Agro-Wastes in Alcaligenes sp. and Pseudomonas sp.
Abstract
1. Introduction
2. Results
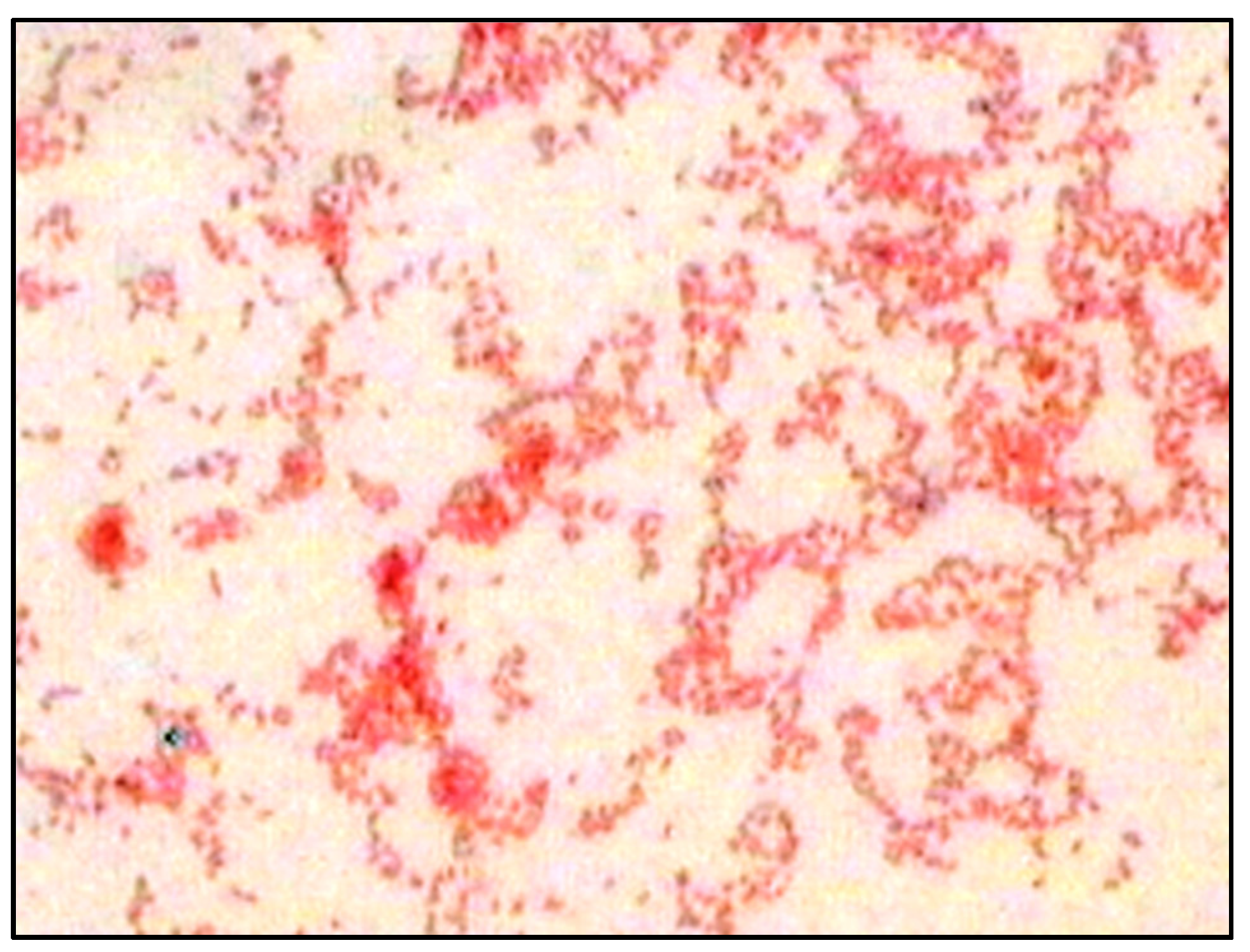
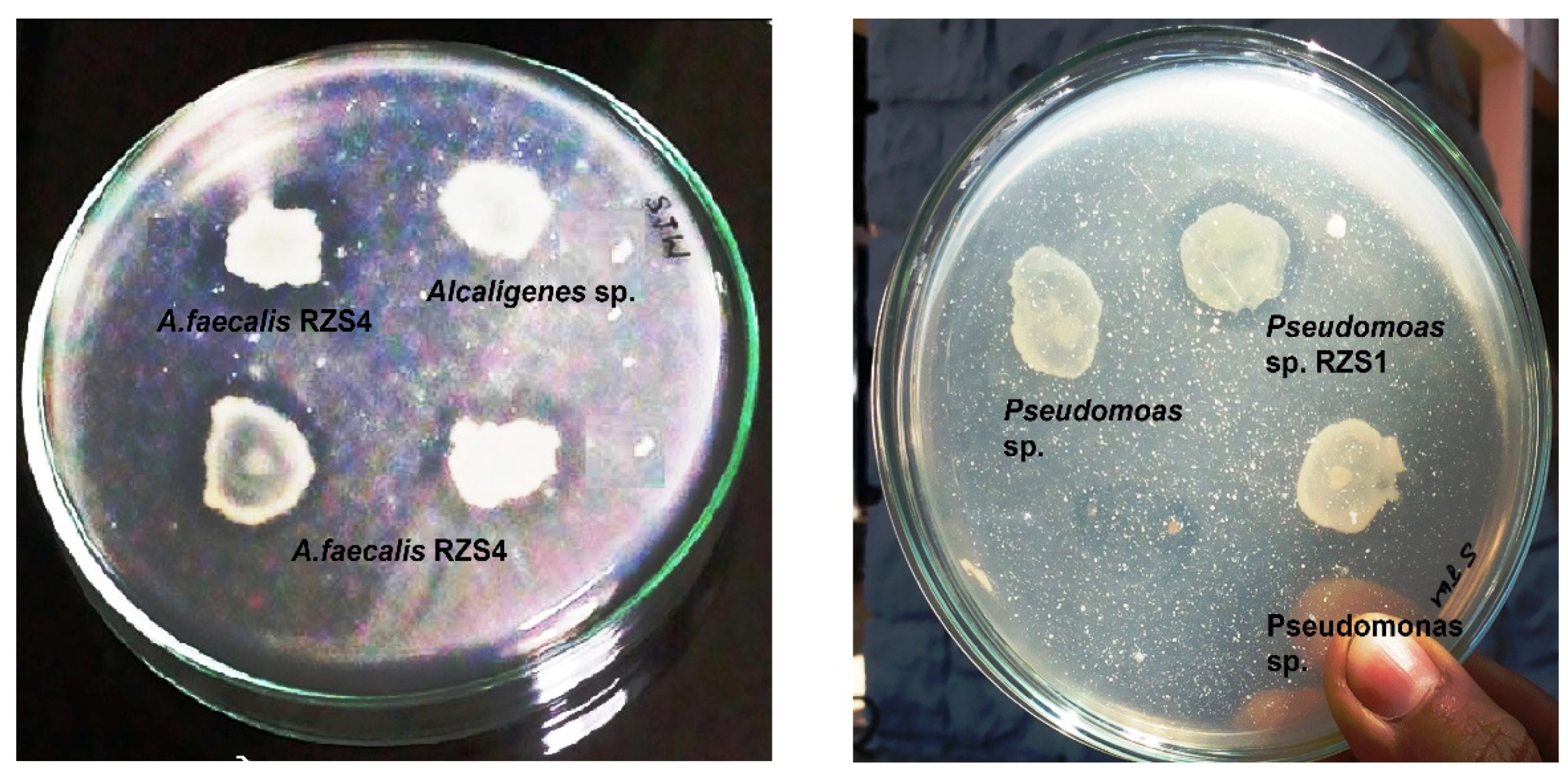
2.1. Screening, Detection, Production, and Estimation of PHB
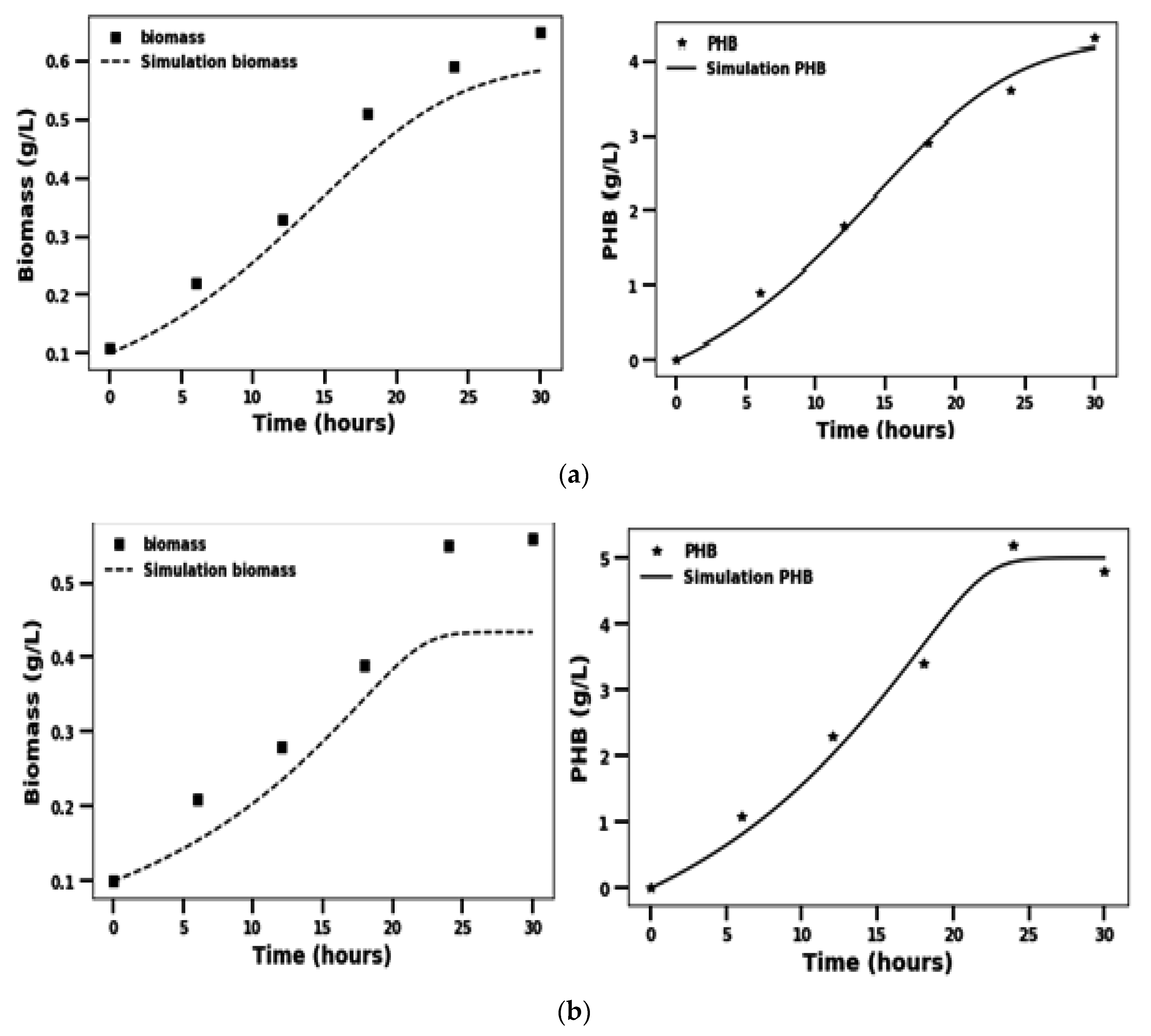
2.2. Growth and PHB Accumulation as a Function of Time
2.3. Kinetics of Biomass Growth and PHB Formation
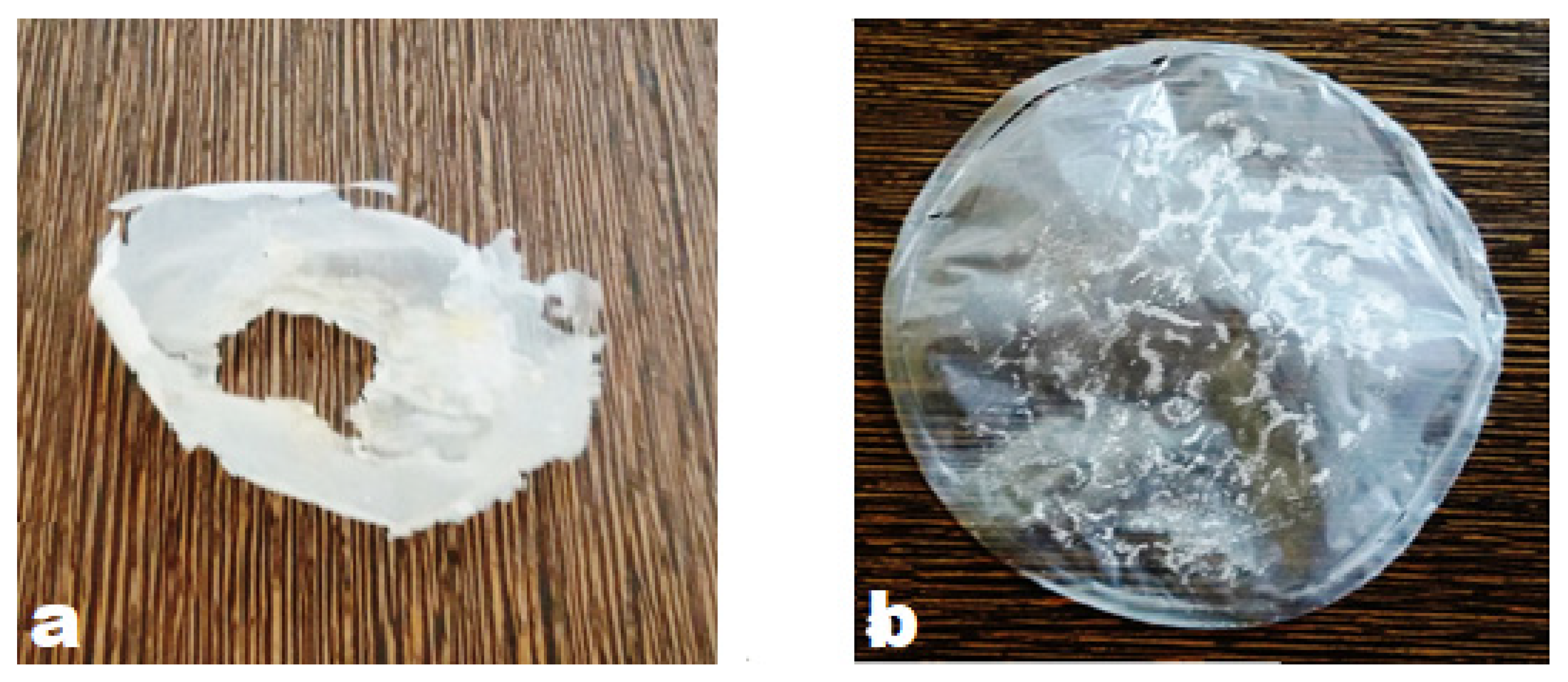
2.4. Extraction and Estimation of PHB and Cell Mass
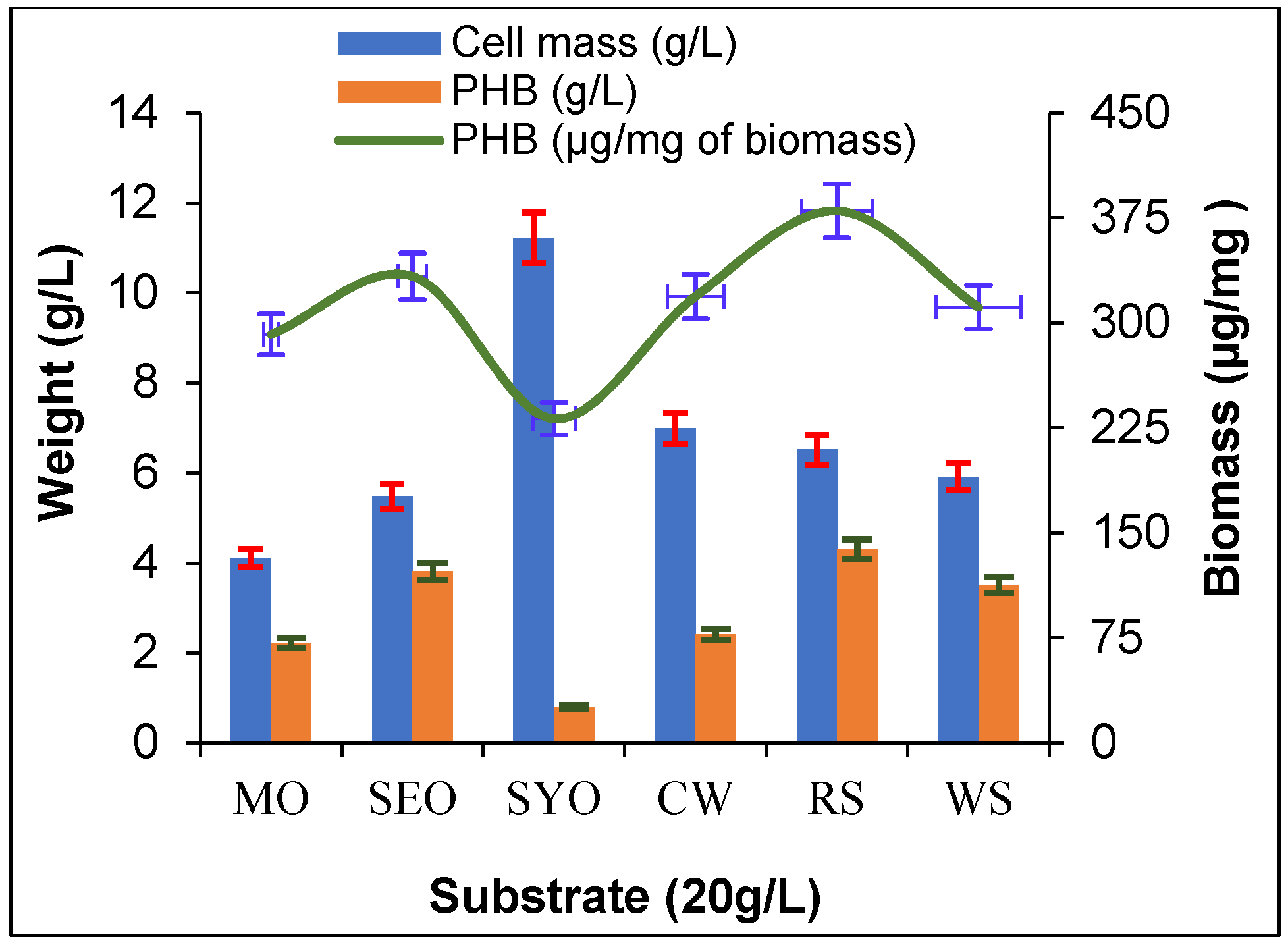
2.5. Production of PHB from Agro-Waste
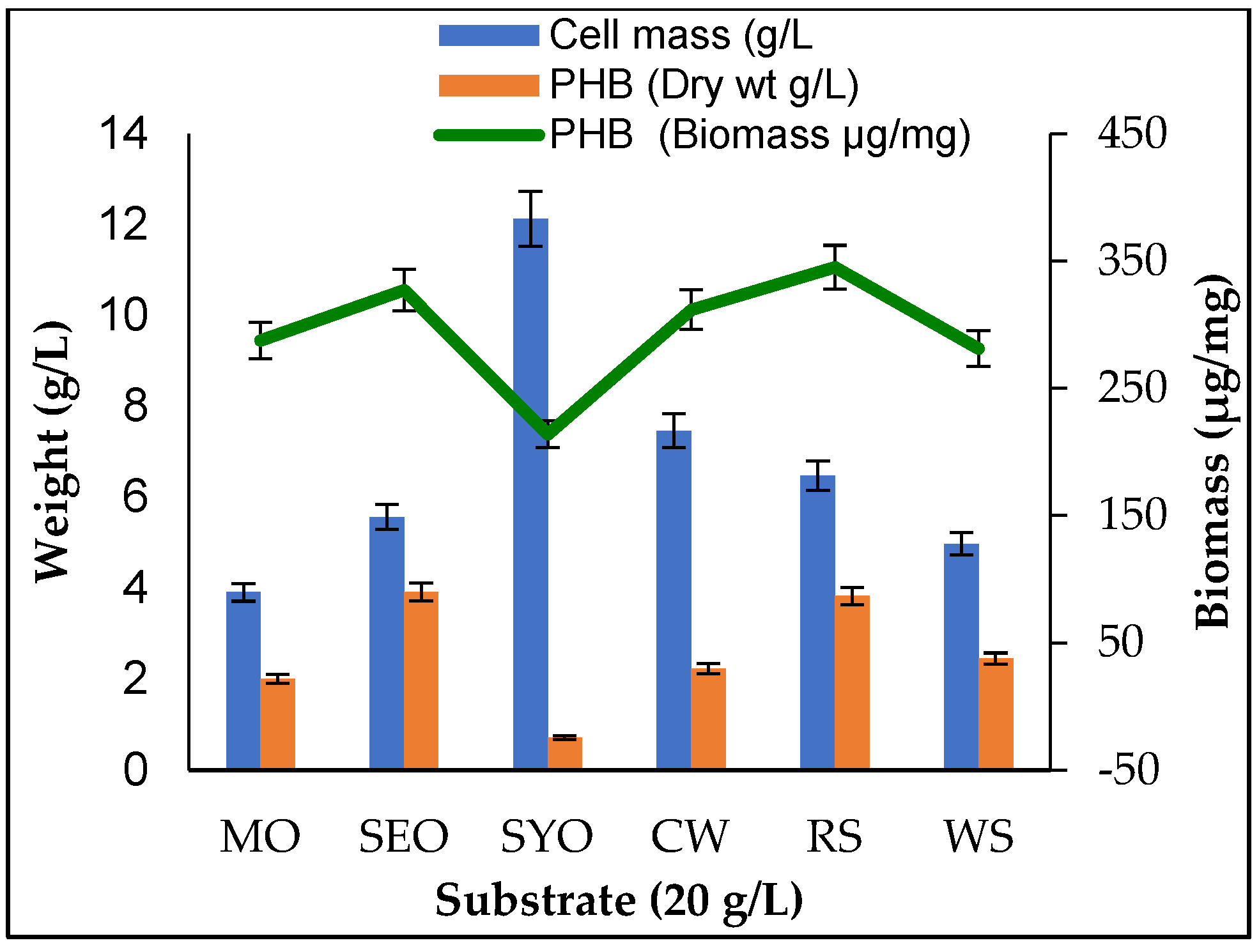
2.6. Production of PHB from Crude Vegetable Oils
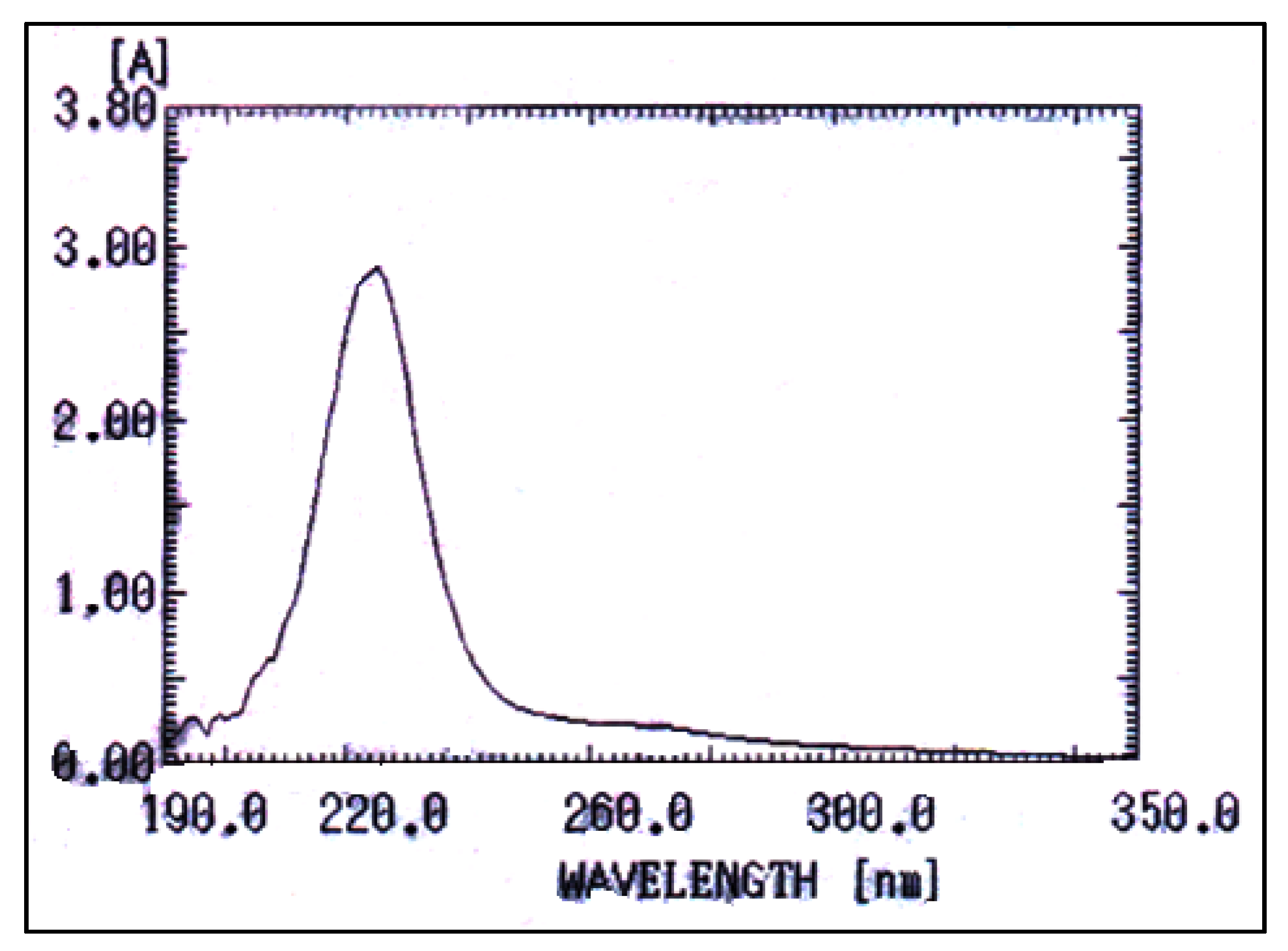
2.7. Characterization of PHB
UV–Visible Spectrophotometry
2.8. Assessment of Biodegradation of PHB
2.9. Assay of PHB Depolymerase Enzyme
3. Discussion
3.1. Screening and Detection of PHB Production
3.2. Growth and PHB Accumulation as a Function of Time
3.3. Kinetics of Biomass Growth and PHB Formation
3.4. Extraction and Estimation of PHB and Cell Mass
3.5. Production of PHB from Agro-Waste and Vegetable Oils
3.6. Characterization of PHB
3.7. Assessment of Biodegradation of PHB
3.8. Assay of PHB Depolymerase Enzyme
4. Materials and Methods
4.1. Source of Culture
4.2. Culture Media and Growth Conditions
4.3. Source of Agro-Wastes and Vegetable Oils
4.4. Screening for PHB Production
4.5. Detection of PHB by Sudan Black B Staining
4.6. Growth and PHB Accumulation as a Function of Time
4.7. Kinetic Study of Growth and PHB Formation
4.8. Extraction and Estimation of PHB and Measurement of Growth
4.9. Production of PHB from Agro-Waste
4.10. Production of PHB from Crude Vegetable Oils
4.11. Characterization of PHB Extracts
UV–Vis Spectrophotometry
4.12. Assessment of Biodegradation of PHB
4.12.1. Plate Assay
4.12.2. Shake Flask Method
4.12.3. Assay of PHB Depolymerase Enzyme
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Blanco, I. Lifetime Prediction of Polymers: To Bet, or Not to Bet—Is This the Question? Materials 2018, 11, 1383. [Google Scholar] [CrossRef]
- D’Anna, A.; Arrigo, R.; Frache, A. PLA/PHB Blends: Biocompatibilizer Effects. Polymers 2019, 11, 1416. [Google Scholar] [CrossRef] [PubMed]
- Siracusa, V.; Blanco, I. Bio-Polyethylene (Bio-PE), Bio-Polypropylene (Bio-PP) and Bio-Poly(ethylene terephthalate) (Bio-PET): Recent Developments in Bio-Based Polymers Analogous to Petroleum-Derived Ones for Packaging and Engineering Applications. Polymers 2020, 12, 1641. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.J.; Sayyed, R.Z. Production, efficient recovery and partial characterization of biodegradable polymer produced by soil Streptomyces. Indian J. Biotechnol. 2016, 15, 127–129. [Google Scholar]
- Sayyed, R.Z.; Wani, S.J.; Alyousef, A.A.; Alqasim, A.; Syed, A.; El-Enshasy, H.A. Purification and kinetics of the PHB depolymerase of Microbacterium paraoxydans RZS6 isolated from a dumping yard. PLoS ONE 2019, 14, e0212324. [Google Scholar] [CrossRef]
- Gangurde, N.S.; Sayyed, R.Z.; Gulati, A.; Kiran, S. Development of eco-friendly bioplastic like PHB by distillery effluent microorganisms. Environ. Sci. Poll. Res. 2013, 20, 488–497. [Google Scholar] [CrossRef]
- Sayyed, R.Z.; Wani, S.J.; Alarfaj, A.A.; Syed, A.; El-Enshasy, H.A. Production, purification, and evaluation of biodegradation potential of PHB depolymerase of Stenotrophomonas sp. RZS7. PLoS ONE 2019, 15, e0220095. [Google Scholar] [CrossRef]
- Obruca, S.; Petrik, S.; Benesova, P.; Svoboda, Z.; Eremka, L.; Marova, I. Utilization of oil extracted from spent coffee grounds for sustainable production of polyhydroxyalkanoates. Appl. Microbiol. Biotechnol. 2014, 98, 5883–5890. [Google Scholar] [CrossRef]
- Taniguchi, I.; Kagotani, K.; Kimura, Y. Microbial production of poly (hydroxyl alkanoates)s from waste edible oils. Green Chem. 2003, 5, 545–548. [Google Scholar] [CrossRef]
- Chaudhary, W.; Jamil, N.; Ali, I.; Ayaz, M.; Hasnain, S. Screening for polyhydroxyalkanoate (PHA)-producing bacterial strains and comparison of PHA production from various inexpensive carbon sources. Ann. Microbiol. 2011, 61, 623–629. [Google Scholar] [CrossRef]
- Riedel, S.L.; Bader, J.; Brigham, C.J.; Budde, C.F.; Yusof, Z.A.M.; Rha, C.; Sinskey, A.J. Production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) by Ralstonia eutropha in high cell density palm oil fermentation. Biotechnol. Bioeng. 1987, 109, 74–83. [Google Scholar] [CrossRef]
- Razaif-Mazinah, M.R.M.; Anis, S.N.S.; Harun, H.I.; Rashid, K.A.; Annuar, M.S.M. Unusual poly(3-hydroxyalkanoate) (PHA) biosynthesis behavior of Pseudomonas putida Bet001 and Delftiat suruhatensis Bet002 isolated from palm oil mill effluent. Biotechnol. Appl. Biochem. 2017, 64, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Ben, M.; Kennes, C.; Veiga, M.C. Optimization of polyhydroxyalkanoate storage using mixed cultures and brewery wastewater. J. Chem. Technol. Biotechnol. 2016, 91, 2817–2826. [Google Scholar] [CrossRef]
- Pakalapati, H.; Chang, C.; Show. P., K.; Arumugasamy, S.K.; Lan, J.C. Development of polyhydroxyalkanoates production from waste feed-stocks and applications. J. Biosci. Bioeng. 2018, 126, 282–292. [Google Scholar] [CrossRef]
- Sayyed, R.Z.; Gangurde, N.S.; Chincholkar, S.B. Hypochlorite digestion method for efficient recovery of PHB from Alcaligenes faecalis. Indian J. Microbiol. 2009, 49, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Sayyed, R.Z.; Chincholkar, S.B. Production of poly -β-hydroxy butyrate from Alcaligenes faecalis. Indian J. Microbiol. 2004, 44, 269–272. [Google Scholar]
- Maier, R.M.; Pepper, I.L. Bacterial growth. Environ. Microbiol. 2015, 37–56. [Google Scholar] [CrossRef]
- Panikov, N.S. Kinetics, microbial growth. Encyclopedia of Industrial Biotechnology: Bioprocess. Biosep. Cell Technol. 2009, 1–34. [Google Scholar] [CrossRef]
- Wang, N.S. Experiment No. 9C Measurements of Cell Biomass Concentration; Department of Chemical & Bio-molecular Engineering University of Maryland College Park: College Park, MD, USA, 2009; pp. 2074–2111. [Google Scholar]
- Dong, Y.; Yan, X.; Lu, F.; Guo, M.; Zhuang, Y. Development and Optimization of an Unstructured Kinetic Model for Sodium Gluconate Fermentation Process, Comput. Mater. Continua 2015, 48, 43–55. [Google Scholar] [CrossRef]
- Monod, J. The growth of bacterial cultures. Annu. Rev. Microbiol. 1949, 3, 371–394. [Google Scholar] [CrossRef]
- Alqahtani, R.T. Modelling of Biological Wastewater Treatment. Ph.D. Thesis, School of Mathematics and Applied Mathematics, University of Wollongong, Singapore, 2013. [Google Scholar]
- Gharibzahedi, S.M.T.; Razavi, S.H.; Mousavi, M. Kinetic analysis and mathematical modeling of cell growth and canthaxanthin biosynthesis by Dietzia natronolimnaea HS-1 on waste molasses hydrolysate. R. Soc. Chem. 2013, 3, 23495–23502. [Google Scholar] [CrossRef]
- Sharifahmadian, A. Numerical Models for Submerged Breakwaters: Coastal Hydrodynamics and Morphodynamics; Butterworth-Heinemann: Oxford, UK, 2015. [Google Scholar]
- Juan, M.L.; Gonzalez, L.W.; Walker, G.C. A novel screening method for isolating exo-polysaccharide deficient mutants. Appl. Environ. Microbiol. 1998, 64, 4600–4602. [Google Scholar]
- Mohamed, T.; Shaaban, M.A.; Azza, S.T.; Eman, I.M. Production of some biopolymers by some selective Egyptian soil bacterial isolates. J. Appl. Sci. Res. 2012, 8, 94–105. [Google Scholar]
- Phanse, N.; Chincholikar, A.; Patel, B.; Rathore, P.; Vyas. P.; Patel, M. Screening of PHA (polyhydroxyalkanoate) producing bacteria from diverse sources. Intl. J. Biosci. 2011, 1, 27–32. [Google Scholar]
- Wani, S.J.; Shaikh, S.S.; Tabassum, B.; Thakur, A.; Gulati, A. Stenotrophomonas sp. RZS7. A novel PHB degrader isolated from plastic contaminated soil in Shahada, Maharashtra, Western India. 3 Biotechnol. 2017, 6, 179. [Google Scholar]
- Matavulj, M.; Molitoris, H.P. Fungal degradation of polyhydroxyalkanoates and a semiquantitative assay for screening their degradation by terrestrial fungi. FEMS Microbiol. Rev. 1992, 103, 323–331. [Google Scholar] [CrossRef]
- Yüksekdaǧ, N.Z.; Aslim, B.; Beyatli, Y.; Mercan, N. Effect of carbon and nitrogen sources and incubation times on poly-beta-hydroxybutyrate (PHB) synthesis by Bacillus subtilis 25 and Bacillus megaterium 12. Afr. J. Biotechnol. 2004, 3, 63–66. [Google Scholar]
- Singh, G.; Kumari, A.; Mittal, A.; Yadav, A.; Aggarwal, N.K. Poly β-Hydroxybutyrate Production by Bacillus subtilis NG220 Using Sugar Industry Waste Water. BioMed Res. Intl. 2013, 952641. [Google Scholar] [CrossRef]
- Akthiselvan, P.; Madhumathi, R. Kinetic evaluation on cell growth and biosynthesis of polyhydroxybutyrate (PHB) by Bacillus safensis EBT1 from sugarcane bagasse. Eng. Agric. Environ. Food 2018, 11, 145–152. [Google Scholar] [CrossRef]
- Tripathi, A.D.; Srivastava, S.K. Kinetic Study of Biopolymer (PHB) Synthesis in Alcaligenes sp. in submerged fermentation process Using TEM. J. Polym Environ. 2019, 19, 732. [Google Scholar] [CrossRef]
- Yellore, V.; Desai, A. Production of native poly-β-hydroxybutyrate from lactose and whey by Methylobacterium sp. ZP24. Lett. Appl. Microbiol. 1998, 26, 391–394. [Google Scholar] [CrossRef]
- Rawte, T.; Mavinkurve, S. A rapid hypochlorite method for the extraction of polyhydroxyalkonoates from bacterial cells. Indian J. Exp. Biol. 2002, 40, 924–929. [Google Scholar]
- Sayyed, R.Z.; Gangurde, N.S. Poly-β-hydroxybutyrate production by Pseudomonas sp. RZS 1 under the aerobic and semi-aerobic conditions. Indian J. Exp. Biol. 2016, 48, 942–947. [Google Scholar]
- Gangurde, N.S.; Patil, Y.P.; Jain, R.; Sayyed, R.Z. Poly-β-hydroxybutyrate biodegradation by mixed culture population vis-à-vis single culture population under varying environmental conditions: A new approach. Indian J. Exp. Biol. 2017, 55, 311–320. [Google Scholar]
- Hann, S.K.; Chang, Y.K.; Kim, B.S.; Lee, K.M.; Chang, H.N. Optimization of microbial (3-hydroxybutyrate) recovery using dispersion of sodium hypochlorite solution and chloroform. Biotechnol. Bioeng. 1994, 44, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Gowda, V.; Shivakumar, S. Agro-waste-based Polyhydroxyalkanoate (PHA) production using the hydrolytic potential of Bacillus thuringiensis IAM 12077. Braz. Arch. Biol. Technol. 2014, 57, 1. [Google Scholar] [CrossRef]
- Thapa, C.; Shakya, P.; Shrestha, R.; Pal, S.; Manandhar, P. Isolation of Polyhydroxybutyrate (PHB) Producing bacteria, optimization of culture conditions for PHB production, extraction and characterization of PHB. Nepal J. Biotechnol. 2019, 6, 62–68. [Google Scholar] [CrossRef]
- Oliviera, F.C.; Freire, D.M.G.; Castilho, L.R. Production of poly (3- hydroxybutyrate) by solid-state fermentation with Ralstonia eutropha. Biores. Technol. 2004, 25, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Ahn, W.S.; Park, S.J.; Lee, S.Y. Production of poly (3-hydroxybutyrate) from whey by cell recycle fed-batch culture of recombinant Escherichia coli. Biotechnol Lett. 2001, 23, 235–240. [Google Scholar] [CrossRef]
- Kulkarni, S.O.; Kanekar, P.P.; Jog, J.P.; Sarnaik, S.S.; Nilegaonkar, S.S. Production of the copolymer, poly (hydroxybutyrate-co-hydroxy valerate) by Halomonas campisalis MCM B-1027 using agrowastes. Intl. J. Biol. Macromol. 2015, 72, 784–789. [Google Scholar] [CrossRef]
- Getachew, A.; Woldesenbet, F. Production of biodegradable plastic by polyhydroxybutyrate (PHB) accumulating bacteria using low-cost agricultural waste material. BMC Res. Notes 2016, 9, 509. [Google Scholar] [CrossRef] [PubMed]
- Alshehrei, F. Production of Polyhydroxybutyrate (PHB) by Bacteria isolated from soil of Saudi Arabia. J. Pure Appl. Microbiol. 2019, 13, 13–897. [Google Scholar] [CrossRef]
- Arun, A.; Murrugappan, R.M.; Ravindran, A.D.D.; Veeramanikandan, V.; Shanmuga, B. Utilization of various industrial wastes for the production of poly-β-hydroxybutyrate (PHB) by Alcaligenes eutrophus. African J. Biotechnol. 2006, 5, 1524–1527. [Google Scholar]
- Song, J.-H.; Jeon, C.-O.; Choi, M.-H.; Yoon, S.-C.; Park, W.-J. Polyhydroxyalkanoate (PHA) production using waste vegetable oil by Pseudomonas sp. strain DR2. J. Microbiol. Biotechnol. 2008, 18, 1408–1415. [Google Scholar]
- Thakor, N.; Trivedi, U.; Patel, K.C. Biosynthesis of medium chain length Poly(3-hydroxy alkanoates) (mcl-PHAs) by Comamonas testosteroni during cultivation on vegetable oils. Biores. Technol. 2005, 96, 1843–1850. [Google Scholar] [CrossRef]
- Mikkili, I.; Karlapudi, A.P.; Venkateswarulu, T.C.; Nath, S.B.; Kodali, V.P. Isolation, screening and extraction of polyhydroxybutyrate (PHB) producing bacteria from sewage sample. Intl. J. PharmTech Res. 2014, 6, 850–857. [Google Scholar]
- Mergaert, J.; Webb, A.; Anderson, C.; Wouters, A.; Swings, J. Microbial degradation of poly (3-hydroxybutyrate) and poly (3-F hydroxybutyrate-co–3-hydroxybutyrate) in compost, J. Environ. Polym. Degrad. 1993, 2, 177–183. [Google Scholar] [CrossRef]
- Elbanna, K.; Lutke-Eversloh, T.; Jendrossek, D.; Luftmann, H.; Steinbuchel, A. Studies on the biodegradability of polythioester copolymers and homopolymers by polyhydroxyalkanoate (PHA) degrading bacteria and PHA depolymerases. Arch. Microbiol. 2004, 182, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wang, Z.; Chen, S.; Liu, D.; Xia, H. Purification and characterization of extracellular poly-(β-hydroxybutyrate) depolymerase from Penicillium sp. DS9701-D2. Polym. Plast. Technol. Eng. 2008, 48, 58–63. [Google Scholar] [CrossRef]
- Kim, B.S.; Lee, S.C.; Lee, S.Y.; Chang, H.N.; Chang, Y.K.; Woo, S.I. Production of poly (3-hydroxy butyric acid co-3- hyroxyvaleric acid) by fed batch culture of Alcaligenes eutrophus with substrate feeding using online glucose analyzer. Biotechnol. Bioeng. 2014, 43, 892–898. [Google Scholar] [CrossRef]
- Rapske, R.; Rapske, A.C. Quantitative requirements for exponential growth of Alcaligenes eutrophus. Appl. Environ. Microbiol. 1976, 32, 585–591. [Google Scholar] [CrossRef]
- Schlegel, H.G.; Lafferty, R.; Krauss, I. The isolation of mutants not accumulating poly-beta-hydroxybutyric acid, Arch. Microbiol. 1970, 71, 283–294. [Google Scholar]
- Law, J.H.; Slepecky, R.A. Assay of poly-β-hydroxybutyric acid. J. Bacteriol. 1961, 82, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Papaneophytou, C.P.; Pantazaki, A.A.; Kyriakidis, D.A. An extracellular polyhydroxybutyrate depolymerase in Thermus thermophilus hb8. Appl. Microbiol. Biotechnol. 2009, 83, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.E. Continuous distribution: Tests of significance. In Introductory Statistics for Biology, 2nd ed.; Parker, R.E., Ed.; Cambridge University Press: London, UK, 1979; pp. 18–42. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sayyed, R.Z.; Shaikh, S.S.; Wani, S.J.; Rehman, M.T.; Al Ajmi, M.F.; Haque, S.; El Enshasy, H.A. Production of Biodegradable Polymer from Agro-Wastes in Alcaligenes sp. and Pseudomonas sp. Molecules 2021, 26, 2443. https://doi.org/10.3390/molecules26092443
Sayyed RZ, Shaikh SS, Wani SJ, Rehman MT, Al Ajmi MF, Haque S, El Enshasy HA. Production of Biodegradable Polymer from Agro-Wastes in Alcaligenes sp. and Pseudomonas sp. Molecules. 2021; 26(9):2443. https://doi.org/10.3390/molecules26092443
Chicago/Turabian StyleSayyed, R. Z., S. S. Shaikh, S. J. Wani, Md Tabish Rehman, Mohammad F. Al Ajmi, Shafiul Haque, and Hesham Ali El Enshasy. 2021. "Production of Biodegradable Polymer from Agro-Wastes in Alcaligenes sp. and Pseudomonas sp." Molecules 26, no. 9: 2443. https://doi.org/10.3390/molecules26092443
APA StyleSayyed, R. Z., Shaikh, S. S., Wani, S. J., Rehman, M. T., Al Ajmi, M. F., Haque, S., & El Enshasy, H. A. (2021). Production of Biodegradable Polymer from Agro-Wastes in Alcaligenes sp. and Pseudomonas sp. Molecules, 26(9), 2443. https://doi.org/10.3390/molecules26092443









