Anti-Cancer Effects of Carnosine—A Dipeptide Molecule
Abstract
1. Introduction
2. Results
2.1. Carnosine Inhibits Cancer Cell Proliferation
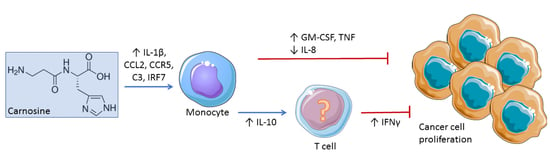
2.2. Carnosine Upregulates Expression of Proinflammatory Molecules in U937 Cell
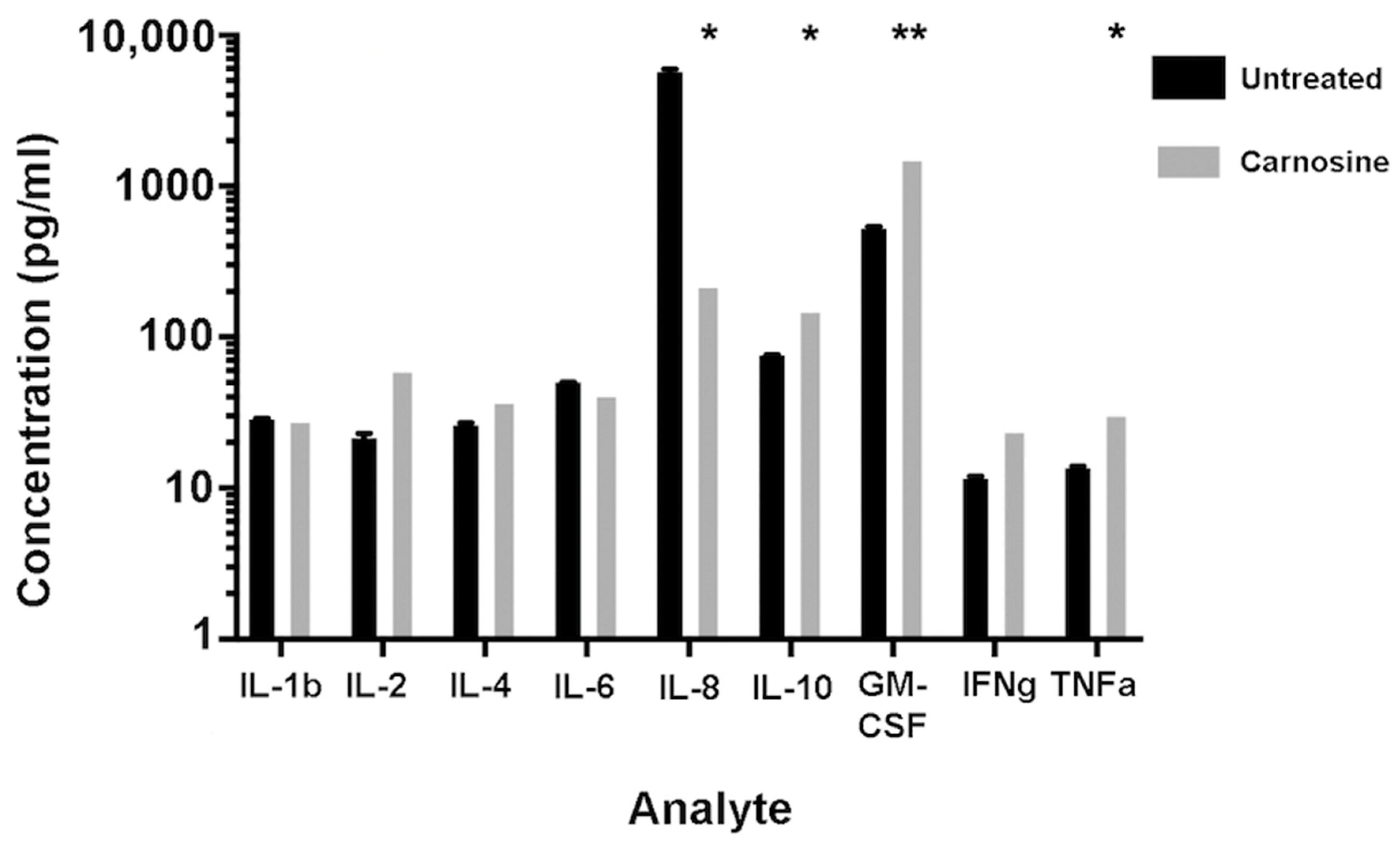
2.3. Carnosine Modulates Cytokine Secretion from U937 Cells
2.4. Carnosine Alters U937 Differentiation and Phenotype
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Proliferation Assay
4.3. Gene Array
4.4. Cytokine Secretion
4.5. Flow Cytometry
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sundstrom, C.; Nilsson, K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 1976, 17, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Riddy, D.M.; Goy, E.; Delerive, P.; Summers, R.J.; Sexton, P.M.; Langmead, C.J. Comparative genotypic and phenotypic analysis of human peripheral blood monocytes and surrogate monocyte-like cell lines commonly used in metabolic disease research. PLoS ONE 2018, 13, e0197177. [Google Scholar] [CrossRef]
- Chanput, W.; Peters, V.; Wichers, H. THP-1 and U937 Cells. In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 147–159. [Google Scholar]
- Baye, E.; Ukropcova, B.; Ukropec, J.; Hipkiss, A.; Aldini, G.; De Courten, B. Physiological and therapeutic effects of carnosine on cardiometabolic risk and disease. Amino Acids 2016, 48, 1131–1149. [Google Scholar] [CrossRef]
- Hipkiss, A.R.; Preston, J.E.; Himsworth, D.T.M.; Worthington, V.C.; Keown, M.; Michaelis, J.; Lawrence, J.; Mateen, A.; Allende, L.; Eagles, P.A.; et al. Pluripotent protective effects of carnosine, a naturally occurring dipeptide. Ann. N. Y. Acad. Sci. 1998, 854, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Derave, W.; De Courten, B.; Baba, S.P. An update on carnosine and anserine research. Amino Acids 2019, 51, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Holliday, R.; McFarland, G.A. Inhibition of the growth of transformed and neoplastic cells by the dipeptide carnosine. Br. J. Cancer 1996, 73, 966–971. [Google Scholar] [CrossRef]
- McFarland, G.A.; Holliday, R. Retardation of the senescence of cultured human diploid fibroblasts by carnosine. Exp. Cell Res. 1994, 212, 167–175. [Google Scholar] [CrossRef]
- Gaunitz, F.; Hipkiss, A.R. Carnosine and cancer: A perspective. Amino Acids 2012, 43, 135–142. [Google Scholar] [CrossRef]
- Renner, C.; Seyffarth, A.; de Arriba, S.G.; Meixensberger, J.; Gebhardt, R.; Gaunitz, F. Carnosine Inhibits Growth of Cells Isolated from Human Glioblastoma Multiforme. Int. J. Pept. Res. Ther. 2008, 14, 127–135. [Google Scholar] [CrossRef]
- Iovine, B.; Iannella, M.L.; Nocella, F.; Pricolo, M.R.; Bevilacqua, M.A. Carnosine inhibits KRAS-mediated HCT116 proliferation by affecting ATP and ROS production. Cancer Lett. 2012, 315, 122–128. [Google Scholar] [CrossRef]
- Shen, Y.; Yang, J.; Li, J.; Shi, X.; Ouyang, L.; Tian, Y.; Lu, J. Carnosine Inhibits the Proliferation of Human Gastric Cancer SGC-7901 Cells through Both of the Mitochondrial Respiration and Glycolysis Pathways. PLoS ONE 2014, 9, e104632. [Google Scholar] [CrossRef]
- Mikula-Pietrasik, J.; Ksiazek, K. L-Carnosine Prevents the Pro-cancerogenic Activity of Senescent Peritoneal Mesothelium Towards Ovarian Cancer Cells. Anticancer Res. 2016, 36, 665–671. [Google Scholar]
- Hipkiss, A.R.; Baye, E.; de Courten, B. Carnosine and the processes of ageing. Maturitas 2016, 93, 28–33. [Google Scholar] [CrossRef]
- Kohen, R.; Yamamoto, Y.; Cundy, K.C.; Ames, B.N. Antioxidant activity of carnosine, homocarnosine, and anserine present in muscle and brain. Proc. Natl. Acad. Sci. USA 1988, 85, 3175–3179. [Google Scholar] [CrossRef] [PubMed]
- Boldyrev, A.; Stvolinsky, S.L.; Fedorova, T.N.; Suslina, Z.A. Carnosine as a natural antioxidant and geroprotector: From molecular mechanisms to clinical trials. Rejuvenation Res. 2010, 13, 156–158. [Google Scholar] [CrossRef] [PubMed]
- Baraniuk, J.N.; El-Amin, S.; Corey, R.; Rayhan, R.U.; Timbol, C.R. Carnosine treatment for gulf war illness: A randomized controlled trial. Glob. J. Health Sci. 2013, 5, 69. [Google Scholar] [CrossRef] [PubMed]
- De Courten, B.; Jakubova, M.; De Courten, M.P.; Kukurova, I.J.; Vallova, S.; Krumpolec, P.; Valkovic, L.; Kurdiova, T.; Garzon, D.; Barbaresi, S.; et al. Effects of carnosine supplementation on glucose metabolism: Pilot clinical trial. Obesity 2016, 24, 1027–1034. [Google Scholar] [CrossRef]
- Haabeth, O.A.; Lorvik, K.B.; Yagita, H.; Bogen, B.; Corthay, A. Interleukin-1 is required for cancer eradication mediated by tumor-specific Th1 cells. Oncoimmunology 2016, 5, e1039763. [Google Scholar] [CrossRef]
- Li, M.; Knight, D.A.; Snyder, L.A.; Smyth, M.J.; Stewart, T.J. A role for CCL2 in both tumor progression and immunosurveillance. Oncoimmunology 2013, 2, e25474. [Google Scholar] [CrossRef]
- Singh, S.K.; Mishra, M.K.; Eltoum IE, A.; Bae, S.; Lillard, J.W.; Singh, R. CCR5/CCL5 axis interaction promotes migratory and invasiveness of pancreatic cancer cells. Sci. Rep. 2018, 8, 1323. [Google Scholar] [CrossRef]
- Gonzalez-Martin, A.; Mira, E.; Manes, S. CCR5 in cancer immunotherapy: More than an “attractive” receptor for T cells. Oncoimmunology 2012, 1, 106–108. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Martin, A.; Mira, E.; Manes, S. CCR5 as a potential target in cancer therapy: Inhibition or stimulation? Anticancer Agents Med. Chem. 2012, 12, 1045–1057. [Google Scholar] [CrossRef]
- Kourtzelis, I.; Rafail, S. The dual role of complement in cancer and its implication in anti-tumor therapy. Ann. Transl. Med. 2016, 4, 265. [Google Scholar] [CrossRef] [PubMed]
- Bidwell, B.N.; Slaney, C.Y.; Withana, N.P.; Forster, S.; Cao, Y.; Loi, S.; Andrews, D.; Mikeska, T.; Mangan, N.E.; Samarajiwa, S.A.; et al. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat. Med. 2012, 18, 1224–1231. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, W.; Zhu, W.; Meng, H.; Chen, J.; Zhang, J. Overexpression of Interferon Regulatory Factor 7 (IRF7) Reduces Bone Metastasis of Prostate Cancer Cells in Mice. Oncol. Res. 2017, 25, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.M.; Ojcius, D.M.; Garaud, F.; Roth, C.; Maxwell, E.; Li, Z.; Rong, H.; Chen, J.; Wang, X.Y.; Catino, J.J.; et al. Interleukin-10 inhibits tumor metastasis through an NK cell-dependent mechanism. J. Exp. Med. 1996, 184, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Jackson, M.J.; Kundu, N.; Fulton, A.M. Interleukin-10 gene transfer activates interferon-gamma and the interferon-gamma-inducible genes Gbp-1/Mag-1 and Mig-1 in mammary tumors. Int. J. Cancer 1999, 80, 624–629. [Google Scholar] [CrossRef]
- Groux, H.; Cottrez, F.; Rouleau, M.; Mauze, S.; Antonenko, S.; Hurst, S.; McNeil, T.; Bigler, M.; Roncarolo, M.-G.; Coffman, R.L. A transgenic model to analyze the immunoregulatory role of IL-10 secreted by antigen-presenting cells. J. Immunol. 1999, 162, 1723–1729. [Google Scholar]
- Mumm, J.B.; Emmerich, J.; Zhang, X.; Chan, I.; Wu, L.; Mauze, S.; Blaisdell, S.; Basham, B.; Dai, J.; Grein, J.; et al. IL-10 elicits IFNgamma-dependent tumor immune surveillance. Cancer Cell 2011, 20, 781–796. [Google Scholar] [CrossRef]
- Yan, W.L.; Shen, K.Y.; Tien, C.Y.; Chen, Y.A.; Liu, S.J. Recent progress in GM-CSF-based cancer immunotherapy. Immunotherapy 2017, 9, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Josephs, S.F.; Ichim, T.E.; Prince, S.M.; Kesari, S.; Marincola, F.M.; Escobedo, A.R.; Jafri, A. Unleashing endogenous TNF-alpha as a cancer immunotherapeutic. J. Transl. Med. 2018, 16, 242. [Google Scholar] [CrossRef] [PubMed]
- Yuan, A.; Chen, J.J.; Yao, P.L.; Yang, P.C. The role of interleukin-8 in cancer cells and microenvironment interaction. Front. Biosci. 2005, 10, 853–865. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of Cell Viability by the MTT Assay. Cold Spring Harb. Protoc. 2018, 2018, pdb.prot095505. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prakash, M.D.; Fraser, S.; Boer, J.C.; Plebanski, M.; de Courten, B.; Apostolopoulos, V. Anti-Cancer Effects of Carnosine—A Dipeptide Molecule. Molecules 2021, 26, 1644. https://doi.org/10.3390/molecules26061644
Prakash MD, Fraser S, Boer JC, Plebanski M, de Courten B, Apostolopoulos V. Anti-Cancer Effects of Carnosine—A Dipeptide Molecule. Molecules. 2021; 26(6):1644. https://doi.org/10.3390/molecules26061644
Chicago/Turabian StylePrakash, Monica D., Sarah Fraser, Jennifer C. Boer, Magdalena Plebanski, Barbora de Courten, and Vasso Apostolopoulos. 2021. "Anti-Cancer Effects of Carnosine—A Dipeptide Molecule" Molecules 26, no. 6: 1644. https://doi.org/10.3390/molecules26061644
APA StylePrakash, M. D., Fraser, S., Boer, J. C., Plebanski, M., de Courten, B., & Apostolopoulos, V. (2021). Anti-Cancer Effects of Carnosine—A Dipeptide Molecule. Molecules, 26(6), 1644. https://doi.org/10.3390/molecules26061644