Conditionally Activated (“Caged”) Oligonucleotides
Abstract
1. Introduction
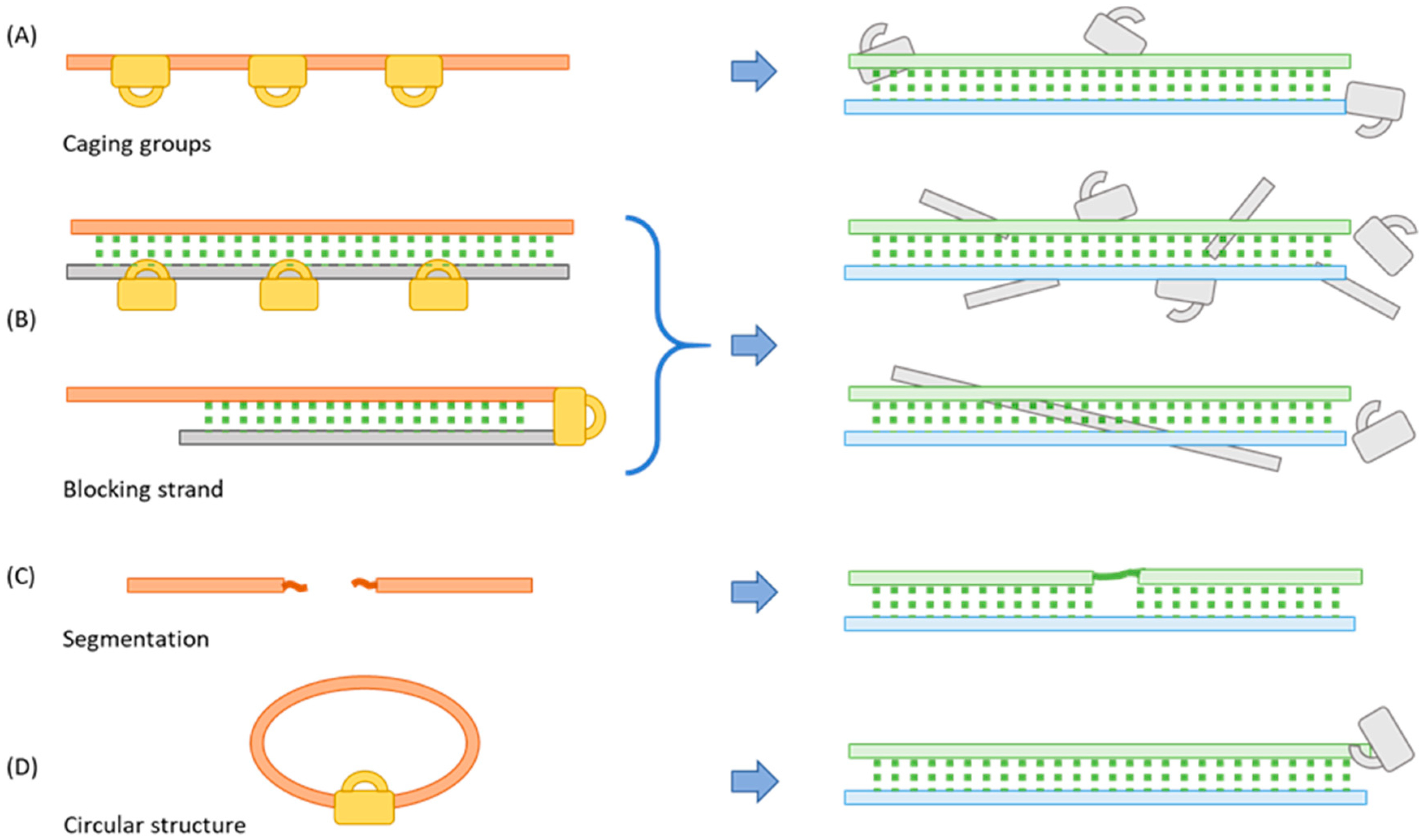
2. Methods for Blocking the Function of Oligos
2.1. Incorporating a Caging Group
2.2. Binding to a Complementary Blocking Strand
2.3. Segmentation
2.4. Cyclization
3. Activation Stimuli
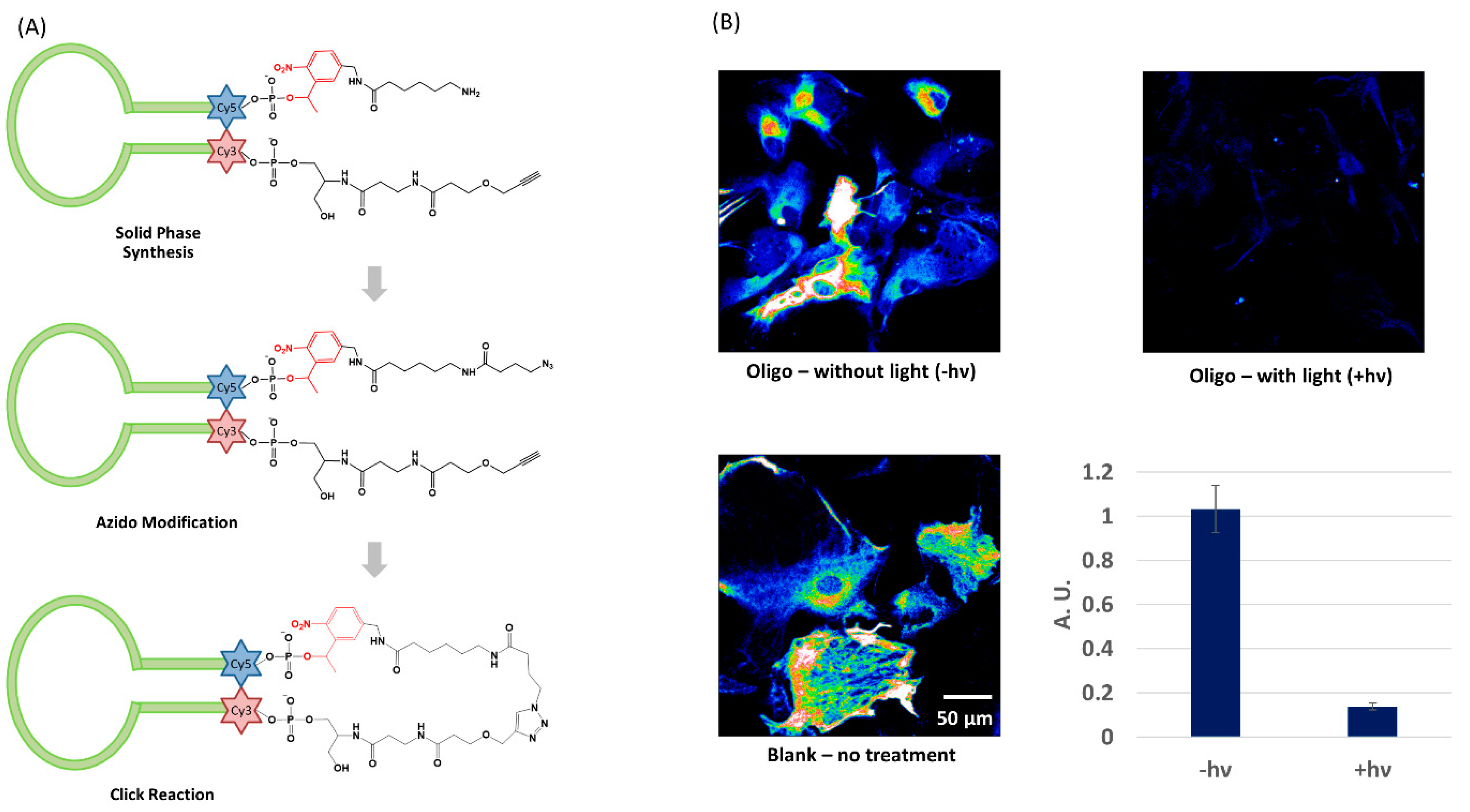
3.1. Light Activation
3.2. Cellular Oxidation and Reduction (Redox) Activation
3.3. Enzyme Activation
3.4. Heat Activation
4. Recent Applications of Caged Oligos
4.1. Circular Light-Activated Antisense DNA
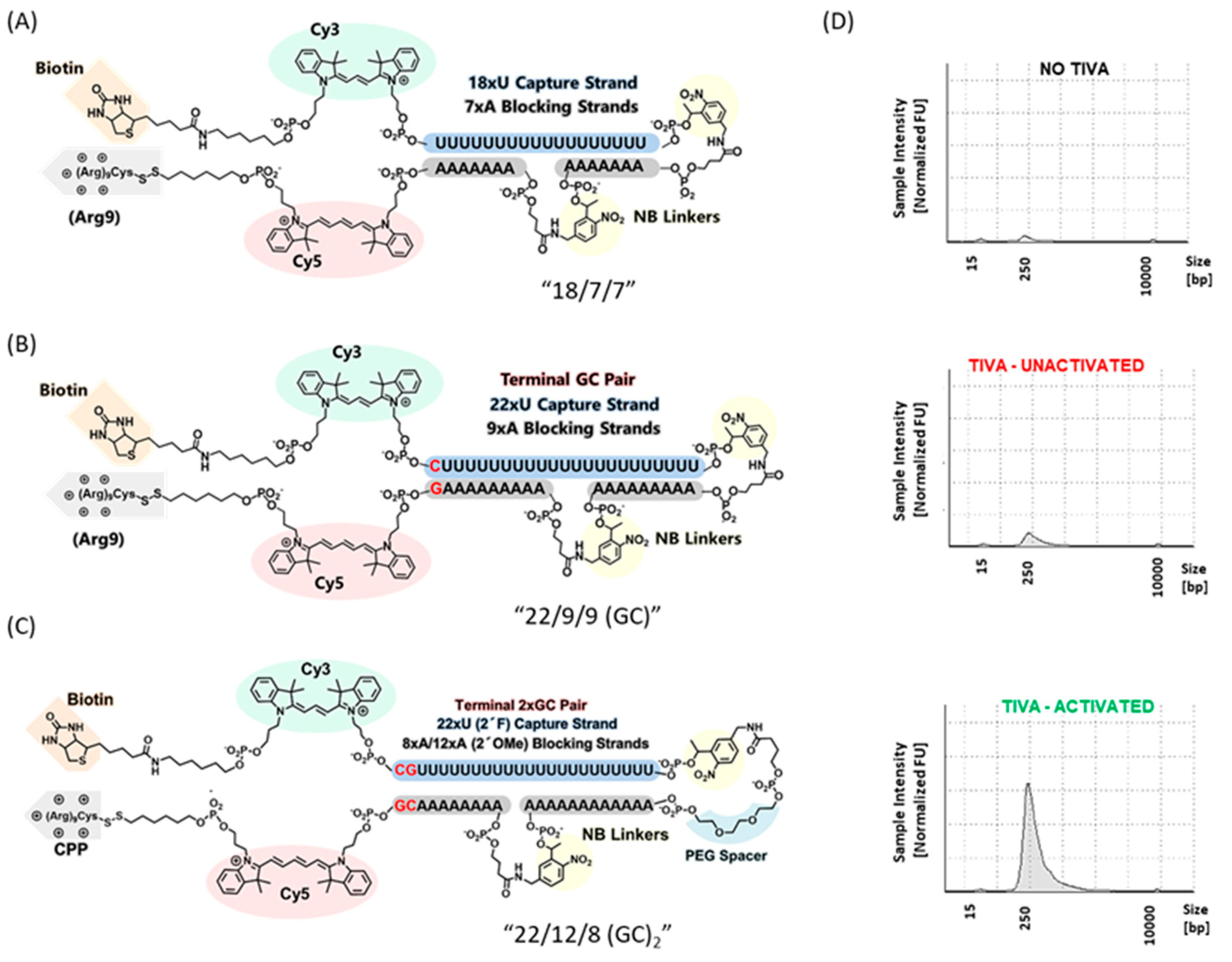
4.2. Transcriptome In Vivo Analysis (TIVA)
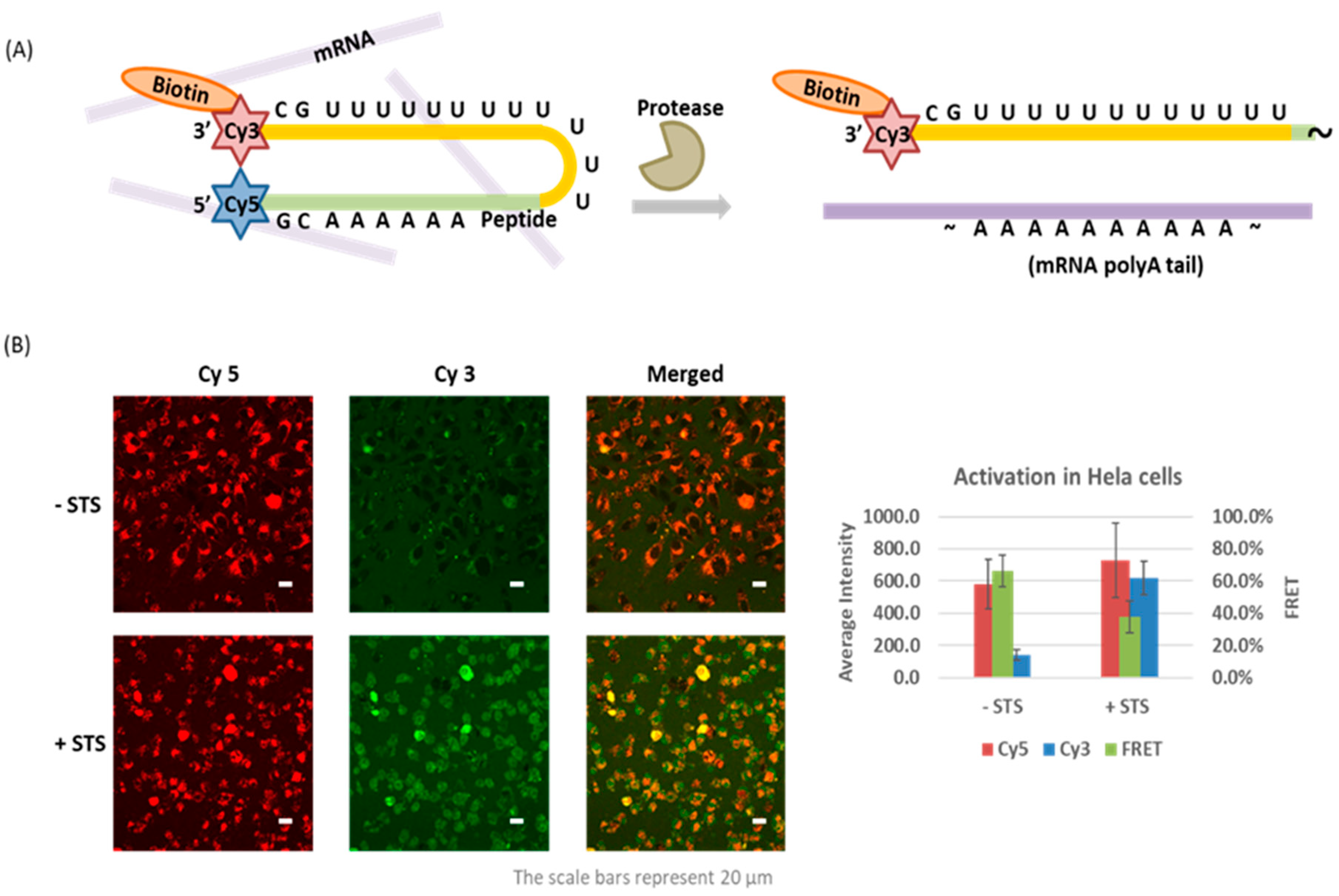
4.3. Caspase-Activated Probe
5. Summary
Funding
Acknowledgments
Conflicts of Interest
References
- Beaucage, S.L.; Caruthers, M.H. Deoxynucleoside phosphoramidites—A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981, 22, 1859–1862. [Google Scholar] [CrossRef]
- Matteucci, M.D.; Caruthers, M.H. Nucleotide chemistry. 4. Synthesis of deoxyoligonucleotides on a polymer support. J. Am. Chem. Soc. 1981, 103, 3185–3191. [Google Scholar] [CrossRef]
- Nowakowski, T.J.; Bhaduri, A.; Pollen, A.A.; Alvarado, B.; Mostajo-Radji, M.A.; Di Lullo, E.; Haeussler, M.; Sandoval-Espinosa, C.; Liu, S.J.; Velmeshev, D.; et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 2017, 358, 1318–1323. [Google Scholar] [CrossRef]
- van der Heijden, M.; Miedema, D.M.; Waclaw, B.; Veenstra, V.L.; Lecca, M.C.; Nijman, L.E.; van Dijk, E.; van Neerven, S.M.; Lodestijn, S.C.; Lenos, K.J.; et al. Spatiotemporal regulation of clonogenicity in colorectal cancer xenografts. Proc. Natl. Acad. Sci. USA 2019, 116, 6140–6145. [Google Scholar] [CrossRef]
- Li, S.C.; Kabeer, M.H. Spatiotemporal switching signals for cancer stem cell activation in pediatric origins of adulthood cancer: Towards a watch-and-wait lifetime strategy for cancer treatment. World J. Stem Cells 2018, 10, 15–22. [Google Scholar] [CrossRef]
- Debart, F.; Dupouy, C.; Vasseur, J.-J. Stimuli-responsive oligonucleotides in prodrug-based approaches for gene silencing. Beilstein. J. Org. Chem. 2018, 14, 436–469. [Google Scholar] [CrossRef] [PubMed]
- Ankenbruck, N.; Courtney, T.; Naro, Y.; Deiters, A. Optochemical control of biological processes in cells and animals. Angew. Chem. Int. Ed. Engl. 2018, 57, 2768–2798. [Google Scholar] [CrossRef] [PubMed]
- Ruble, B.K.; Yeldell, S.B.; Dmochowski, I.J. Caged oligonucleotides for studying biological systems. J. Inorg. Biochem. 2015, 150, 182–188. [Google Scholar] [CrossRef]
- Dmochowski, I.J.; Tang, X. Taking control of gene expression with light-activated oligonucleotides. BioTechniques 2007, 43, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Dmochowski, I.J. Regulating gene expression with light-activated oligonucleotides. Mol. BioSyst. 2007, 3, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Dmochowski, I.J. Phototriggering of caged fluorescent oligodeoxynucleotides. Org. Lett. 2005, 7, 279–282. [Google Scholar] [CrossRef]
- Tang, X.; Richards, J.L.; Peritz, A.E.; Dmochowski, I.J. Photoregulation of DNA polymerase I (Klenow) with caged fluorescent oligodeoxynucleotides. Bioorg. Med. Chem. Lett. 2005, 15, 5303–5306. [Google Scholar] [CrossRef] [PubMed]
- Hobartner, C.; Silverman, S.K. Modulation of RNA tertiary folding by incorporation of caged nucleotides. Angew. Chem. Int. Ed. Engl. 2005, 44, 7305–7309. [Google Scholar] [CrossRef]
- Rodrigues-Correia, A.; Koeppel, M.B.; Schafer, F.; Joshi, K.B.; Mack, T.; Heckel, A. Comparison of the duplex-destabilizing effects of nucleobase-caged oligonucleotides. Anal. Bioanal. Chem. 2011, 399, 441–447. [Google Scholar] [CrossRef]
- Young, D.D.; Lively, M.O.; Deiters, A. Activation and deactivation of DNAzyme and antisense function with light for the photochemical regulation of gene expression in mammalian cells. J. Am. Chem. Soc. 2010, 132, 6183–6193. [Google Scholar] [CrossRef] [PubMed]
- Anhäuser, L.; Klöcker, N.; Muttach, F.; Mäsing, F.; Špaček, P.; Studer, A.; Rentmeister, A. A benzophenone-based photocaging strategy for the N7 position of guanosine. Angew. Chem. Int. Ed. Engl. 2020, 59, 3161–3165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhou, C.Y.; Busby, K.N.; Alexander, S.C.; Devaraj, N.K. Light-activated control of translation by enzymatic covalent mRNA labeling. Angew. Chem. Int. Ed. Engl. 2018, 57, 2822–2826. [Google Scholar] [CrossRef]
- Ando, H.; Furuta, T.; Tsien, R.Y.; Okamoto, H. Photo-mediated gene activation using caged RNA/DNA in zebrafish embryos. Nat. Genet. 2001, 28, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Monroe, W.T.; McQuain, M.M.; Chang, M.S.; Alexander, J.S.; Haselton, F.R. Targeting expression with light using caged DNA. J. Biol. Chem. 1999, 274, 20895–20900. [Google Scholar] [CrossRef]
- Shah, S.; Rangarajan, S.; Friedman, S.H. Light-activated RNA interference. Angew. Chem. Int. Ed. Engl. 2005, 44, 1328–1332. [Google Scholar] [CrossRef] [PubMed]
- Chaulk, S.G.; MacMillan, A.M. Caged RNA: Photo-control of a ribozyme reaction. Nucleic Acids Res. 1998, 26, 3173–3178. [Google Scholar] [CrossRef] [PubMed]
- Kadina, A.; Kietrys, A.M.; Kool, E.T. RNA cloaking by reversible acylation. Angew. Chem. Int. Ed. Engl. 2018, 57, 3059–3063. [Google Scholar] [CrossRef]
- Velema, W.A.; Kietrys, A.M.; Kool, E.T. RNA control by photoreversible acylation. J. Am. Chem. Soc. 2018, 140, 3491–3495. [Google Scholar] [CrossRef]
- Park, H.S.; Kietrys, A.M.; Kool, E.T. Simple alkanoyl acylating agents for reversible RNA functionalization and control. Chem. Commun. 2019, 55, 5135–5138. [Google Scholar] [CrossRef] [PubMed]
- Daniher, A.T.; Xie, J.; Mathur, S.; Bashkin, J.K. Modulation of RNase H activity by modified DNA probes: Major groove vs minor groove effects. Bioorg. Med. Chem. 1997, 5, 1037–1042. [Google Scholar] [CrossRef]
- Ji, Y.; Yang, J.; Wu, L.; Yu, L.; Tang, X. Photochemical regulation of gene expression using caged siRNAs with single terminal Vitamin E modification. Angew. Chem. Int. Ed. Engl. 2016, 55, 2152–2156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, C.; Fan, X.; Tang, X. Photomodulating gene expression by using caged siRNAs with single-aptamer modification. ChemBioChem 2018, 19, 1259–1263. [Google Scholar] [CrossRef]
- Yu, L.; Liang, D.; Chen, C.; Tang, X. Caged siRNAs with single cRGD modification for photoregulation of exogenous and endogenous gene expression in cells and mice. Biomacromolecules 2018, 19, 2526–2534. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Jing, N.; Yang, Z.; Zhang, L.; Tang, X. Caged siRNAs with single folic acid modification of antisense RNA for photomodulation of exogenous and endogenous gene expression in cells. Org. Biomol. Chem. 2018, 16, 7029–7035. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, C.; Tang, X. Cholesterol-modified caged siRNAs for photoregulating exogenous and endogenous gene expression. Bioconjug. Chem. 2018, 29, 1010–1015. [Google Scholar] [CrossRef]
- Chen, C.; Wang, Z.; Zhang, J.; Fan, X.; Xu, L.; Tang, X. Dextran-conjugated caged siRNA nanoparticles for photochemical regulation of RNAi-induced gene silencing in cells and mice. Bioconjug. Chem. 2019, 30, 1459–1465. [Google Scholar] [CrossRef]
- Acevedo-Jake, A.; Ball, A.T.; Galli, M.; Kukwikila, M.; Denis, M.; Singleton, D.G.; Tavassoli, A.; Goldup, S.M. AT-CuAAC synthesis of mechanically interlocked oligonucleotides. J. Am. Chem. Soc. 2020, 142, 5985–5990. [Google Scholar] [CrossRef]
- Richards, J.L.; Tang, X.; Turetsky, A.; Dmochowski, I.J. RNA bandages for photoregulating in vitro protein synthesis. Bioorg. Med. Chem. Lett. 2008, 18, 6255–6258. [Google Scholar] [CrossRef]
- Ruble, B.K.; Richards, J.L.; Cheung-Lau, J.C.; Dmochowski, I.J. Mismatch discrimination and efficient photomodulation with split 10–23 DNAzymes. Inorganica Chim. Acta. 2012, 380, 386–391. [Google Scholar] [CrossRef]
- Tang, X.; Maegawa, S.; Weinberg, E.S.; Dmochowski, I.J. Regulating gene expression in zebrafish embryos using light-activated, negatively charged peptide nucleic acids. J. Am. Chem. Soc. 2007, 129, 11000–11001. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Dmochowski, I.J. Controlling RNA digestion by RNase H with a light-activated DNA hairpin. Angew. Chem. Int. Ed. Engl. 2006, 45, 3523–3526. [Google Scholar] [CrossRef]
- Griepenburg, J.C.; Ruble, B.K.; Dmochowski, I.J. Caged oligonucleotides for bidirectional photomodulation of let-7 miRNA in zebrafish embryos. Bioorg. Med. Chem. 2013, 21, 6198–6204. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Dmochowski, I.J. Synthesis of light-activated antisense oligodeoxynucleotide. Nat. Protoc. 2006, 1, 3041–3048. [Google Scholar] [CrossRef]
- Tang, X.; Swaminathan, J.; Gewirtz, A.M.; Dmochowski, I.J. Regulating gene expression in human leukemia cells using light-activated oligodeoxynucleotides. Nucleic Acids Res. 2008, 36, 559–569. [Google Scholar] [CrossRef]
- Jain, P.K.; Ramanan, V.; Schepers, A.G.; Dalvie, N.S.; Panda, A.; Fleming, H.E.; Bhatia, S.N. Development of light-activated CRISPR using guide RNAs with photocleavable protectors. Angew. Chem. Int. Ed. Engl. 2016, 55, 12440–12444. [Google Scholar] [CrossRef]
- Tan, Z.; Feagin, T.A.; Heemstra, J.M. Temporal control of aptamer biosensors using covalent self-caging to shift equilibrium. J. Am. Chem. Soc. 2016, 138, 6328–6331. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Zhang, X.; Lake, R.J.; Pawel, G.T.; Guo, Z.; Pei, R.; Lu, Y. A photo-regulated aptamer sensor for spatiotemporally controlled monitoring of ATP in the mitochondria of living cells. Chem. Sci. 2020, 11, 713–720. [Google Scholar] [CrossRef]
- Maruyama, H.; Nakashima, Y.; Shuto, S.; Matsuda, A.; Ito, Y.; Abe, H. An intracellular buildup reaction of active siRNA species from short RNA fragments. Chem. Commun. 2014, 50, 1284–1287. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Shu, Z.; Ito, M.; Abe, N.; Nakamoto, K.; Tomoike, F.; Shuto, S.; Ito, Y.; Abe, H. Intracellular build-up RNAi with single-strand circular RNAs as siRNA precursors. Chem. Commun. 2020, 56, 466–469. [Google Scholar] [CrossRef]
- Richards, J.L.; Seward, G.K.; Wang, Y.H.; Dmochowski, I.J. Turning the 10–23 DNAzyme on and off with light. ChemBioChem 2010, 11, 320–324. [Google Scholar] [CrossRef]
- Tang, X.; Su, M.; Yu, L.; Lv, C.; Wang, J.; Li, Z. Photomodulating RNA cleavage using photolabile circular antisense oligodeoxynucleotides. Nucleic Acids Res. 2010, 38, 3848–3855. [Google Scholar] [CrossRef]
- Seyfried, P.; Eiden, L.; Grebenovsky, N.; Mayer, G.; Heckel, A. Photo-tethers for the (multi-)cyclic, conformational caging of long oligonucleotides. Angew. Chem. Int. Ed. Engl. 2017, 56, 359–363. [Google Scholar] [CrossRef]
- Yamazoe, S.; Shestopalov, I.A.; Provost, E.; Leach, S.D.; Chen, J.K. Cyclic caged morpholinos: Conformationally gated probes of embryonic gene function. Angew. Chem. Int. Ed. Engl. 2012, 51, 6908–6911. [Google Scholar] [CrossRef]
- O’Connor, M.J.; Beebe, L.L.; Deodato, D.; Ball, R.E.; Page, A.T.; VanLeuven, A.J.; Harris, K.T.; Park, S.; Hariharan, V.; Lauderdale, J.D.; et al. Bypassing glutamic acid decarboxylase 1 (gad1) induced craniofacial defects with a photoactivatable translation blocker morpholino. ACS Chem. Neurosci. 2019, 10, 266–278. [Google Scholar] [CrossRef]
- Deodato, D.; Dore, T.M. Practical synthesis of quinoline-protected morpholino oligomers for light-triggered regulation of gene function. Molecules 2020, 25, 2078. [Google Scholar] [CrossRef]
- Zhang, L.; Liang, D.; Chen, C.; Wang, Y.; Amu, G.; Yang, J.; Yu, L.; Dmochowski, I.J.; Tang, X. Circular siRNAs for reducing off-target effects and enhancing long-term gene silencing in cells and mice. Mol. Ther. Nucleic Acids 2018, 10, 237–244. [Google Scholar] [CrossRef]
- Kaplan, J.H.; Forbush, B., 3rd; Hoffman, J.F. Rapid photolytic release of adenosine 5′-triphosphate from a protected analogue: Utilization by the Na:K pump of human red blood cell ghosts. Biochemistry 1978, 17, 1929–1935. [Google Scholar] [CrossRef]
- Walbert, S.; Pfleiderer, W.; Steiner, U.E. Photolabile protecting groups for nucleosides: Mechanistic studies of the 2-(2-nitrophenyl)ethyl group. Helv. Chim. Acta 2001, 84, 1601–1611. [Google Scholar] [CrossRef]
- Connelly, C.M.; Uprety, R.; Hemphill, J.; Deiters, A. Spatiotemporal control of microRNA function using light-activated antagomirs. Mol. Biosyst. 2012, 8, 2987–2993. [Google Scholar] [CrossRef]
- Momotake, A.; Lindegger, N.; Niggli, E.; Barsotti, R.J.; Ellis-Davies, G.C.R. The nitrodibenzofuran chromophore: A new caging group for ultra-efficient photolysis in living cells. Nat. Methods 2006, 3, 35–40. [Google Scholar] [CrossRef]
- Ordoukhanian, P.; Taylor, J.-S. Design and synthesis of a versatile photocleavable DNA building block. Application to phototriggered hybridization. J. Am. Chem. Soc. 1995, 117, 9570–9571. [Google Scholar] [CrossRef]
- Weyel, X.M.M.; Fichte, M.A.H.; Heckel, A. A two-photon-photocleavable linker for triggering light-induced strand breaks in oligonucleotides. ACS Chem. Biol. 2017, 12, 2183–2190. [Google Scholar] [CrossRef]
- Zhao, J.; Gao, J.; Xue, W.; Di, Z.; Xing, H.; Lu, Y.; Li, L. Upconversion luminescence-activated DNA nanodevice for ATP sensing in living cells. J. Am. Chem. Soc. 2018, 140, 578–581. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, Y.; Yu, M.; Gu, Z.; Li, L.; Zhao, Y. Time-resolved activation of pH sensing and imaging in vivo by a remotely controllable DNA nanomachine. Nano Lett. 2020, 20, 874–880. [Google Scholar] [CrossRef]
- Zhao, J.; Chu, H.; Zhao, Y.; Lu, Y.; Li, L. A NIR light gated DNA nanodevice for spatiotemporally controlled imaging of microRNA in cells and animals. J. Am. Chem. Soc. 2019, 141, 7056–7062. [Google Scholar] [CrossRef]
- Yang, Z.; Loh, K.Y.; Chu, Y.-T.; Feng, R.; Satyavolu, N.S.R.; Xiong, M.; Nakamata Huynh, S.M.; Hwang, K.; Li, L.; Xing, H.; et al. Optical control of metal ion probes in cells and zebrafish using highly selective DNAzymes conjugated to upconversion nanoparticles. J. Am. Chem. Soc. 2018, 140, 17656–17665. [Google Scholar] [CrossRef]
- Rapp, T.L.; Phillips, S.R.; Dmochowski, I.J. Kinetics and photochemistry of ruthenium bisbipyridine diacetonitrile complexes: An interdisciplinary inorganic and physical chemistry laboratory exercise. J. Chem. Educ. 2016, 93, 2101–2105. [Google Scholar] [CrossRef]
- Griepenburg, J.C.; Rapp, T.L.; Carroll, P.J.; Eberwine, J.; Dmochowski, I.J. Ruthenium-caged antisense morpholinos for regulating gene expression in zebrafish embryos. Chem. Sci. 2015, 6, 2342–2346. [Google Scholar] [CrossRef]
- Rapp, T.L.; Highley, C.B.; Manor, B.C.; Burdick, J.A.; Dmochowski, I.J. Ruthenium-crosslinked hydrogels with rapid, visible-light degradation. Chem. Eur. J. 2018, 24, 2328–2333. [Google Scholar] [CrossRef]
- Rapp, T.L.; Wang, Y.; Delessio, M.A.; Gau, M.R.; Dmochowski, I.J. Designing photolabile ruthenium polypyridyl crosslinkers for hydrogel formation and multiplexed, visible-light degradation. RSC Adv. 2019, 9, 4942–4947. [Google Scholar] [CrossRef]
- Korman, A.; Sun, H.; Hua, B.; Yang, H.; Capilato, J.N.; Paul, R.; Panja, S.; Ha, T.; Greenberg, M.M.; Woodson, S.A. Light-controlled twister ribozyme with single-molecule detection resolves RNA function in time and space. Proc. Natl. Acad. Sci. USA 2020, 117, 12080–12086. [Google Scholar] [CrossRef]
- Zhou, W.; Brown, W.; Bardhan, A.; Delaney, M.; Ilk, A.S.; Rauen, R.R.; Kahn, S.I.; Tsang, M.; Deiters, A. Spatiotemporal control of CRISPR/Cas9 function in cells and zebrafish using light-activated guide RNA. Angew. Chem. Int. Ed. Engl. 2020, 59, 8998–9003. [Google Scholar] [CrossRef]
- Liu, Y.; Zou, R.S.; He, S.; Nihongaki, Y.; Li, X.; Razavi, S.; Wu, B.; Ha, T. Very fast CRISPR on demand. Science 2020, 368, 1265–1269. [Google Scholar] [CrossRef]
- Mori, S.; Morihiro, K.; Okuda, T.; Kasahara, Y.; Obika, S. Hydrogen peroxide-triggered gene silencing in mammalian cells through boronated antisense oligonucleotides. Chem. Sci. 2018, 9, 1112–1118. [Google Scholar] [CrossRef]
- Ochi, Y.; Nakagawa, O.; Sakaguchi, K.; Wada, S.-I.; Urata, H. A post-synthetic approach for the synthesis of 2′-O-methyldithiomethyl-modified oligonucleotides responsive to a reducing environment. Chem. Commun. 2013, 49, 7620–7622. [Google Scholar] [CrossRef]
- Ochi, Y.; Imai, M.; Nakagawa, O.; Hayashi, J.; Wada, S.-I.; Urata, H. Gene silencing by 2′-O-methyldithiomethyl-modified siRNA, a prodrug-type siRNA responsive to reducing environment. Bioorg. Med. Chem. Lett. 2016, 26, 845–848. [Google Scholar] [CrossRef]
- Hayashi, J.; Ochi, Y.; Morita, Y.; Soubou, K.; Ohtomo, Y.; Nishigaki, M.; Tochiyama, Y.; Nakagawa, O.; Wada, S.-i.; Urata, H. Syntheses of prodrug-type 2′-O-methyldithiomethyl oligonucleotides modified at natural four nucleoside residues and their conversions into natural 2′-hydroxy oligonucleotides under reducing condition. Bioorg. Med. Chem. 2018, 26, 5838–5844. [Google Scholar] [CrossRef]
- Hayashi, J.; Samezawa, Y.; Ochi, Y.; Wada, S.-I.; Urata, H. Syntheses of prodrug-type phosphotriester oligonucleotides responsive to intracellular reducing environment for improvement of cell membrane permeability and nuclease resistance. Bioorg. Med. Chem. Lett. 2017, 27, 3135–3138. [Google Scholar] [CrossRef]
- Hayakawa, Y.; Banno, A.; Kitagawa, H.; Higashi, S.; Kitade, Y.; Shibata, A.; Ikeda, M. Reduction-responsive DNA duplex containing O6-nitrobenzyl-guanine. ACS Omega. 2018, 3, 9267–9275. [Google Scholar] [CrossRef]
- Ikeda, M.; Kamimura, M.; Hayakawa, Y.; Shibata, A.; Kitade, Y. Reduction-responsive guanine incorporated into G-quadruplex-forming DNA. ChemBioChem 2016, 17, 1304–1307. [Google Scholar] [CrossRef] [PubMed]
- Saneyoshi, H.; Yamamoto, Y.; Kondo, K.; Hiyoshi, Y.; Ono, A. Conjugatable and bioreduction cleavable linker for the 5′-functionalization of oligonucleotides. J. Org. Chem. 2017, 82, 1796–1802. [Google Scholar] [CrossRef]
- Saneyoshi, H.; Iketani, K.; Kondo, K.; Saneyoshi, T.; Okamoto, I.; Ono, A. Synthesis and characterization of cell-permeable oligonucleotides bearing reduction-activated protecting groups on the internucleotide linkages. Bioconjug. Chem. 2016, 27, 2149–2156. [Google Scholar] [CrossRef]
- Yamazoe, S.; McQuade, L.E.; Chen, J.K. Nitroreductase-activatable morpholino oligonucleotides for in vivo gene silencing. ACS Chem. Biol. 2014, 9, 1985–1990. [Google Scholar] [CrossRef]
- Meade, B.R.; Gogoi, K.; Hamil, A.S.; Palm-Apergi, C.; Berg, A.V.D.; Hagopian, J.C.; Springer, A.D.; Eguchi, A.; Kacsinta, A.D.; Dowdy, C.F.; et al. Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat. Biotechnol. 2014, 32, 1256–1261. [Google Scholar] [CrossRef]
- Cieślak, J.; Grajkowski, A.; Livengood, V.; Beaucage, S.L. Thermolytic 4-methylthio-1-butyl group for phosphate/thiophosphate protection in solid-phase synthesis of DNA oligonucleotides. J. Org. Chem. 2004, 69, 2509–2515. [Google Scholar] [CrossRef] [PubMed]
- Grajkowski, A.; Pedras-Vasconcelos, J.; Wang, V.; Ausín, C.; Hess, S.; Verthelyi, D.; Beaucage, S.L. Thermolytic CpG-containing DNA oligonucleotides as potential immunotherapeutic prodrugs. Nucleic. Acids. Res. 2005, 33, 3550–3560. [Google Scholar] [CrossRef]
- Ausín, C.; Kauffman, J.S.; Duff, R.J.; Shivaprasad, S.; Beaucage, S.L. Assessment of heat-sensitive thiophosphate protecting groups in the development of thermolytic DNA oligonucleotide prodrugs. Tetrahedron 2010, 66, 68–79. [Google Scholar] [CrossRef]
- Madaoui, M.; Meyer, A.; Vasseur, J.-J.; Morvan, F. Thermolytic reagents to synthesize 5′- or 3′-mono(thio)phosphate oligodeoxynucleotides or 3′-modified oligodeoxynucleotides. Eur. J. Org. Chem. 2019, 2019, 2832–2842. [Google Scholar] [CrossRef]
- Knutson, S.D.; Sanford, A.A.; Swenson, C.S.; Korn, M.M.; Manuel, B.A.; Heemstra, J.M. Thermoreversible control of nucleic acid structure and function with glyoxal caging. J. Am. Chem. Soc. 2020, 142, 17766–17781. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Kim, H.B.; Sul, J.Y.; Yeldell, S.B.; Eberwine, J.H.; Dmochowski, I.J. Efficient synthesis of light-triggered circular antisense oligonucleotides targeting cellular protein expression. ChemBioChem. 2018, 19, 1250–1254. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; Khoo, W.H.; Moran, I.; Croucher, P.I.; Phan, T.G. Single cell RNA sequencing of rare immune cell populations. Front. Immunol. 2018, 9, 1553. [Google Scholar] [CrossRef] [PubMed]
- Iacono, G.; Massoni-Badosa, R.; Heyn, H. Single-cell transcriptomics unveils gene regulatory network plasticity. Genome Biol. 2019, 20, 110. [Google Scholar] [CrossRef]
- Kester, L.; van Oudenaarden, A. Single-cell transcriptomics meets lineage tracing. Cell Stem Cell 2018, 23, 166–179. [Google Scholar] [CrossRef] [PubMed]
- Lovatt, D.; Ruble, B.K.; Lee, J.; Dueck, H.; Kim, T.K.; Fisher, S.; Francis, C.; Spaethling, J.M.; Wolf, J.A.; Grady, M.S.; et al. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat. Methods 2014, 11, 190–196. [Google Scholar] [CrossRef]
- Yeldell, S.B.; Ruble, B.K.; Dmochowski, I.J. Oligonucleotide modifications enhance probe stability for single cell transcriptome in vivo analysis (TIVA). Org. Biomol. Chem. 2017, 15, 10001–10009. [Google Scholar] [CrossRef]
- Yeldell, S.B.; Yang, L.; Lee, J.; Eberwine, J.H.; Dmochowski, I.J. Oligonucleotide probe for transcriptome in vivo analysis (TIVA) of single neurons with minimal background. ACS Chem. Biol. 2020, 15, 2714–2721. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Eberwine, J.H.; Dmochowski, I.J. Caspase-activated oligonucleotide probe. Bioconjug. Chem. 2020, 31, 2172–2178. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Dmochowski, I.J. Conditionally Activated (“Caged”) Oligonucleotides. Molecules 2021, 26, 1481. https://doi.org/10.3390/molecules26051481
Yang L, Dmochowski IJ. Conditionally Activated (“Caged”) Oligonucleotides. Molecules. 2021; 26(5):1481. https://doi.org/10.3390/molecules26051481
Chicago/Turabian StyleYang, Linlin, and Ivan J. Dmochowski. 2021. "Conditionally Activated (“Caged”) Oligonucleotides" Molecules 26, no. 5: 1481. https://doi.org/10.3390/molecules26051481
APA StyleYang, L., & Dmochowski, I. J. (2021). Conditionally Activated (“Caged”) Oligonucleotides. Molecules, 26(5), 1481. https://doi.org/10.3390/molecules26051481