Hydrogen Bonding in Natural and Unnatural Base Pairs—A Local Vibrational Mode Study
Abstract
1. Introduction
2. Methodologies
2.1. The Local Vibrational Mode Analysis
2.2. QTAIM and NBO Analysis
2.3. Computational Methods
3. Results and Discussion
3.1. Internal HB Strength
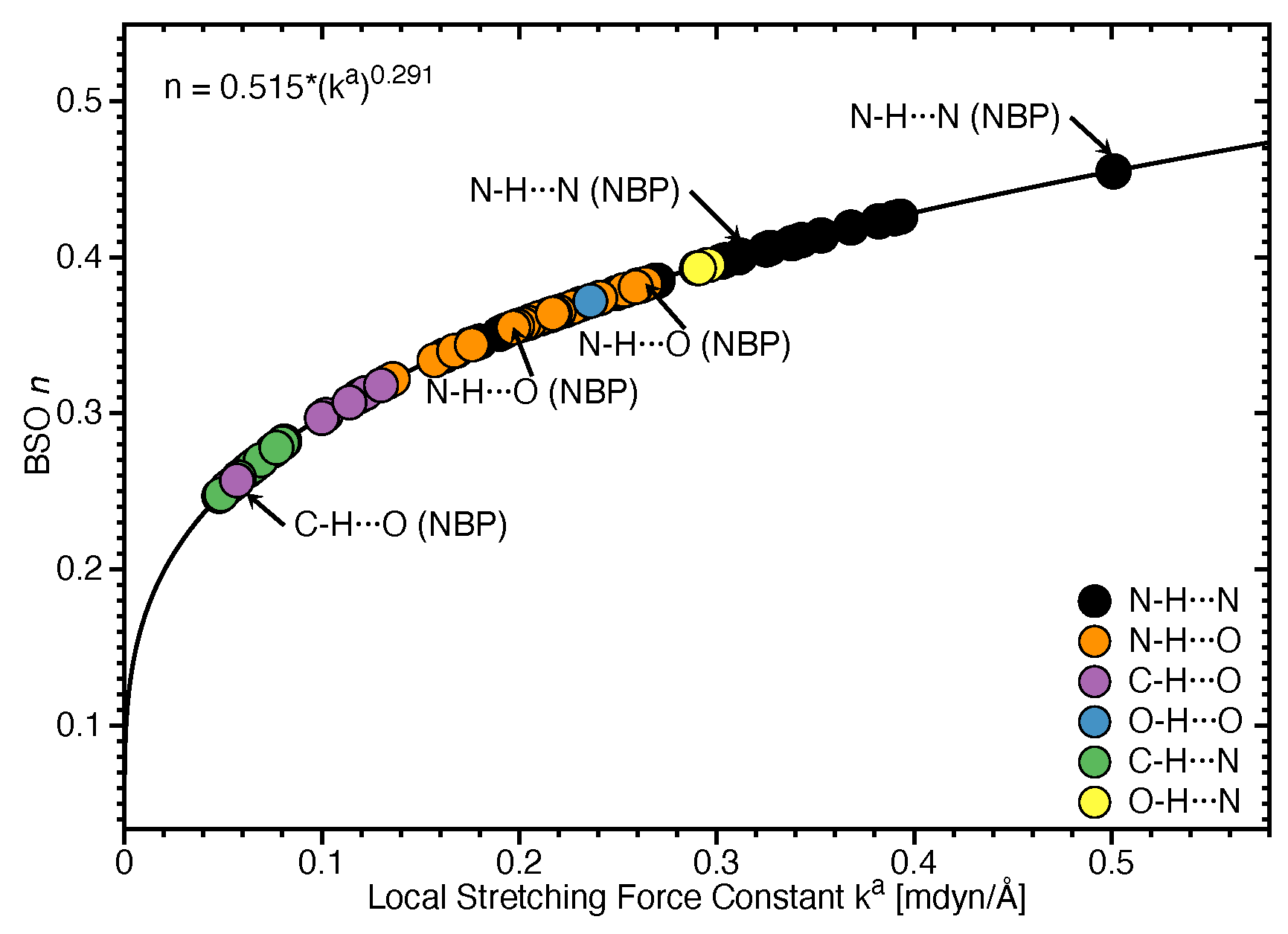
3.2. Significance of Non-Classical HBs
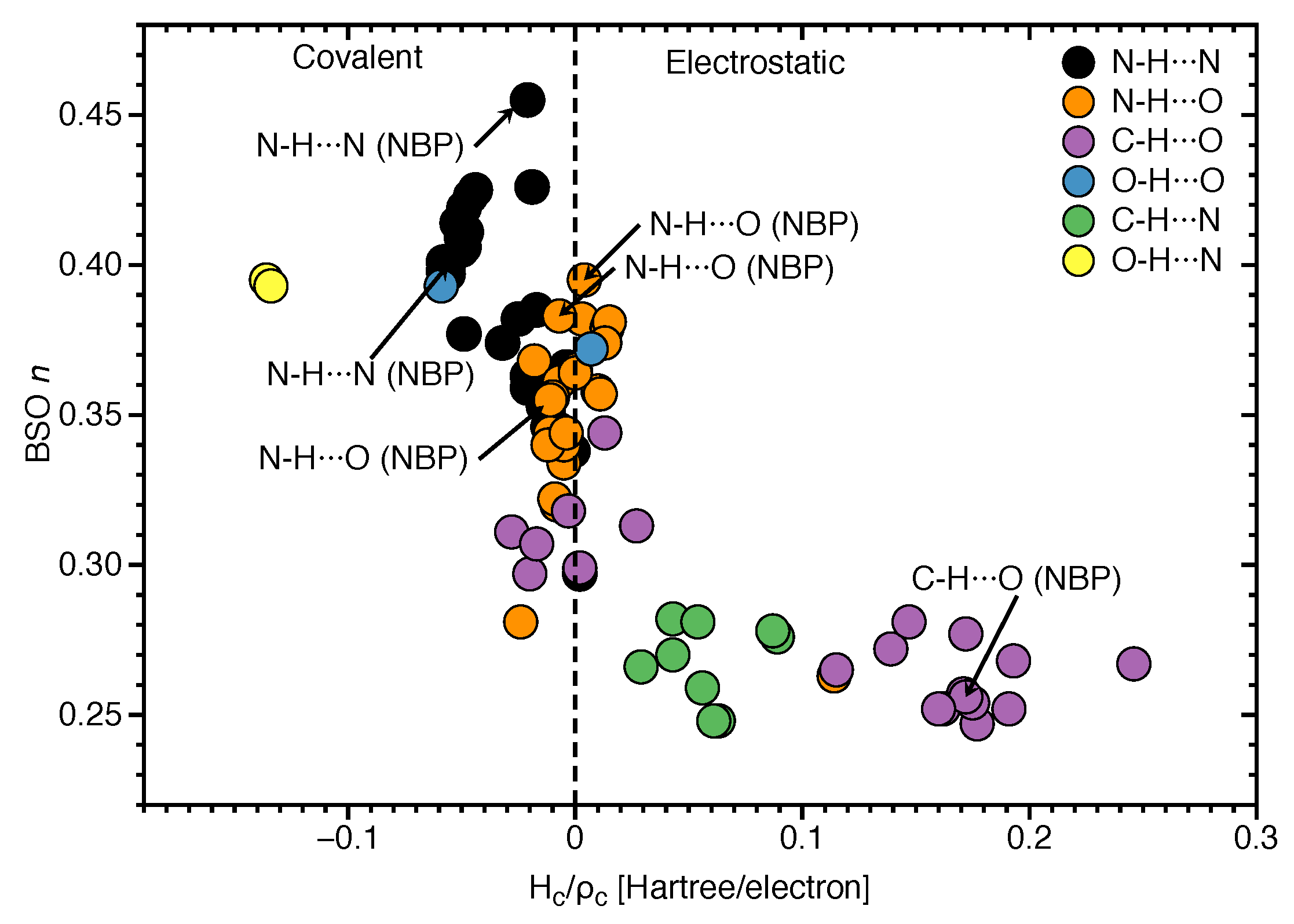
3.3. Covalent Character of HBs
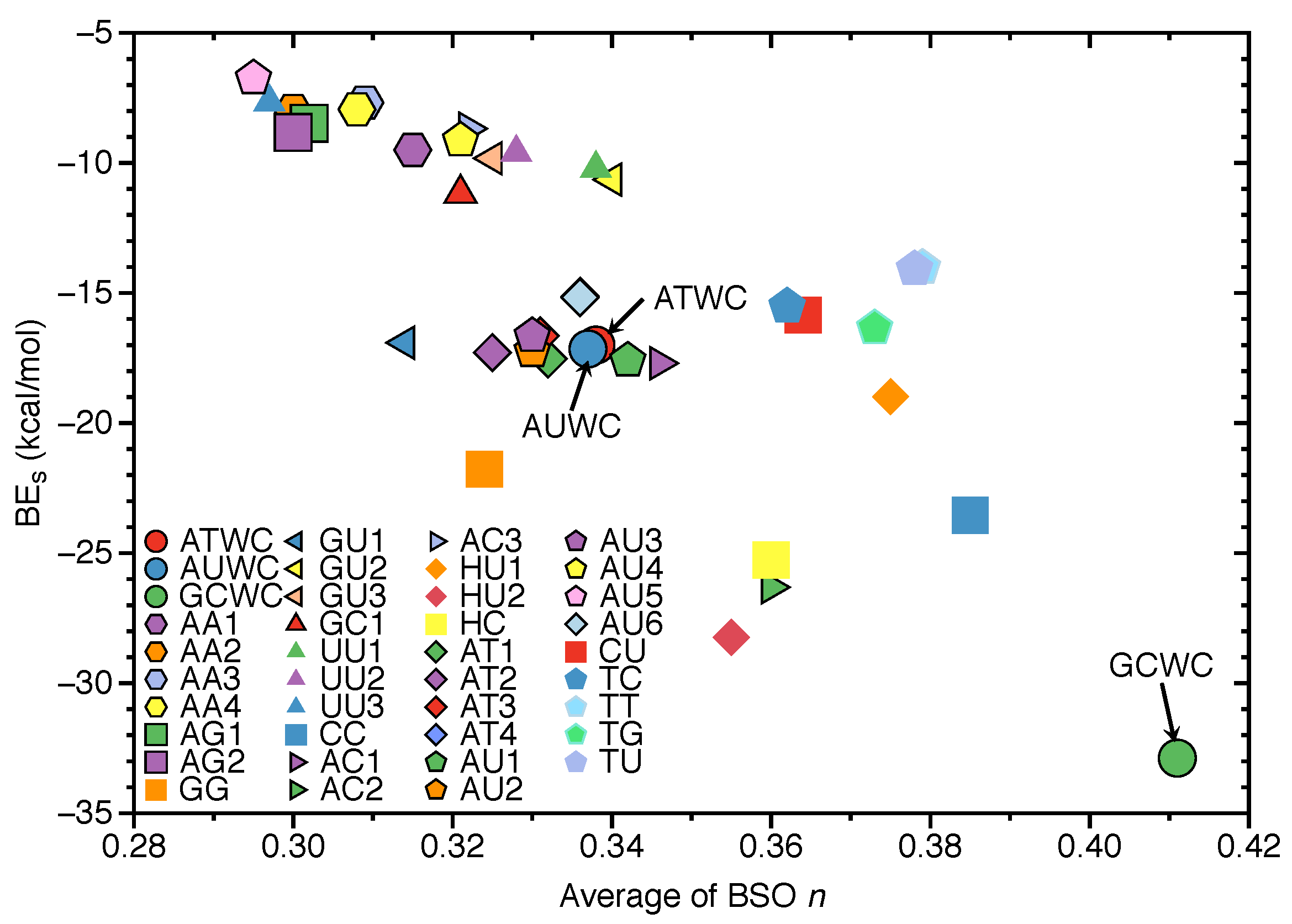
3.4. Intrinsic HB Strength and BEs
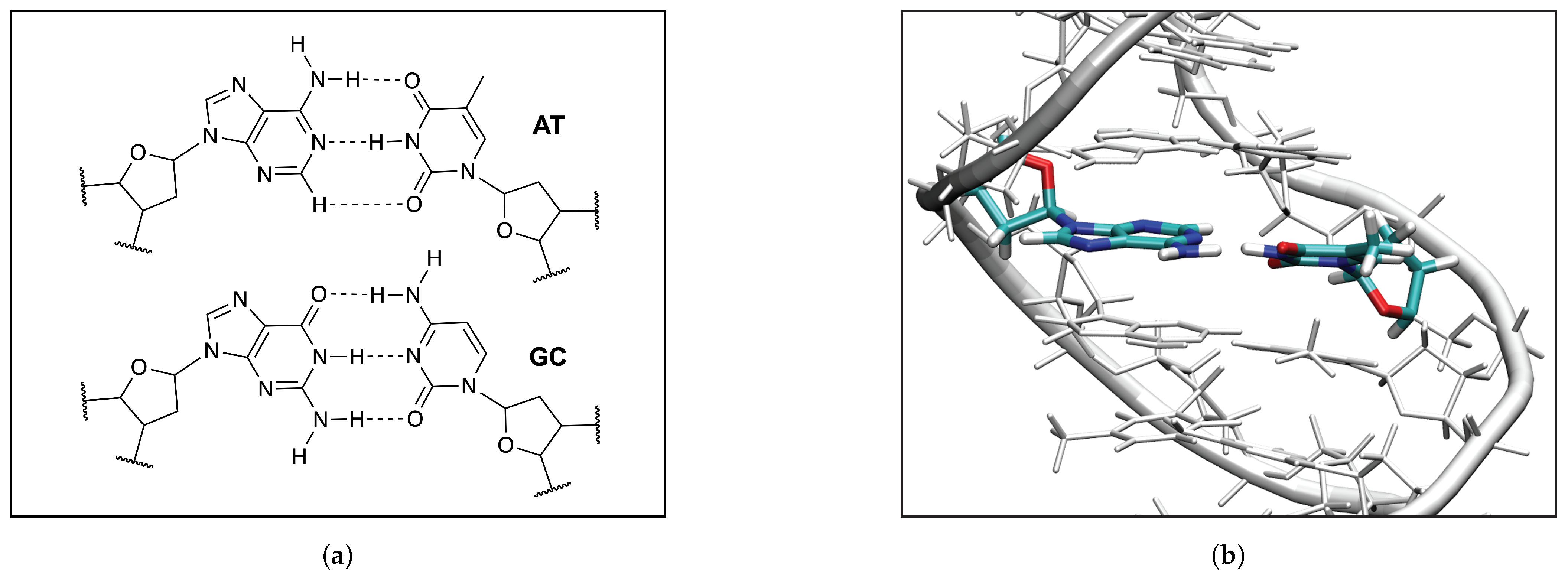
3.5. HBs in the DNA Environment
4. Conclusions and Outlook
- Hydrogen bonding in Watson Crick base pairs is not exceptionally strong and the N–H⋯N bond is the most favorable hydrogen bond in both unnatural and natural base pairs while O–H⋯N/O bonds are the less favorable in unnatural base pairs and not found at all in natural base pairs.
- In addition, non-classical C–H⋯N/O bonds play an important role for the stabilization of base pairs, especially C–H⋯O bonds in Watson Crick base pairs. This suggests that Nature’s choice to combine classical and non-classical hydrogen bonding should also be copied in the design of new unnatural base pair combinations.
- Hydrogen bonding in Watson Crick base pairs modeled in the DNA via a QM/MM approach showed that the DNA environment increases the strength of the central N–H⋯N bond and the C–H⋯O bonds, and at the same time decreases the strength of the N–H⋯O bond. However, the general trends observed in the gas phase calculations remain unchanged reflecting that electrostatic interactions with the environment are a less important factor determining the intermolecular hydrogen bond strength; an important validation of the gas phase model applied in this work.
- Natural base pairs do not possess larger binding energies than their unnatural counterparts. We also did not find a significant correlation between hydrogen bond strengths and binding energies, i.e., BSO n and BE values, as expected because these two quantities cannot directly be compared.
- We expect that the presence of base pairs with more nonclassical, i.e., weaker HBs in DNA will make the environment less covalent. During electron transfer these bonds will couple with specific vibrational modes of the DNA strand changing the electronic properties of the DNA. It has been documented [155,156,157,158] that these changes can stretch over 10 to 80 nucleobases accompanied by a decrease of the corresponding normal frequencies. When the DNA body lengthens, it becomes more mobile and less rigid. The experiment can only acquire normal vibrational frequencies of the backbone and the bases of DNA molecules characterized by coupled vibrational modes, while we can capture via LMA individual local frequencies from low to high and decode specific atomic motions, leading to more comprehensive and deeper insights into the stability of the DNA strand, which we will further explore in future work.
- The stability of the DNA double helix is mainly determined by (i) non-covalent interactions involving hydrogen bonds between A-T and G-C base pairs, (ii) stacking interactions between adjacent bases along the helix, and (iii) cross-interactions between base pairs [159]. Interactions outside the DNA double helix generally play a less important role [160]. The interplay between hydrogen bonding and stacking interactions in DNA has been the subject of several experimental [161,162,163,164,165] and theoretical investigations [159,166,167,168,169,170,171,172,173]. Based on DNA melting and energetics of the double helix [174], it has been recently suggested that in accordance with previous experiments [165,175] the stability of DNA double strands depends mainly on G-C base pair rich sequences. This is completely in line with our results identifying the hydrogen bonds of the G-C base pairs as one of the strongest. The local mode analysis can also quantitatively assess the strength of the stacking interactions between adjacent DNA bases along the helix, which is currently under investigation.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HB | Hydrogen bond |
| NBP | natural base pair |
| UBP | Unnatural base pair |
References
- Minchin, S.; Lodge, J. Understanding biochemistry: Structure and function of nucleic acids. Essays Biochem. 2020, 63, 433–456. [Google Scholar] [CrossRef]
- Chevizovich, D.; Michieletto, D.; Mvogo, A.; Zakiryanov, F.; Zdravkovic, S. A review on nonlinear DNA physics. R. Soc. Open Sci. 2020, 7, 200774. [Google Scholar] [CrossRef] [PubMed]
- Varela, S.; Montañes, B.; López, F.; Berche, B.; Guillot, B.; Mujica, V.; Medina, E. Intrinsic Rashba Coupling Due to Hydrogen Bonding in DNA. J. Chem. Phys. 2019, 151, 125102. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, V. A Proposed Structure for the Nucleic Acids (1953) by Linus Pauling and Robert Brainard Corey. Embryo Project Encyclopedia, SSN: 1940-5030. Available online: http://embryo.asu.edu/handle/10776/13121 (accessed on 26 August 2019).
- Watson, J. The Double Helix: A Personal Account of the Discovery of the Structure of DNA; Weidenfeld and Nicolson: London, UK, 2012. [Google Scholar]
- Holmes, F.L. Meselson, Stahl, and the Replication of DNA: A History of ‘The Most BeautifulExperiment in Biology’; Yale University Press: London, UK, 2001. [Google Scholar]
- Cobb, M. A Speculative History of DNA: What If Oswald Avery Had Died in 1934? PLoS Biol. 2016, 14, e2001197. [Google Scholar] [CrossRef] [PubMed]
- Pauling, L.; Corey, R.B. A Proposed Structure For The Nucleic Acids. Proc. Natl. Acad. Sci. USA 1953, 39, 84–97. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H. Molecular Structure of Nucleic Acids. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Durmaz, A.A.; Karaca, E.; Demkow, U.; Toruner, G.; Schoumans, J.; Cogulu, O. Evolution of Genetic Techniques: Past, Present, and Beyond. BioMed Res. Int. 2015, 2015, 461524. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D. The DNA Story: A Documentary History of Gene Cloning; WH Freeman and Co.: New York, NY, USA, 1981. [Google Scholar]
- Watson, J.D.; Crick, F.H. Genetical Implications of the Structure of Deoxyribonucleic Acid. Nature 1953, 171, 964–967. [Google Scholar] [CrossRef]
- Fonseca Guerra, C.; Bickelhaupt, F.M.; Snijders, J.G.; Baerends, E.J. The Nature of the Hydrogen Bond in DNA Base Pairs: The Role of Charge Transfer and Resonance Assistance. Chem. Eur. J. 1999, 5, 3581–3594. [Google Scholar] [CrossRef]
- Heinemann, U.; Roske, Y. Symmetry in Nucleic-Acid Double Helices. Symmetry 2020, 12, 737. [Google Scholar] [CrossRef]
- Raguseo, F.; Chowdhury, S.; Minard, A.; Di Antonio, M. Chemical-biology Approaches to Probe DNA and RNA G-quadruplex Structures in The Genome. ChemComm 2020, 56, 1317–1324. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, H.; Gatenby, R.A. Evolutionary advantage of anti-parallel strand orientation of duplex DNA. Sci. Rep. 2020, 10, 9883. [Google Scholar] [CrossRef] [PubMed]
- Ramezani, H.; Dietz, H. Building Machines with DNA Molecules. Nat. Rev. Genet. 2020, 21, 5–26. [Google Scholar] [CrossRef]
- Nie, P.; Bai, Y.; Mei, H. Synthetic Life with Alternative Nucleic Acids as Genetic Materials. Molecules 2020, 25, 3483. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Datta, L.P.; Govindaraju, T. Molecular Architectonics of DNA for Functional Nanoarchitectures. Beilstein J. Nanotechnol. 2020, 11, 124–140. [Google Scholar] [CrossRef] [PubMed]
- Romesberg, F.E. Synthetic Biology: The Chemist’s Approach. Isr. J. Chem. 2019, 59, 91–94. [Google Scholar] [CrossRef]
- Hamashima, K.; Soong, Y.T.; Matsunaga, K.I.; Kimoto, M.; Hirao, I. DNA Sequencing Method Including Unnatural Bases for DNA Aptamer Generation by Genetic Alphabet Expansion. ACS Synth. Biol. 2019, 8, 1401–1410. [Google Scholar] [CrossRef]
- Devine, K.G.; Jheeta, S. De Novo Nucleic Acids: A Review of Synthetic Alternatives to DNA and RNA That Could Act as Bio-Information Storage Molecules. Life 2020, 10, 346. [Google Scholar] [CrossRef]
- Feldman, A.W.; Ledbetter, M.P.; Zhang, Y.; Romesberg, F.E. Reply to Hettinger: Hydrophobic Unnatural Base Pairs and the Expansion of the Genetic Alphabet. Proc. Natl. Acad. Sci. USA 2017, 114, E6478–E6479. [Google Scholar] [CrossRef]
- Feldman, A.W.; Dien, V.T.; Karadeema, R.J.; Fischer, E.C.; You, Y.; Anderson, B.A.; Krishnamurthy, R.; Chen, J.S.; Li, L.; Romesberg, F.E. Optimization of Replication, Transcription, and Translation in a Semi-Synthetic Organism. J. Am. Chem. Soc. 2019, 141, 10644–10653. [Google Scholar] [CrossRef]
- Weaver, J. Expanding the genetic alphabet. BioTechniques 2017, 62, 252–253. [Google Scholar] [CrossRef]
- Biondi, E.; Benner, S.A. Artificially Expanded Genetic Information Systems for New Aptamer Technologies. Biomedicines 2018, 6, 53. [Google Scholar] [CrossRef]
- Eggert, F.; Kurscheidt, K.; Hoffmann, E.; Kath-Schorr, S. Towards Reverse Transcription with an Expanded Genetic Alphabet. ChemBioChem 2019, 20, 1642–1645. [Google Scholar] [CrossRef] [PubMed]
- Dien, V.T.; Holcomb, M.; Romesberg, F.E. Eight-Letter DNA. Biochemistry 2019, 58, 2581–2583. [Google Scholar] [CrossRef] [PubMed]
- Jahiruddin, S.; Datta, A. What Sustains the Unnatural Base Pairs (UBPs) with No Hydrogen Bonds. J. Phys. Chem. B 2015, 119, 5839–5845. [Google Scholar] [CrossRef] [PubMed]
- Jahiruddin, S.; Mandal, N.; Datta, A. Structure and Electronic Properties of Unnatural Base Pairs: The Role of Dispersion Interactions. ChemPhysChem 2018, 19, 67–74. [Google Scholar] [CrossRef]
- Hernandez, A.R.; Shao, Y.; Hoshika, S.; Yang, Z.; Shelke, S.A.; Herrou, J.; Kim, H.J.; Kim, M.J.; Piccirilli, J.A.; Benner, S.A. A Crystal Structure of a Functional RNA Molecule Containing an Artificial Nucleobase Pair. Angew. Chem. Int. Ed. 2015, 127, 9991–9994. [Google Scholar] [CrossRef]
- Hoshika, S.; Leal, N.A.; Kim, M.J.; Kim, M.S.; Karalkar, N.B.; Kim, H.J.; Bates, A.M.; Watkins, N.E.; SantaLucia, H.A.; Meyer, A.J. Hachimoji DNA and RNA: A Genetic System with Eight Building Blocks. Science 2019, 363, 884–887. [Google Scholar] [CrossRef] [PubMed]
- Beck, K.M.; Krogh, M.B.; Hornum, M.; Ludford, P.T., III; Tor, Y.; Nielsen, P. Double-headed nucleotides as xeno nucleic acids: Information storage and polymerase recognition. Org. Biomol. Chem. 2020, 18, 7213–7223. [Google Scholar] [CrossRef] [PubMed]
- Hamashima, K.; Kimoto, M.; Hirao, I. Creation of Unnatural Base Pairs for Genetic Alphabet Expansion Toward Synthetic Xenobiology. Curr. Opin. Chem. Biol. 2018, 46, 108–114. [Google Scholar] [CrossRef]
- Feldman, A.W.; Dien, V.T.; Romesberg, F.E. Chemical Stabilization of Unnatural Nucleotide Triphosphates for the in Vivo Expansion of the Genetic Alphabet. J. Am. Chem. Soc. 2017, 139, 2464–2467. [Google Scholar] [CrossRef]
- Dien, V.T.; Holcomb, M.; Feldman, A.W.; Fischer, E.C.; Dwyer, T.J.; Romesberg, F.E. Progress Toward a Semi-Synthetic Organism with an Unrestricted Expanded Genetic Alphabet. J. Am. Chem. Soc. 2018, 140, 16115–16123. [Google Scholar] [CrossRef]
- Taylor, A.I.; Houlihan, G.; Holliger, P. Beyond DNA and RNA: The Expanding Toolbox of Synthetic Genetics. Cold Spring Harb. Perspect. Biol. 2019, 11, a032490. [Google Scholar] [CrossRef] [PubMed]
- Kubyshkin, V.; Budisa, N. Anticipating Alien Cells with Alternative Genetic Codes: Away from the Alanine World! Curr. Opin. Chem. Biol. 2019, 60, 242–249. [Google Scholar] [CrossRef]
- Whitford, C.M.; Dymek, S.; Kerkhoff, D.; März, C.; Schmidt, O.; Edich, M.; Droste, J.; Pucker, B.; Rückert, C.; Kalinowski, J. Auxotrophy to Xeno-DNA: An exploration of combinatorial mechanisms for a high-fidelity biosafety system for synthetic biology applications. J. Biol. Eng. 2018, 12, 13. [Google Scholar] [CrossRef]
- Arunan, E. One Hundred Years After The Latimer and Rodebush Paper, Hydrogen Bonding Remains an Elephant! Indian J. Sci. 2020, 100, 249–255. [Google Scholar] [CrossRef]
- Gibb, B.C. The Centenary (maybe) of The Hydrogen Bond. Nat. Chem. 2020, 12, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Scheiner, S. The Hydrogen Bond: A Hundred Years and Counting. J. Indian Inst. Sci. 2020, 100, 61–76. [Google Scholar] [CrossRef]
- Scheiner, S. Comparison of Bifurcated Halogen with Hydrogen Bonds. Molecules 2021, 26, 350. [Google Scholar] [CrossRef] [PubMed]
- Wain-Hobson, S. The Third Bond. Nature 2006, 439, 539. [Google Scholar] [CrossRef]
- Jayaraman, A. 100th Anniversary of Macromolecular Science Viewpoint: Modeling and Simulation of Macromolecules with Hydrogen Bonds: Challenges, Successes, and Opportunities. ACS Macro Lett. 2020, 2, 656–665. [Google Scholar] [CrossRef]
- Pauling, L.; Corey, R.B. Two hydrogen-bonded spiral configurations of the polypeptide chain. J. Am. Chem. Soc. 1950, 72, 5349. [Google Scholar] [CrossRef]
- Pauling, L.; Corey, R.B. The structure of proteins: Two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. USA 1951, 37, 205–211. [Google Scholar] [CrossRef]
- Pauling, L.; Corey, R.B. Configurations of polypeptide chains with favored orientations around single bonds: Two new pleated sheets. Proc. Natl. Acad. Sci. USA 1951, 37, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Nobel Prize for Chemistry: Prof. Linus Pauling, For. Mem. R.S. Nature 1954, 174, 907–908. [CrossRef]
- Pauling, L. The Nature of the Chemical Bond; Cornell University Press: Ithaca, NY, USA, 1960; Volume 260. [Google Scholar]
- Oswald, S.; Suhm, M.A. Soft Experimental Constraints for Soft Interactions: A Spectroscopic Benchmark Data Set For Weak and Strong Hydrogen Bonds. Phys. Chem. Chem. Phys. 2019, 21, 18799–18810. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, S.J. Understanding Hydrogen Bonds: Theoretical and Experimental Views, 1st ed.; Royal Society of Chemistry: London, UK, 2020. [Google Scholar]
- Karas, L.J.; Wu, C.H.; Das, R.; Wu, J.I.C. Hydrogen bond design principles. WIREs Comput. Mol. Sci. 2020, 10, e1477. [Google Scholar] [CrossRef]
- Van der Lubbe, S.C.C.; Guerra, C.F. The Nature of Hydrogen Bonds: A Delineation of the Role of Different Energy Components on Hydrogen Bond Strengths and Lengths. Chem. Asian J. 2019, 14, 2760–2769. [Google Scholar] [PubMed]
- Williams-Ashman, G. Review of Horizons in Biochemistry. Perspect. Biol. Med. 1963, 6, 264–267. [Google Scholar] [CrossRef]
- Hettinger, T.P. Helix Instability and Self-Pairing Prevent Unnatural Base Pairs From Expanding the Genetic Alphabet. PNAS 2017, 114, E6476–E6477. [Google Scholar] [CrossRef]
- Czyznikowska, Z.; Góra, R.; Zalesny, R.; Lipkowski, P.; Jarzembska, K.; Dominiak, P.; Leszczynski, J. Structural Variability and the Nature of Intermolecular Interactions in Watson- Crick B-DNA Base Pairs. J. Phys. Chem. B 2010, 114, 9629–9644. [Google Scholar] [CrossRef]
- Brovarets’, O.O.; Yurenko, Y.P.; Hovorun, D.M. Intermolecular CH⋯O/N H-bonds in the Biologically Important Pairs of Natural Nucleobases: A Thorough Quantum-Chemical Study. J. Biomol. Struct. Dyn. 2014, 32, 993–1022. [Google Scholar] [CrossRef] [PubMed]
- Stasyuk, O.A.; Solà, M.; Swart, M.; Fonseca Guerra, C.; Krygowski, T.M.; Szatylowicz, H. Effect of Alkali Metal Cations on Length and Strength of Hydrogen Bonds in DNA Base Pairs. ChemPhysChem 2020, 21, 2112–2126. [Google Scholar] [CrossRef]
- Cerón-Carrasco, J.P.; Cerezo, J.; Jacquemin, D. How DNA Is Damaged by External Electric Fields: Selective Mutation vs. Random Degradation. Phys. Chem. Chem. Phys. 2014, 16, 8243–8246. [Google Scholar] [CrossRef] [PubMed]
- Cerón-Carrasco, J.P.; Jacquemin, D. Electric-Field Induced Mutation of DNA: A Theoretical Investigation of the GC Base Pair. Phys. Chem. Chem. Phys. 2013, 15, 4548–4553. [Google Scholar] [CrossRef] [PubMed]
- Kounovsky-Shafer, K.L.; Hernandez-Ortiz, J.P.; Potamousis, K.; Tsvid, G.; Place, M.; Ravindran, P.; Jo, K.; Zhou, S.; Odijk, T.; De Pablo, J.J.; et al. Electrostatic Confinement and Manipulation of DNA Molecules for Genome Analysis. Proc. Natl. Acad. Sci. USA 2017, 114, 13400–13405. [Google Scholar] [CrossRef]
- Slocombe, L.; Al-Khalili, J.; Sacchi, M. Quantum and Classical Effects in DNA Point Mutations: Watson–Crick Tautomerism in AT and GC BBase Pairs. Phys. Chem. Chem. Phys. 2021, 23, 4141–4150. [Google Scholar] [CrossRef]
- Florián, J.; Leszczyński, J. Spontaneous DNA Mutations Induced by Proton Transfer in the Guanine? Cytosine Base Pairs: An Energetic Perspective. J. Am. Chem. Soc. 1996, 118, 3010–3017. [Google Scholar] [CrossRef]
- Konkoli, Z.; Cremer, D. A New Way of Analyzing Vibrational Spectra. I. Derivation of Adiabatic Internal Modes. Int. J. Quantum Chem. 1998, 67, 1–9. [Google Scholar] [CrossRef]
- Konkoli, Z.; Larsson, J.A.; Cremer, D. A New Way of Analyzing Vibrational Spectra. II. Comparison of Internal Mode Frequencies. Int. J. Quantum Chem. 1998, 67, 11–27. [Google Scholar] [CrossRef]
- Konkoli, Z.; Cremer, D. A New Way of Analyzing Vibrational Spectra. III. Characterization of Normal Vibrational Modes in terms of Internal Vibrational Modes. Int. J. Quantum Chem. 1998, 67, 29–40. [Google Scholar] [CrossRef]
- Konkoli, Z.; Larsson, J.A.; Cremer, D. A New Way of Analyzing Vibrational Spectra. IV. Application and Testing of Adiabatic Modes within the Concept of the Characterization of Normal Modes. Int. J. Quantum Chem. 1998, 67, 41–55. [Google Scholar] [CrossRef]
- Cremer, D.; Larsson, J.A.; Kraka, E. New Developments in the Analysis of Vibrational Spectra on the Use of Adiabatic Internal Vibrational Modes. In Theoretical and Computational Chemistry; Parkanyi, C., Ed.; Elsevier: Amsterdam, The Netherlands, 1998; pp. 259–327. [Google Scholar]
- Mohajeri, A.; Nobandegani, F.F. Detection and Evaluation of Hydrogen Bond Strength in Nucleic Acid Base Pairs. J. Phys. Chem. A 2008, 112, 281–295. [Google Scholar] [CrossRef]
- Bader, R. Atoms in Molecules: A Quantum Theory; Clarendron Press: Oxford, UK, 1995. [Google Scholar]
- Popelier, P. Atoms in Molecules: An Introduction; Prentice-Hall: Harlow, UK, 2000. [Google Scholar]
- Kraka, E.; Zou, W.; Tao, Y. Decoding Chemical Information from Vibrational Spectroscopy Data: Local Vibrational Mode Theory. WIREs Comput. Mol. Sci. 2020, 10, 1480. [Google Scholar] [CrossRef]
- Wilson, E.B.; Decius, J.C.; Cross, P.C.M. Molecular Vibrations. The Theory of Infrared and Raman Vibrational Spectra; McGraw-Hill: New York, NY, USA, 1955; pp. 59–136. [Google Scholar]
- Wilson, E.B.; Decius, J.C.; Cross, P.C.; Sundheim, B.R. Molecular Vibrations: The Theory of Infrared and Raman Vibrational Spectra. J. Electrochem. Soc. 1955, 102, 235C. [Google Scholar] [CrossRef]
- Zou, W.; Cremer, D. C2 in a Box: Determining its Intrinsic Bond Strength for the X1Σ+g Ground State. Chem. Eur. J. 2016, 22, 4087–4097. [Google Scholar] [CrossRef]
- Oomens, J.; Kraka, E.; Nguyen, M.K.; Morton, T.M. Structure, Vibrational Spectra, and Unimolecular Dissociation of Gaseous 1-Fluoro-1-phenethyl Cations. J. Phys. Chem. A 2008, 112, 10774–10783. [Google Scholar] [CrossRef]
- Zou, W.; Kalescky, R.; Kraka, E.; Cremer, D. Relating Normal Vibrational Modes to Local Vibrational Modes: Benzene and Naphthalene. J. Mol. Model. 2012, 19, 2865–2877. [Google Scholar] [CrossRef] [PubMed]
- Kalescky, R.; Kraka, E.; Cremer, D. Identification of the Strongest Bonds in Chemistry. J. Phys. Chem. A 2013, 117, 8981–8995. [Google Scholar] [CrossRef] [PubMed]
- Kalescky, R.; Kraka, E.; Cremer, D. Description of Aromaticity with the Help of Vibrational Spectroscopy: Anthracene and Phenanthrene. J. Phys. Chem. A 2013, 118, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Kalescky, R.; Kraka, E.; Cremer, D. New Approach to Tolman’s Electronic Parameter Based on Local Vibrational Modes. Inorg. Chem. 2013, 53, 478–495. [Google Scholar] [CrossRef]
- Kalescky, R.; Kraka, E.; Cremer, D. Are Carbon-Halogen Double and Triple Bonds Possible? Int. J. Quantum Chem. 2014, 114, 1060–1072. [Google Scholar] [CrossRef]
- Kalescky, R.; Zou, W.; Kraka, E.; Cremer, D. Quantitative Assessment of the Multiplicity of Carbon-Halogen Bonds: Carbenium and Halonium Ions with F, Cl, Br, and I. J. Phys. Chem. A 2014, 118, 1948–1963. [Google Scholar] [CrossRef]
- Humason, A.; Zou, W.; Cremer, D. 11,11-Dimethyl-1,6-methano[10]annulene—An Annulene with an Ultralong CC Bond or a Fluxional Molecule? J. Phys. Chem. A 2014, 119, 1666–1682. [Google Scholar] [CrossRef]
- Sethio, D.; Lawson Daku, L.M.; Hagemann, H.; Kraka, E. Quantitative Assessment of B−B−B, B−Hb−B, and B−Ht Bonds: From BH3 to B12H122−. ChemPhysChem 2019, 20, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Makoś, M.Z.; Freindorf, M.; Sethio, D.; Kraka, E. New Insights into Fe–H2 and Fe–H− Bonding of a [NiFe] Hydrogenase Mimic—A Local Vibrational Mode Study. Theor. Chem. Acc. 2019, 138, 76. [Google Scholar] [CrossRef]
- Makoś, M.Z.; Zou, W.; Freindorf, M.; Kraka, E. Metal-Ring Interactions in Actinide Sandwich Compounds: A Combined Normalized Elimination of the Small Component and Local Vibrational Mode Study. Mol. Phys. 2020, 118, e1768314. [Google Scholar] [CrossRef]
- Verma, N.; Tao, Y.; Zou, W.; Chen, X.; Chen, X.; Freindorf, M.; Kraka, E. A Critical Evaluation of Vibrational Stark Effect (VSE) Probes with the Local Vibrational Mode Theory. Sensors 2020, 20, 2358. [Google Scholar] [CrossRef]
- Freindorf, M.; Kraka, E. Critical Assessment of the FeC and CO Bond strength in Carboxymyoglobin—A QM/MM Local Vibrational Mode Study. J. Mol. Model. 2020, 26, 281. [Google Scholar] [CrossRef]
- Kraka, E.; Freindorf, M. Characterizing the Metal Ligand Bond Strength via Vibrational Spectroscopy: The Metal Ligand Electronic Parameter (MLEP). In Topics in Organometallic Chemistry—New Directions in the Modeling of Organometallic Reactions; Lledós, A., Ujaque, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2020; Volume 67, pp. 1–43. [Google Scholar]
- Freindorf, M.; Kraka, E.; Cremer, D. A Comprehensive Analysis of Hydrogen Bond Interactions Based on Local Vibrational Modes. Int. J. Quantum Chem. 2012, 112, 3174–3187. [Google Scholar] [CrossRef]
- Kalescky, R.; Zou, W.; Kraka, E.; Cremer, D. Local Vibrational Modes of the Water Dimer—Comparison of Theory and Experiment. Chem. Phys. Lett. 2012, 554, 243–247. [Google Scholar] [CrossRef]
- Kalescky, R.; Kraka, E.; Cremer, D. Local Vibrational Modes of the Formic Acid Dimer—The Strength of the Double H-Bond. Mol. Phys. 2013, 111, 1497–1510. [Google Scholar] [CrossRef]
- Kraka, E.; Freindorf, M.; Cremer, D. Chiral Discrimination by Vibrational Spectroscopy Utilizing Local Modes. Chirality 2013, 25, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Setiawan, D.; Kraka, E.; Cremer, D. Description of Pnicogen Bonding with the help of Vibrational Spectroscopy—The Missing Link Between Theory and Experiment. Chem. Phys. Lett. 2014, 614, 136–142. [Google Scholar] [CrossRef]
- Setiawan, D.; Kraka, E.; Cremer, D. Strength of the Pnicogen Bond in Complexes Involving Group VA Elements N, P, and As. J. Phys. Chem. A 2014, 119, 1642–1656. [Google Scholar] [CrossRef] [PubMed]
- Setiawan, D.; Kraka, E.; Cremer, D. Hidden Bond Anomalies: The Peculiar Case of the Fluorinated Amine Chalcogenides. J. Phys. Chem. A 2015, 119, 9541–9556. [Google Scholar] [CrossRef]
- Kraka, E.; Setiawan, D.; Cremer, D. Re-Evaluation of the Bond Length-Bond Strength Rule: The Stronger Bond Is not Always the Shorter Bond. J. Comput. Chem. 2015, 37, 130–142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dai, H.; Yan, H.; Zou, W.; Cremer, D. B-H π Interaction: A New Type of Nonclassical Hydrogen Bonding. J. Am. Chem. Soc. 2016, 138, 4334–4337. [Google Scholar] [CrossRef]
- Setiawan, D.; Cremer, D. Super-Pnicogen Bonding in the Radical Anion of the Fluorophosphine Dimer. Chem. Phys. Lett. 2016, 662, 182–187. [Google Scholar] [CrossRef]
- Oliveira, V.; Kraka, E.; Cremer, D. The Intrinsic Strength of the Halogen Bond: Electrostatic and Covalent Contributions Described by Coupled Cluster Theory. Phys. Chem. Chem. Phys. 2016, 18, 33031–33046. [Google Scholar] [CrossRef]
- Oliveira, V.; Kraka, E.; Cremer, D. Quantitative Assessment of Halogen Bonding Utilizing Vibrational Spectroscopy. Inorg. Chem. 2016, 56, 488–502. [Google Scholar] [CrossRef]
- Tao, Y.; Zou, W.; Jia, J.; Li, W.; Cremer, D. Different Ways of Hydrogen Bonding in Water—Why Does Warm Water Freeze Faster than Cold Water? J. Chem. Theory Comput. 2017, 13, 55–76. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, V.; Cremer, D. Transition from Metal-Ligand Bonding to Halogen Bonding Involving a Metal as Halogen Acceptor: A Study of Cu, Ag, Au, Pt, and Hg Complexes. Chem. Phys. Lett. 2017, 681, 56–63. [Google Scholar] [CrossRef]
- Oliveira, V.; Cremer, D.; Kraka, E. The Many Facets of Chalcogen Bonding: Described by Vibrational Spectroscopy. J. Phys. Chem. A 2017, 121, 6845–6862. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, V.; Kraka, E. Systematic Coupled Cluster Study of Noncovalent Interactions Involving Halogens, Chalcogens, and Pnicogens. J. Phys. Chem. A 2017, 121, 9544–9556. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Zhang, X.; Dai, H.; Yan, H.; Cremer, D.; Kraka, E. Description of an Unusual Hydrogen Bond Between Carborane and a Phenyl Group. J. Organometall. Chem. 2018, 856, 114–127. [Google Scholar] [CrossRef]
- Yannacone, S.; Oliveira, V.; Verma, N.; Kraka, E. A Continuum from Halogen Bonds to Covalent Bonds: Where Do λ3 Iodanes Fit? Inorganics 2019, 7, 47. [Google Scholar] [CrossRef]
- Lyu, S.; Beiranvand, N.; Freindorf, M.; Kraka, E. Interplay of Ring Puckering and Hydrogen Bonding in Deoxyribonucleosides. J. Phys. Chem. A 2019, 123, 7087–7103. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, V.P.; Marcial, B.L.; Machado, F.B.C.; Kraka, E. Metal-Halogen Bonding Seen through the Eyes of Vibrational Spectroscopy. Materials 2020, 13, 55. [Google Scholar] [CrossRef]
- Tao, Y.; Qiu, Y.; Zou, W.; Nanayakkara, S.; Yannacone, S.; Kraka, E. In Situ Assessment of Intrinsic Strength of X-I⋯OA Type Halogen Bonds in Molecular Crystals with Periodic Local Vibrational Mode Theory. Molecules 2020, 25, 1589. [Google Scholar] [CrossRef]
- Yannacone, S.; Sethio, D.; Kraka, E. Quantitative Assessment of Intramolecular Hydrogen Bonds in Neutral Histidine. Theor. Chem. Acc. 2020, 139, 125. [Google Scholar] [CrossRef]
- Martins, J.; Quintino, R.P.; Politi, J.R.S.; Sethio, D.; Gargano, R.; Kraka, E. Computational Analysis of Vibrational Frequencies and Rovibrational Spectroscopic Constants of Hydrogen Sulfide Dimer using MP2 and CCSD(T). Spectrochim. Acta A 2020, 239, 118540-1–118540-9. [Google Scholar] [CrossRef] [PubMed]
- Yannacone, S.; Freindorf, M.; Tao, Y.; Zou, W.; Kraka, E. Local Vibrational Mode Analysis of π-Hole Interactions between Aryl Donors and Small Molecule Acceptors. Crystals 2020, 10, 556. [Google Scholar] [CrossRef]
- Bader, R.F.W. Atoms in Molecules. Acc. Chem. Res. 1985, 18, 9–15. [Google Scholar] [CrossRef]
- Bader, R. Atoms in Molecules: A Quantum Theory; International Series of Monographs on Chemistry; Clarendon Press: Oxford, UK, 1990. [Google Scholar]
- Cremer, D.; Kraka, E. Chemical Bonds without Bonding Electron Density? Does the Difference Electron-Density Analysis Suffice for a Description of the Chemical Bond? Angew. Chem. Int. Ed. 1984, 23, 627–628. [Google Scholar] [CrossRef]
- Cremer, D.; Kraka, E. A Description of the Chemical Bond in Terms of Local Properties of Electron Density and Energy. Croat. Chem. Acta 1984, 57, 1259–1281. [Google Scholar]
- Kraka, E.; Cremer, D. Chemical Implication of Local Features of the Electron Density Distribution. In Theoretical Models of Chemical Bonding. The Concept of the Chemical Bond; Maksic, Z.B., Ed.; Springer: Berlin/Heidelberg, Germany, 1990; Volume 2, pp. 453–542. [Google Scholar]
- Reed, A.; Curtiss, L.; Weinhold, F. Intermolecular Interactions from A Natural Bond Orbital, Donor-Acceptor Viewpoint. Chem. Rev. 1988, 88, 899–926. [Google Scholar] [CrossRef]
- Weinhold, F.; Landis, C.R. Valency and Bonding: A Natural Bond Orbital Donor-Acceptor Perspective; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- Glendening, E.D.; Badenhoop, J.K.; Reed, A.E.; Carpenter, J.E.; Bohmann, J.A.; Morales, C.M.; Landis, C.R.; Weinhold, F. NBO6; Theoretical Chemistry Institute, University of Wisconsin: Madison, WI, USA, 2013. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Chai, J.D.; Head-Gordon, M. Long-Range Corrected Hybrid Density Functionals with Damped Atom–Atom Dispersion Corrections. Phys. Chem. Chem. Phys. 2008, 10, 6615–6620. [Google Scholar] [CrossRef]
- Chai, J.D.; Head-Gordon, M. Systematic Optimization of Long-Range Corrected Hybrid Density Functionals. J. Chem. Phys. 2008, 128, 084106. [Google Scholar] [CrossRef]
- Hehre, W.J.; Ditchfield, R.; Pople, J.A. Self-Consistent Molecular Orbital Methods. XII. Further Extensions of Gaussian? Type Basis Sets for Use in Molecular Orbital Studies of Organic Molecules. J. Chem. Phys. 1972, 56, 2257–2261. [Google Scholar] [CrossRef]
- Ditchfield, R.; Hehre, W.J.; Pople, J.A. Self-Consistent Molecular-Orbital Methods. IX. An Extended Gaussian-Type Basis for Molecular-Orbital Studies of Organic Molecules. J. Chem. Phys. 1971, 54, 724–728. [Google Scholar] [CrossRef]
- Clark, T.; Chandrasekhar, J.; Spitznagel, G.W.; Schleyer, P.V.R. Efficient Diffuse Function-Augmented Basis Sets for Anion Calculations. III. The 3-21+ G Basis Set for First-Row Elements, Li–F. J. Comput. Chem. 1983, 4, 294–301. [Google Scholar] [CrossRef]
- Frisch, M.J.; Pople, J.A.; Binkley, J.S. Self-Consistent Molecular Orbital Methods 25. Supplementary Functions for Gaussian Basis Sets. J. Chem. Phys. 1984, 80, 3265–3269. [Google Scholar] [CrossRef]
- Gräfenstein, J.; Cremer, D. Efficient Density-Functional Theory Integrations by Locally Augmented Radial Grids. J. Chem. Phys. 2007, 127, 164113. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Tao, Y.; Freindorf, M.; Makoś, M.Z.; Verma, N.; Kraka, E. Local Vibrational Mode Analysis (LModeA); Computational and Theoretical Chemistry Group (CATCO), Southern Methodist University: Dallas, TX, USA, 2020. [Google Scholar]
- Keith, T.A. AIMAll (Version 17.01. 25); TK Gristmill Software: Overland Park, KS, USA, 2017. [Google Scholar]
- Boys, S.F.; Bernardi, F. The Calculation of Small Molecular Interactions by the Differences of Separate Total Energies. Some Procedures with Reduced Errors. Mol. Phys. 1970, 19, 553–566. [Google Scholar] [CrossRef]
- Senn, H.M.; Thiel, W. QM/MM Methods for Biomolecular Systems. Angew. Chem. Int. Ed. Engl. 2009, 48, 1198–1229. [Google Scholar] [CrossRef] [PubMed]
- Delgado, J.L.; Vance, N.R.; Kerns, R.J. Crystal Structure of DNA Dodecamer D(CGCGAATTCGCG). National Institutes of Health/National Institute of Allergy and Infectious Diseases. 2018. Available online: http://www.rcsb.org/structure/6CQ3 (accessed on 10 April 2021).
- Chung, L.W.; Sameera, W.M.C.; Ramozzi, R.; Page, A.J.; Hatanaka, M.; Petrova, G.P.; Harris, T.V.; Li, X.; Ke, Z.; Liu, F.; et al. The ONIOM Method and Its Applications. Chem. Rev. 2015, 115, 5678–5796. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Li, H.; He, H.; Zhang, X.; Wang, S. An Electrochemical Impedance Sensor for Simple and Specific Recognition of G-G Mismatches in DNA. Anal. Methods 2016, 8, 7413–7419. [Google Scholar] [CrossRef]
- Sun, H.; Bennett, R.J.; Maizels, N. The Saccharomyces Cerevisiae Sgs1 Helicase Efficiently Unwinds G-G Paired DNAs. Nucleic Acids Res. 1999, 27, 1978–1984. [Google Scholar] [CrossRef]
- Mondal, S.; Bhat, J.; Jana, J.; Mukherjee, M.; Chatterjee, S. Reverse Watson-Crick G-G base pair in G-quadruplex formation. Mol. BioSyst. 2016, 12, 18–22. [Google Scholar] [CrossRef]
- Brown, T.; Woodward, P.; Murphy, C.J.; Bursten, B.E.; LeMay, J.H.E. Chemistry: The Central Science, 11th ed.; Pearson Prentice Hall: Boston, MA, USA, 2009. [Google Scholar]
- Reece, J.B.; Urry, L.A.; Cain, M.L.; Wasserman, S.A.; Minorsky, P.V.; Jackson, R.B. Campbell Biology; Pearson: Boston, MA, USA, 2014; Volume 9. [Google Scholar]
- Arunan, E.; Desiraju, G.R.; Klein, R.A.; Sadlej, J.; Scheiner, S.; Alkorta, I.; Clary, D.C.; Crabtree, R.H.; Dannenberg, J.J.; Hobza, P.; et al. Defining the hydrogen bond: An account (IUPAC Technical Report). Pure Appl. Chem. 2011, 83, 1619–1636. [Google Scholar] [CrossRef]
- Desiraju, G.R. A Bond by Any Other Name. Angew. Chem. Int. Ed. 2011, 50, 52–59. [Google Scholar] [CrossRef]
- Horowitz, S.; Trievel, R.C. Carbon-Oxygen Hydrogen Bonding in Biological Structure and Function. J. Biol. Chem. 2012, 287, 41576–41582. [Google Scholar] [CrossRef]
- Nick Pace, C.; Scholtz, J.M.; Grimsley, G.R. Forces Stabilizing Proteins. FEBS Lett. 2014, 588, 2177–2184. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.; Loire, E.; Fridgen, T.D. Hydrogen Bonding in Alkali Metal Cation-Bound i-Motif-Like Dimers of 1-Methyl Cytosine: An IRMPD Spectroscopic and Computational Study. Phys. Chem. Chem. Phys. 2019, 21, 11103–11110. [Google Scholar] [CrossRef] [PubMed]
- Karas, L.J.; Wu, C.H.; Ottosson, H.; Wu, J.I. Electron-Driven Proton Transfer Relieves Excited-State Antiaromaticity in Photoexcited DNA Base Pairs. Chem. Sci. 2020, 11, 10071–10077. [Google Scholar] [CrossRef]
- Zhang, Y.; de La Harpe, K.; Beckstead, A.A.; Improta, R.; Kohler, B. UV-Induced Proton Transfer Between DNA Strands. J. Am. Chem. Soc. 2015, 137, 7059–7062. [Google Scholar] [CrossRef] [PubMed]
- Lewis, F.D.; Wasielewski, M.R. Dynamics and Efficiency of Photoinduced Charge Transport in DNA: Toward the Elusive Molecular Wire. Pure Appl. Chem. 2013, 85, 1379–1387. [Google Scholar] [CrossRef]
- Kumar, A.; Sevilla, M.D. Proton-Coupled Electron Transfer in DNA on Formation of Radiation-Produced Ion Radicals. Chem. Rev. 2010, 110, 7002–7023. [Google Scholar] [CrossRef] [PubMed]
- Black, P.J.; Bernhard, W.A. Excess Electron Trapping in Duplex DNA: Long Range Transfer via Stacked Adenines. J. Phys. Chem. B 2012, 116, 13211–13218. [Google Scholar] [CrossRef]
- Gorb, L.; Podolyan, Y.; Dziekonski, P.; Sokalski, W.A.; Leszczynski, J. Double-Proton Transfer in Adenine- Thymine and Guanine- Cytosine Base Pairs. A Post-Hartree- Fock ab Initio Study. J. Am. Chem. Soc. 2004, 126, 10119–10129. [Google Scholar] [CrossRef]
- Li, P.; Rangadurai, A.; Al-Hashimi, H.M.; Hammes-Schiffer, S. Environmental Effects on Guanine-Thymine Mispair Tautomerization Explored with Quantum Mechanical/Molecular Mechanical Free Energy Simulations. J. Am. Chem. Soc. 2020, 142, 11183–11191. [Google Scholar] [CrossRef]
- Shekaari, A.; Jafari, M. Modeling the Action of Environment on Proton Tunneling in the Adenine–Thymine Base Pair. Prog. Biophys. Mol. Biol. 2020, 150, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Marvi, M.; Ghadiri, M. A Mathematical Model for Vibration Behavior Analysis of DNA and Using a Resonant Frequency of DNA for Genome Engineering. Sci. Rep. 2020, 10, 3439. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, B.B.; Hettler, M.H.; Schön, G. Influence of Vibrational Modes on the Electronic Properties of DNA. Phys. Rev. B 2007, 75, 115125. [Google Scholar] [CrossRef]
- Jo, S.; Son, J.; Lee, B.H.; Dugasani, S.R.; Park, S.H.; Kim, M.K. Vibrational Characteristics of DNA Nanostructures Obtained Through a Mass-Weighted Chemical Elastic Network Model. RSC Adv. 2017, 7, 47190–47195. [Google Scholar] [CrossRef]
- Guchhait, B.; Liu, Y.; Siebert, T.; Elsaesser, T. Ultrafast Vibrational Dynamics of the DNA Backbone at Different Hydration Levels Mapped by Two-Dimensional Infrared Spectroscopy. Struct. Dyn. 2016, 3, 043202. [Google Scholar] [CrossRef] [PubMed]
- Poater, J.; Swart, M.; Bickelhaupt, F.M.; Fonseca Guerra, C. B-DNA structure and stability: The role of hydrogen bonding, π-π–stacking interactions, twist-angle, and solvation. Org. Biomol. Chem. 2014, 12, 4691–4700. [Google Scholar] [CrossRef]
- Barone, G.; Fonseca Guerra, C.; Bickelhaupt, F.M. B-DNA Structure and Stability as Function of Nucleic Acid Composition: Dispersion-Corrected DFT Study of Dinucleoside Monophosphate Single and Double Strands. ChemistryOpen 2013, 2, 186–193. [Google Scholar] [CrossRef]
- Kool, E.T. Hydrogen Bonding, Base Stacking, and Steric Effects in DNA Replication. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 1–22. [Google Scholar] [CrossRef]
- SantaLucia, J.; Hicks, D. The Thermodynamics of DNA Structural Motifs. Ann. Rev. Biophys. Biomol. Struct. 2004, 33, 415–440. [Google Scholar] [CrossRef]
- Yakovchuk, P.; Protozanova, E.; Frank-Kamenetskii, M.D. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006, 34, 564–574. [Google Scholar] [CrossRef]
- Zhang, T.B.; Zhang, C.L.; Dong, Z.L.; Guan, Y.F. Determination of Base Binding Strength and Base Stacking Interaction of DNA Duplex Using Atomic Force Microscope. Sci. Rep. 2015, 5, 9143. [Google Scholar] [CrossRef] [PubMed]
- Vologodskii, A.; Frank-Kamenetskii, M.D. DNA melting and energetics of the double helix. Phys. Life Rev. 2018, 25, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Florián, J.; Šponer, J.; Warshel, A. Thermodynamic Parameters for Stacking and Hydrogen Bonding of Nucleic Acid Bases in Aqueous Solution: Ab Initio/Langevin Dipoles Study. J. Phys. Chem. B 1999, 103, 884–892. [Google Scholar] [CrossRef]
- Oliva, R.; Cavallo, L.; Tramontano, A. Accurate energies of hydrogen bonded nucleic acid base pairs and triplets in tRNA tertiary interactions. Nucleic Acids Res. 2006, 34, 865–879. [Google Scholar] [CrossRef]
- van Mourik, T.; Hogan, S.W.L. DNA base stacking involving adenine and 2-aminopurine. Struct. Chem. 2016, 27, 145–158. [Google Scholar] [CrossRef]
- Lee, C.; Park, K.H.; Cho, M. Vibrational dynamics of DNA. I. Vibrational basis modes and couplings. J. Chem. Phys. 2006, 125, 114508. [Google Scholar] [CrossRef] [PubMed]
- Svozil, D.; Hobza, P.; Šponer, J. Comparison of Intrinsic Stacking Energies of Ten Unique Dinucleotide Steps in A-RNA and B-DNA Duplexes. Can We Determine Correct Order of Stability by Quantum-Chemical Calculations? J. Phys. Chem. B 2010, 114, 1191–1203. [Google Scholar] [CrossRef]
- Chakraborty, K.; Mantha, S.; Bandyopadhyay, S. Molecular dynamics simulation of a single-stranded DNA with heterogeneous distribution of nucleobases in aqueous medium. J. Chem. Phys. 2013, 139, 075103. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.F.; Andrews, C.T.; Elcock, A.H. Stacking Free Energies of All DNA and RNA Nucleoside Pairs and Dinucleoside-Monophosphates Computed Using Recently Revised AMBER Parameters and Compared with Experiment. J. Chem. Theory Comput. 2015, 11, 2315–2328. [Google Scholar] [CrossRef]
- Karwowski, B.T. The AT Interstrand Cross-Link: Structure, Electronic Properties, and Influence on Charge Transfer in dsDNA. Mol. Ther. Nucl. Acids 2018, 13, 665–685. [Google Scholar] [CrossRef] [PubMed]
- Zacharias, M. Base-Pairing and Base-Stacking Contributions to Double-Stranded DNA Formation. J. Phys. Chem. B 2020, 124, 10345–10352. [Google Scholar] [CrossRef] [PubMed]
- Rice, S.A.; Doty, P. The Thermal Denaturation of Desoxyribose Nucleic Acid. J. Am. Chem. Soc. 1957, 79, 3937–3947. [Google Scholar] [CrossRef]
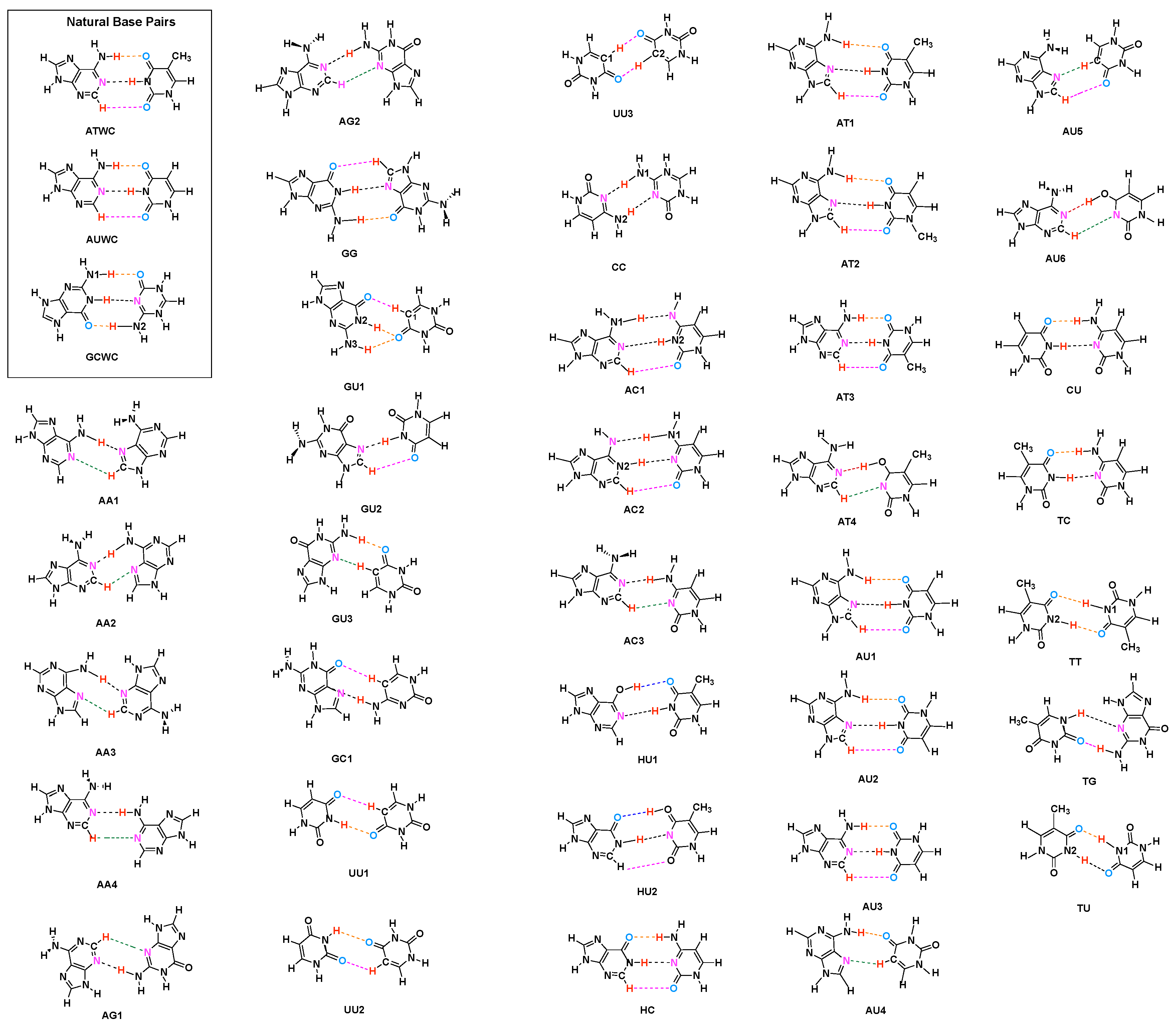
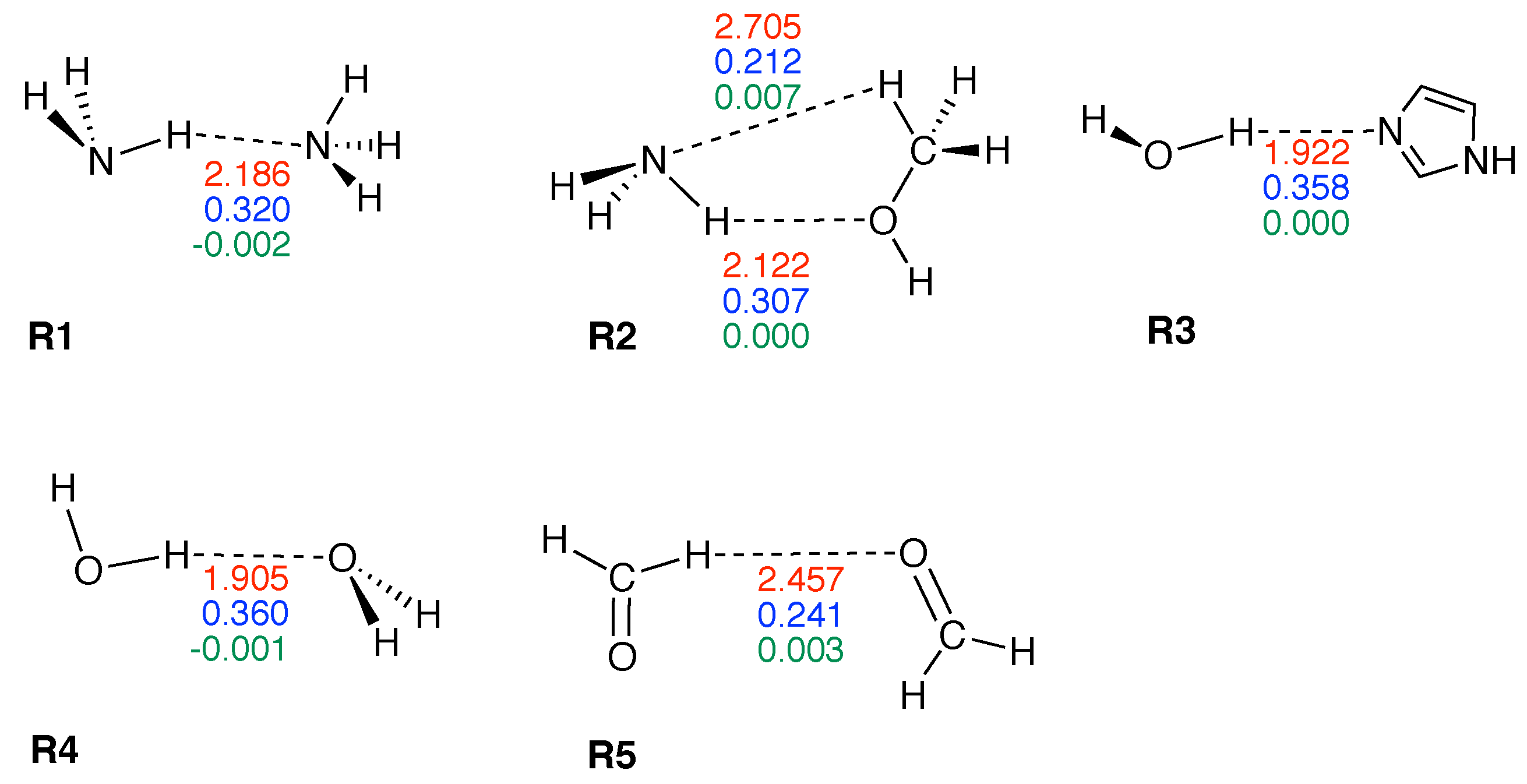
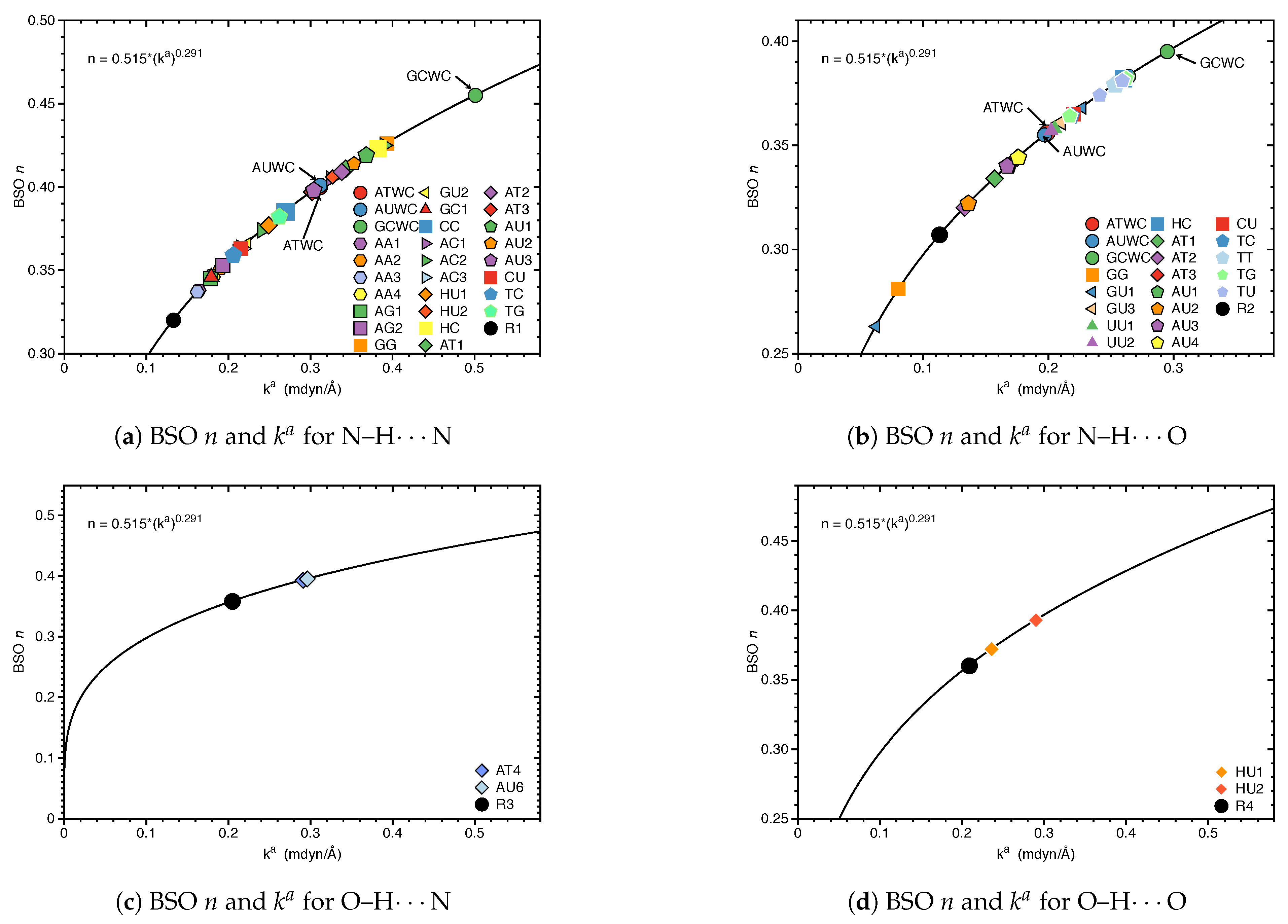
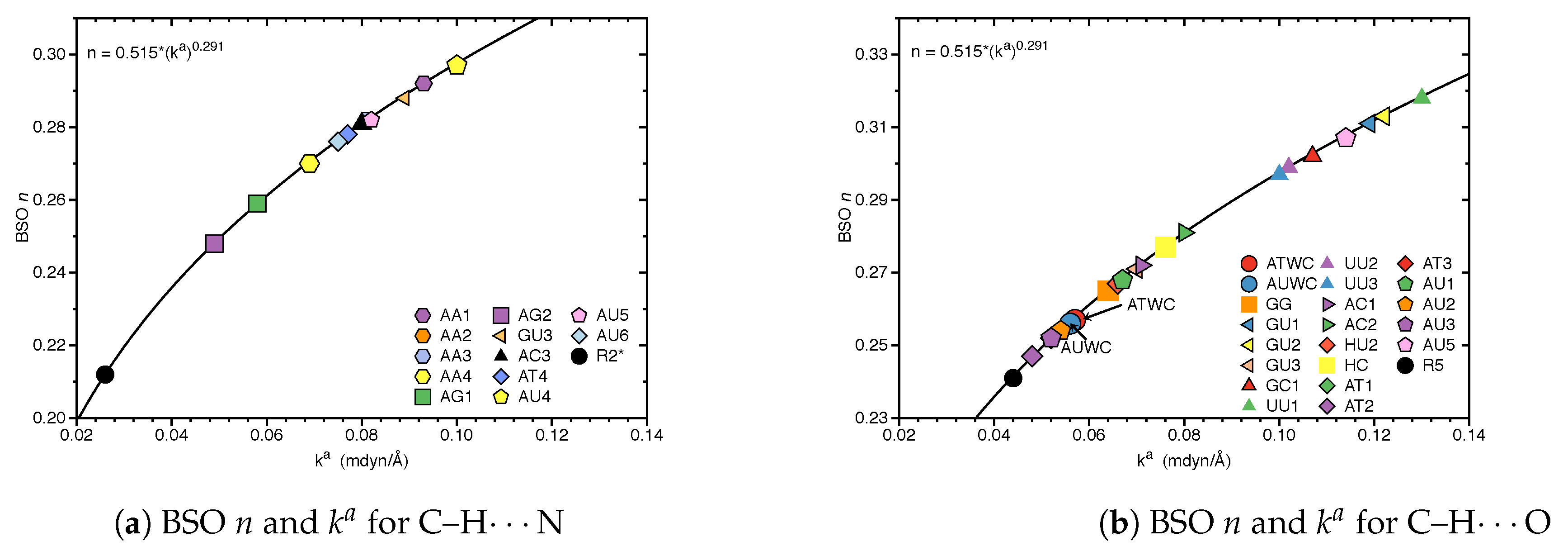
| Molecule | BSO n | R(HB) | / | |||
|---|---|---|---|---|---|---|
| R1 | 0.133 | 0.320 | 2.186 | 0.130 | −0.002 | −0.014 |
| R2 | 0.113 | 0.307 | 2.122 | 0.115 | 0.000 | 0.002 |
| R2* | 0.026 | 0.212 | 2.705 | 0.051 | 0.007 | 0.143 |
| R3 | 0.205 | 0.358 | 1.922 | 0.202 | 0.000 | 0.002 |
| R4 | 0.209 | 0.360 | 1.905 | 0.183 | −0.001 | −0.005 |
| R5 | 0.044 | 0.241 | 2.457 | 0.071 | 0.003 | 0.048 |
| Base Pair | X–H Bond | Y⋯H Bond | X⋯Y Distance | |||
|---|---|---|---|---|---|---|
| d | k | d | k | d | d | |
| (Å) | (mdyn/Å) | (Å) | (mdyn/Å) | (Å) | (Å) | |
| AT1 | ||||||
| N–H⋯O | 1.020 | 6.747 | 2.108 | 0.128 | 3.122 | 3.050 |
| N⋯H–N | 1.050 | 4.496 | 1.764 | 0.383 | 2.812 | 2.776 |
| C–H⋯O | 1.084 | 5.772 | 2.504 | 0.156 | 3.334 | 3.468 |
| AT2 | ||||||
| N–H⋯O | 1.015 | 7.014 | 2.144 | 0.102 | 3.143 | 2.981 |
| N⋯H–N | 1.065 | 3.338 | 1.638 | 0.420 | 2.700 | 2.761 |
| C–H⋯O | 1.085 | 5.701 | 2.385 | 0.141 | 3.234 | 3.475 |
| AT3 | ||||||
| N–H⋯O | 1.022 | 6.554 | 1.903 | 0.201 | 2.921 | - |
| N⋯H–N | 1.047 | 4.673 | 1.777 | 0.318 | 2.824 | - |
| C–H⋯O | 1.087 | 5.635 | 2.738 | 0.060 | 3.570 | - |
| GC1 | ||||||
| N–H⋯O | 1.020 | 6.508 | 1.855 | 0.267 | 2.866 | 2.880 |
| N–H⋯N | 1.035 | 5.508 | 1.831 | 0.511 | 2.851 | 2.912 |
| O⋯H–N | 1.030 | 5.663 | 1.748 | 0.282 | 2.763 | 2.852 |
| GC2 | ||||||
| N–H⋯O | 1.027 | 6.096 | 1.758 | 0.387 | 2.784 | 2.789 |
| N–H⋯N | 1.038 | 5.379 | 1.855 | 0.517 | 2.892 | 2.875 |
| O⋯H–N | 1.025 | 6.241 | 1.868 | 0.230 | 2.888 | 2.839 |
| GC3 | ||||||
| N–H⋯O | 1.022 | 6.418 | 1.866 | 0.264 | 2.888 | - |
| N–H⋯N | 1.033 | 5.695 | 1.891 | 0.486 | 2.924 | - |
| O⋯H–N | 1.034 | 5.496 | 1.750 | 0.283 | 2.785 | - |
| NBO Atomic Charge (e) | H (Hartree/Å) | ||||
|---|---|---|---|---|---|
| Base Pair | q | q | q | X–H | Y⋯H |
| AT1 | |||||
| N–H⋯O | −0.836 | 0.449 | −0.665 | −3.3479 | −0.0047 |
| N⋯H–N | −0.605 | 0.477 | −0.687 | −3.0368 | −0.0189 |
| C–H⋯O | 0.275 | 0.243 | −0.671 | −2.1986 | 0.0054 |
| AT2 | |||||
| N–H⋯O | −0.834 | 0.454 | −0.708 | −3.3701 | −0.0027 |
| N⋯H–N | −0.606 | 0.477 | −0.685 | −2.8613 | −0.0661 |
| C–H⋯O | 0.289 | 0.239 | −0.692 | −2.1885 | 0.0040 |
| AT3 | |||||
| N–H⋯O | −0.829 | 0.458 | −0.672 | −3.2986 | −0.0020 |
| N⋯H–N | −0.615 | 0.477 | −0.688 | −3.0631 | −0.0169 |
| C–H⋯O | 0.271 | 0.237 | −0.668 | −2.1493 | 0.0061 |
| GC1 | |||||
| N–H⋯O | −0.835 | 0.456 | −0.702 | −3.3229 | 0.0007 |
| N–H⋯N | −0.658 | 0.472 | −0.655 | −3.2082 | −0.0074 |
| O⋯H–N | −0.754 | 0.471 | −0.791 | −3.2149 | 0.0027 |
| GC2 | |||||
| N–H⋯O | −0.851 | 0.454 | −0.696 | −3.2736 | 0.0027 |
| N–H⋯N | −0.668 | 0.472 | −0.671 | −3.1751 | −0.0067 |
| O⋯H–N | −0.722 | 0.466 | −0.800 | −3.2716 | 0.0000 |
| GC3 | |||||
| N–H⋯O | −0.860 | 0.458 | −0.711 | −3.2831 | −0.0007 |
| N–H⋯N | −0.677 | 0.467 | −0.661 | −3.2291 | −0.0007 |
| O⋯H–N | −0.686 | 0.468 | −0.810 | −3.1677 | −0.0047 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beiranvand, N.; Freindorf, M.; Kraka, E. Hydrogen Bonding in Natural and Unnatural Base Pairs—A Local Vibrational Mode Study. Molecules 2021, 26, 2268. https://doi.org/10.3390/molecules26082268
Beiranvand N, Freindorf M, Kraka E. Hydrogen Bonding in Natural and Unnatural Base Pairs—A Local Vibrational Mode Study. Molecules. 2021; 26(8):2268. https://doi.org/10.3390/molecules26082268
Chicago/Turabian StyleBeiranvand, Nassim, Marek Freindorf, and Elfi Kraka. 2021. "Hydrogen Bonding in Natural and Unnatural Base Pairs—A Local Vibrational Mode Study" Molecules 26, no. 8: 2268. https://doi.org/10.3390/molecules26082268
APA StyleBeiranvand, N., Freindorf, M., & Kraka, E. (2021). Hydrogen Bonding in Natural and Unnatural Base Pairs—A Local Vibrational Mode Study. Molecules, 26(8), 2268. https://doi.org/10.3390/molecules26082268