Functional Characterization of an Anthocyanin Dimalonyltransferase in Maize
Abstract
1. Introduction
2. Results
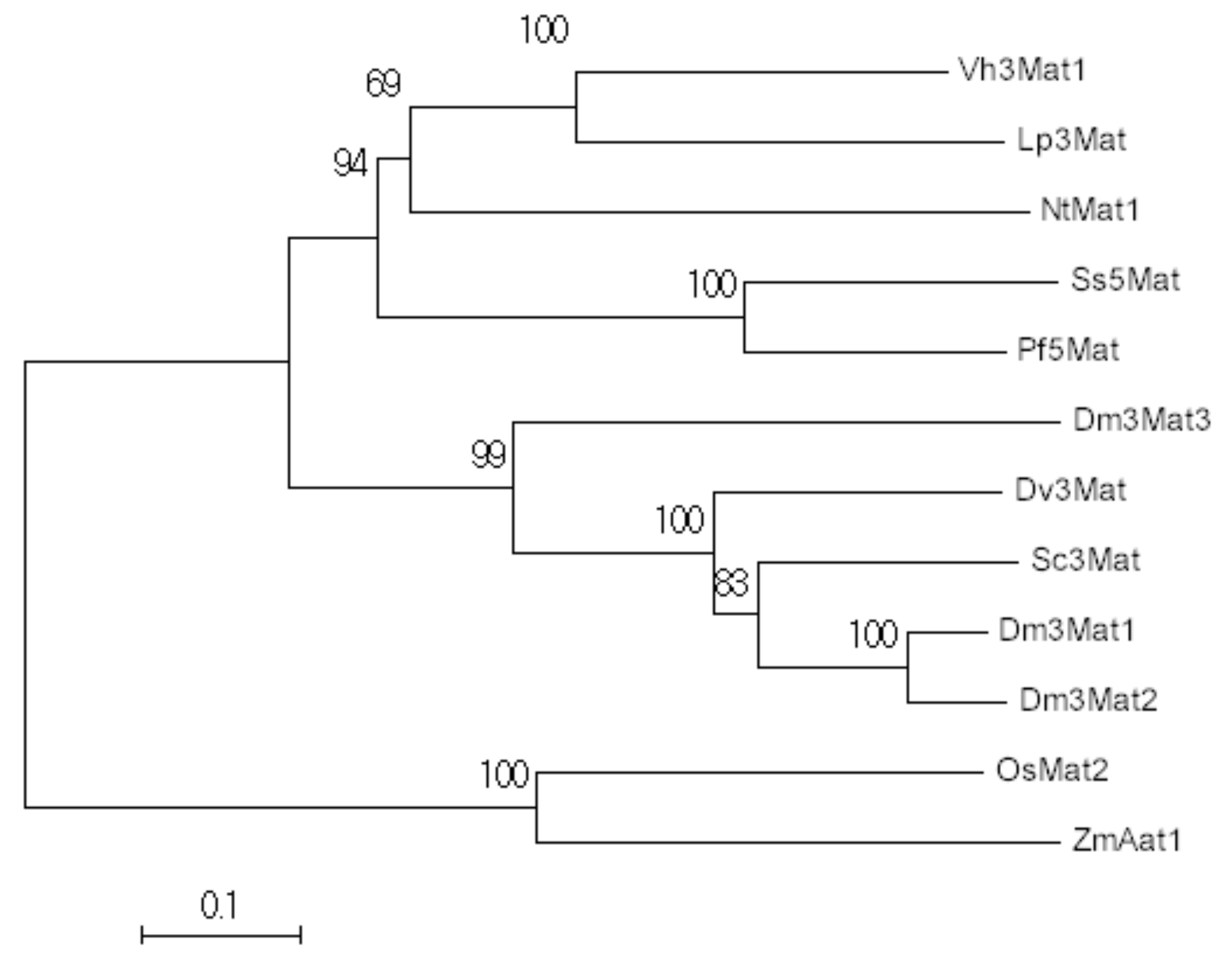
2.1. Phylogenetic Analysis
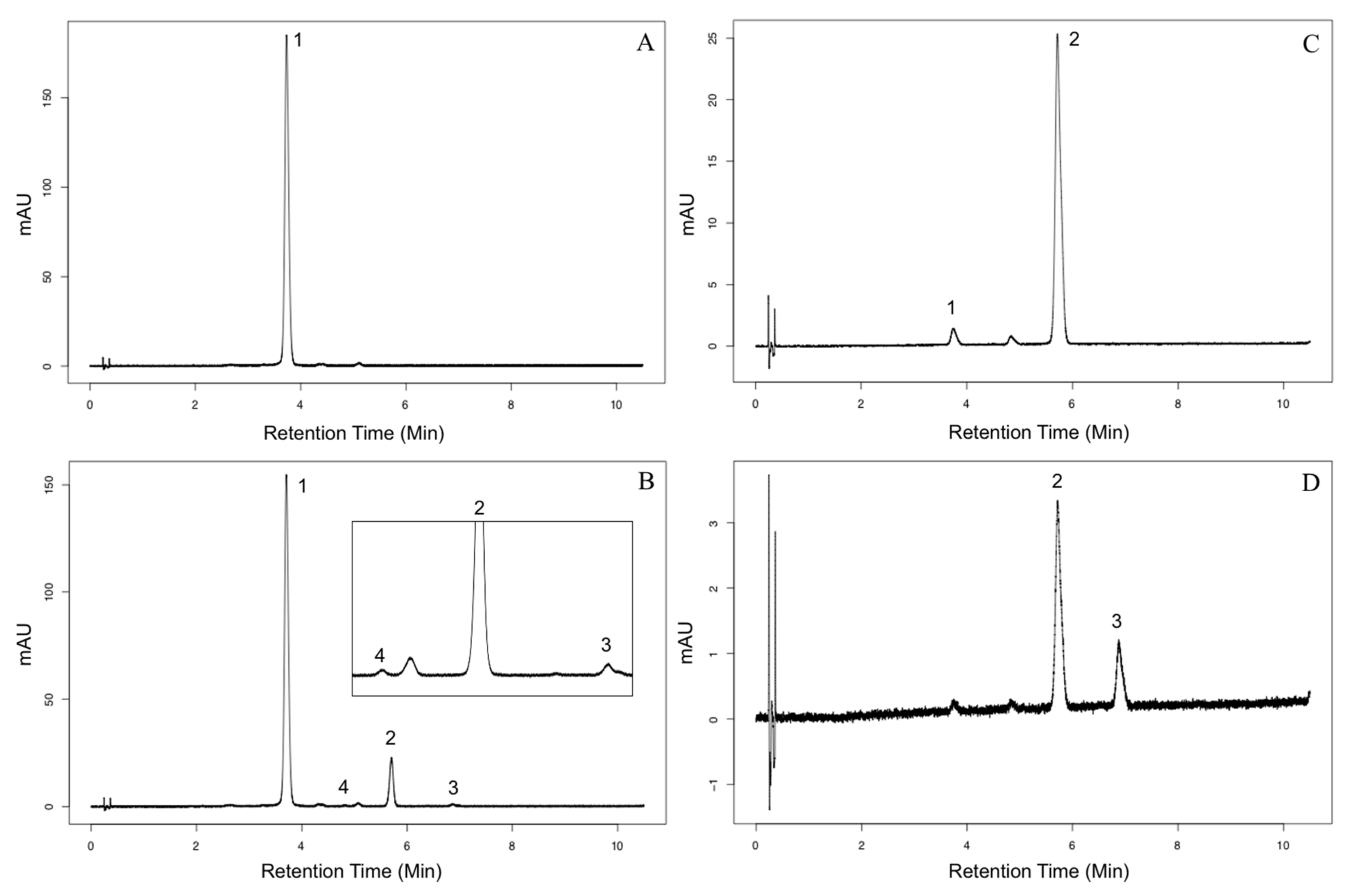
2.2. Characterization of Aat1 Recombinant Protein
3. Materials and Methods
3.1. Substrates
3.2. Cloning
3.3. Affinity Tag Purification
3.4. Enzyme Assays
3.5. Liquid Chromatography and Mass Spectrometry
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Paulsmeyer, M.; Chatham, L.; Becker, T.; West, M.; West, L.; Juvik, J. Survey of Anthocyanin Composition and Concentration in Diverse Maize Germplasms. J. Agric. Food Chem. 2017, 65, 4341–4350. [Google Scholar] [CrossRef]
- Somavat, P.; Li, Q.; de Mejia, E.G.; Liu, W.; Singh, V. Coproduct Yield Comparisons of Purple, Blue and Yellow Dent Corn for Various Milling Processes. Ind. Crops Prod. 2016, 87, 266–272. [Google Scholar] [CrossRef]
- Cevallos-Casals, B.A.; Cisneros-Zevallos, L. Stoichiometric and Kinetic Studies of Phenolic Antioxidants from Andean Purple Corn and Red-Fleshed Sweetpotato. J. Agric. Food Chem. 2003, 51, 3313–3319. [Google Scholar] [CrossRef] [PubMed]
- Lao, F.; Sigurdson, G.T.; Giusti, M.M. Health Benefits of Purple Corn (Zea mays, L.) Phenolic Compounds. Compr. Rev. Food Sci. Food Saf. 2017, 16, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.-L.; Yu, Y.-Q.; Chen, Z.-J.; Wen, G.-S.; Wei, F.-G.; Zheng, Q.; Wang, C.-D.; Xiao, X.-L. Stability-Increasing Effects of Anthocyanin Glycosyl Acylation. Food Chem. 2017, 214, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Paulsmeyer, M.N.; Brown, P.J.; Juvik, J.A. Discovery of Anthocyanin Acyltransferase1 (AAT1) in Maize Using Genotyping-by-Sequencing (GBS). G3 Genes Genomes Genet. 2018, 8, 3669–3678. [Google Scholar] [CrossRef]
- Suzuki, H.; Nakayama, T.; Yonekura-Sakakibara, K.; Fukui, Y.; Nakamura, N.; Yamaguchi, M.; Tanaka, Y.; Kusumi, T.; Nishino, T. CDNA Cloning, Heterologous Expressions, and Functional Characterization of Malonyl-Coenzyme A:Anthocyanidin 3-O-Glucoside-6”-O-Malonyltransferase from Dahlia Flowers. Plant. Physiol. 2002, 130, 2142–2151. [Google Scholar] [CrossRef]
- Zhao, J.; Huhman, D.; Shadle, G.; He, X.-Z.; Sumner, L.W.; Tang, Y.; Dixon, R.A. MATE2 Mediates Vacuolar Sequestration of Flavonoid Glycosides and Glycoside Malonates in Medicago Truncatula. Plant. Cell 2011, 23, 1536–1555. [Google Scholar] [CrossRef] [PubMed]
- Bąkowska-Barczak, A. Acylated Anthocyanins as Stable, Natural Food Colorants. Pol. J. Food Nutr. Sci. 2005, 55, 107–116. [Google Scholar]
- Suzuki, H.; Nakayama, T.; Yamaguchi, M.; Nishino, T. CDNA Cloning and Characterization of Two Dendranthema × Morifolium Anthocyanin Malonyltransferases with Different Functional Activities. Plant. Sci. 2004, 166, 89–96. [Google Scholar] [CrossRef]
- D’Auria, J.C. Acyltransferases in Plants: A Good Time to Be BAHD. Curr. Opin. Plant. Biol. 2006, 9, 331–340. [Google Scholar] [CrossRef]
- Manjasetty, B.A.; Yu, X.-H.; Panjikar, S.; Taguchi, G.; Chance, M.R.; Liu, C.-J. Structural Basis for Modification of Flavonol and Naphthol Glucoconjugates by Nicotiana Tabacum Malonyltransferase (NtMaT1). Planta 2012, 236, 781–793. [Google Scholar] [CrossRef]
- Unno, H.; Ichimaida, F.; Suzuki, H.; Takahashi, S.; Tanaka, Y.; Saito, A.; Nishino, T.; Kusunoki, M.; Nakayama, T. Structural and Mutational Studies of Anthocyanin Malonyltransferases Establish the Features of BAHD Enzyme Catalysis. J. Biol. Chem. 2007, 282, 15812–15822. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Nakayama, T.; Nishino, T. Proposed Mechanism and Functional Amino Acid Residues of Malonyl-CoA:Anthocyanin 5-O-Glucoside-6′’’-O-Malonyltransferase from Flowers of Salvia Splendens, a Member of the Versatile Plant Acyltransferase Family. Biochemistry 2003, 42, 1764–1771. [Google Scholar] [CrossRef]
- Kim, B.-G.; Lee, Y.; Hur, H.-G.; Lim, Y.; Ahn, J.-H. Flavonoid 3′-O-Methyltransferase from Rice: CDNA Cloning, Characterization and Functional Expression. Phytochemistry 2006, 67, 387–394. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, S.K.; Kim, J.-H.; Kim, B.-G.; Ahn, J.-H. Molecular Characterization of Flavonoid Malonyltransferase from Oryza Sativa. Plant. Physiol. Biochem. 2009, 47, 991–997. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Zuckerkandl, E.; Pauling, L. Evolutionary Divergence and Convergence in Proteins. Evol. Genes Proteins 1965, 97, 97–166. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Suzuki, H.; Nakayama, T.; Yonekura-Sakakibara, K.; Fukui, Y.; Nakamura, N.; Nakao, M.; Tanaka, Y.; Yamaguchi, M.-a.; Kusumi, T.; Nishino, T. Malonyl-CoA:Anthocyanin 5-O-Glucoside-6′’’-O-Malonyltransferase from Scarlet Sage (Salvia splendens) Flowers: Enzyme Purification, Gene Cloning, Expression, and Characterization. J. Biol. Chem. 2001, 276, 49013–49019. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Sawada, S.; Yonekura-Sakakibara, K.; Nakayama, T.; Yamaguchi, M.; Nishino, T. Identification of a CDNA Encoding Malonyl-Coenzyme A: Anthocyanidin 3-O-Glucoside 6”-O-Malonyltransferase from Cineraria (Senecio cruentus) Flowers. Plant. Biotechnol. 2003, 20, 229–234. [Google Scholar] [CrossRef][Green Version]
- Hong, Y.; Tang, X.; Huang, H.; Zhang, Y.; Dai, S. Transcriptomic Analyses Reveal Species-Specific Light-Induced Anthocyanin Biosynthesis in Chrysanthemum. BMC Genom. 2015, 16, 202. [Google Scholar] [CrossRef]
- Novagen, Inc. PET System Manual, 11th ed.; EMD Chemicals Inc.: Gibbstown, NJ, USA, 2011. [Google Scholar]
- Christensen, D.G.; Orr, J.S.; Rao, C.V.; Wolfe, A.J. Increasing Growth Yield and Decreasing Acetylation in Escherichia Coli by Optimizing the Carbon-to-Magnesium Ratio in Peptide-Based Media. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Johnson, K.A. New Standards for Collecting and Fitting Steady State Kinetic Data. Beilstein J. Org. Chem. 2019, 15, 16–29. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
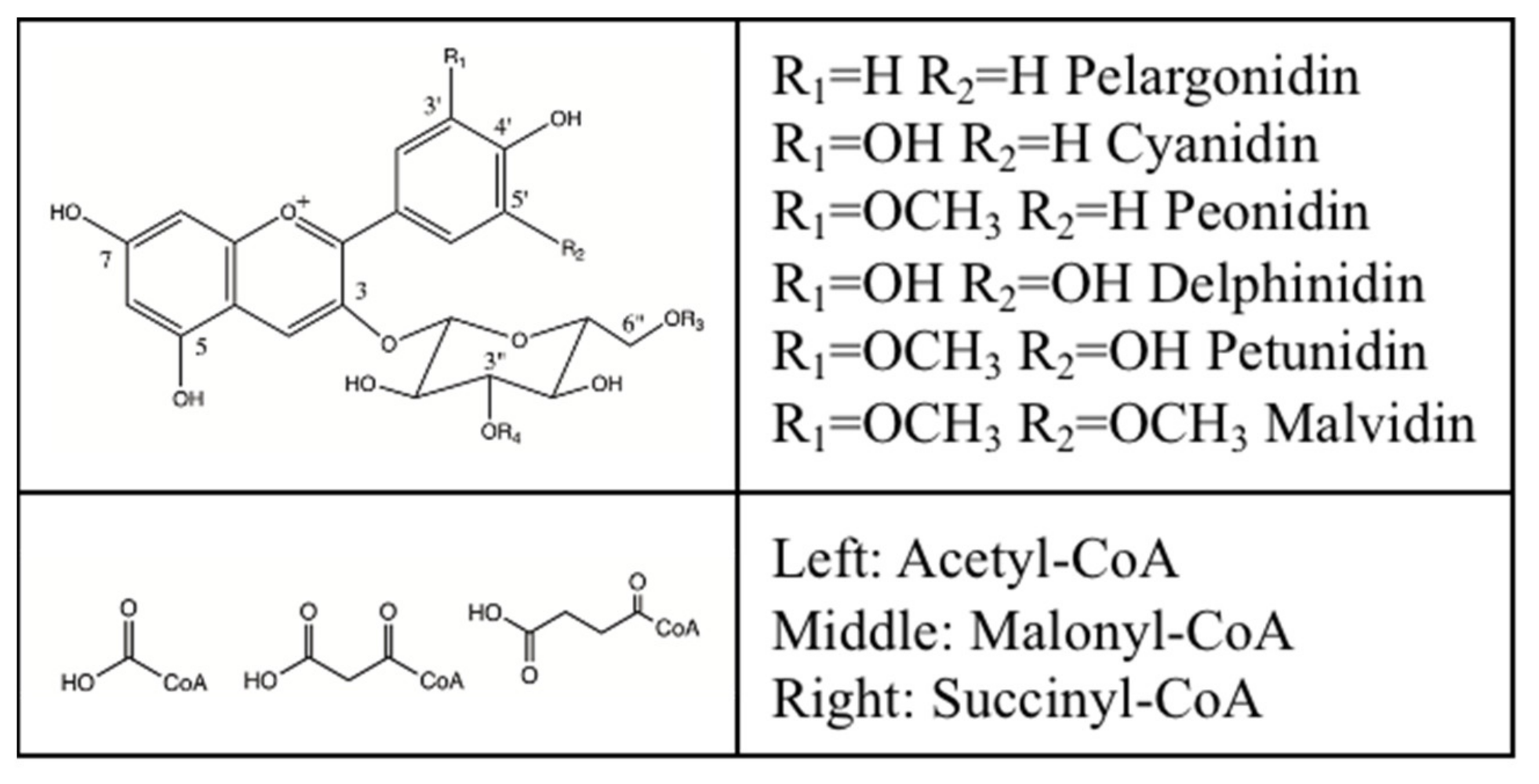
| Compound | Relative Activity a | Km | Kcat | Kcat/Km |
|---|---|---|---|---|
| Units | % | µM | s−1 × 1000 | µM−1·s−1 × 1000 |
| Acyl Acceptors b | ||||
| Cyanidin 3-O-Glucoside | 100 ± 2.07 | 13.06 ± 0.23 | 6.233 ± 0.145 | 0.477 ± 0.034 |
| Pelargonidin 3-O-Glucoside | 89.8 ± 3.24 | 8.92 ± 0.41 | 5.594 ± 0.074 | 0.627 ± 0.030 |
| Peonidin 3-O-Glucoside | 43 ± 1.55 | 5.14 ± 0.93 | 2.596 ± 0.028 | 0.505 ± 0.026 |
| Delphinidin 3-O-Glucoside | 20.1 ± 0.74 | ND | ND | ND |
| Cyanidin 3-O-6”-Malonylglucoside | ND | 1.33 ± 3.97 | 0.725 ± 0.044 | 0.543 ± 0.173 |
| Acyl Donors c | ||||
| Malonyl-CoA | 100 ± 2.07 | 40.12 ± 0.04 | 8.043 ± 0.751 | 0.200 ± 0.026 |
| Succinyl-CoA | 18.5 ± 0.67 | 60.09 ± 0.02 | 1.607 ± 0.194 | 0.027 ± 0.004 |
| Acetyl-CoA | <0.1 | ND | ND | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paulsmeyer, M.; Juvik, J. Functional Characterization of an Anthocyanin Dimalonyltransferase in Maize. Molecules 2021, 26, 2020. https://doi.org/10.3390/molecules26072020
Paulsmeyer M, Juvik J. Functional Characterization of an Anthocyanin Dimalonyltransferase in Maize. Molecules. 2021; 26(7):2020. https://doi.org/10.3390/molecules26072020
Chicago/Turabian StylePaulsmeyer, Michael, and John Juvik. 2021. "Functional Characterization of an Anthocyanin Dimalonyltransferase in Maize" Molecules 26, no. 7: 2020. https://doi.org/10.3390/molecules26072020
APA StylePaulsmeyer, M., & Juvik, J. (2021). Functional Characterization of an Anthocyanin Dimalonyltransferase in Maize. Molecules, 26(7), 2020. https://doi.org/10.3390/molecules26072020





